Submitted:
15 June 2023
Posted:
15 June 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Results
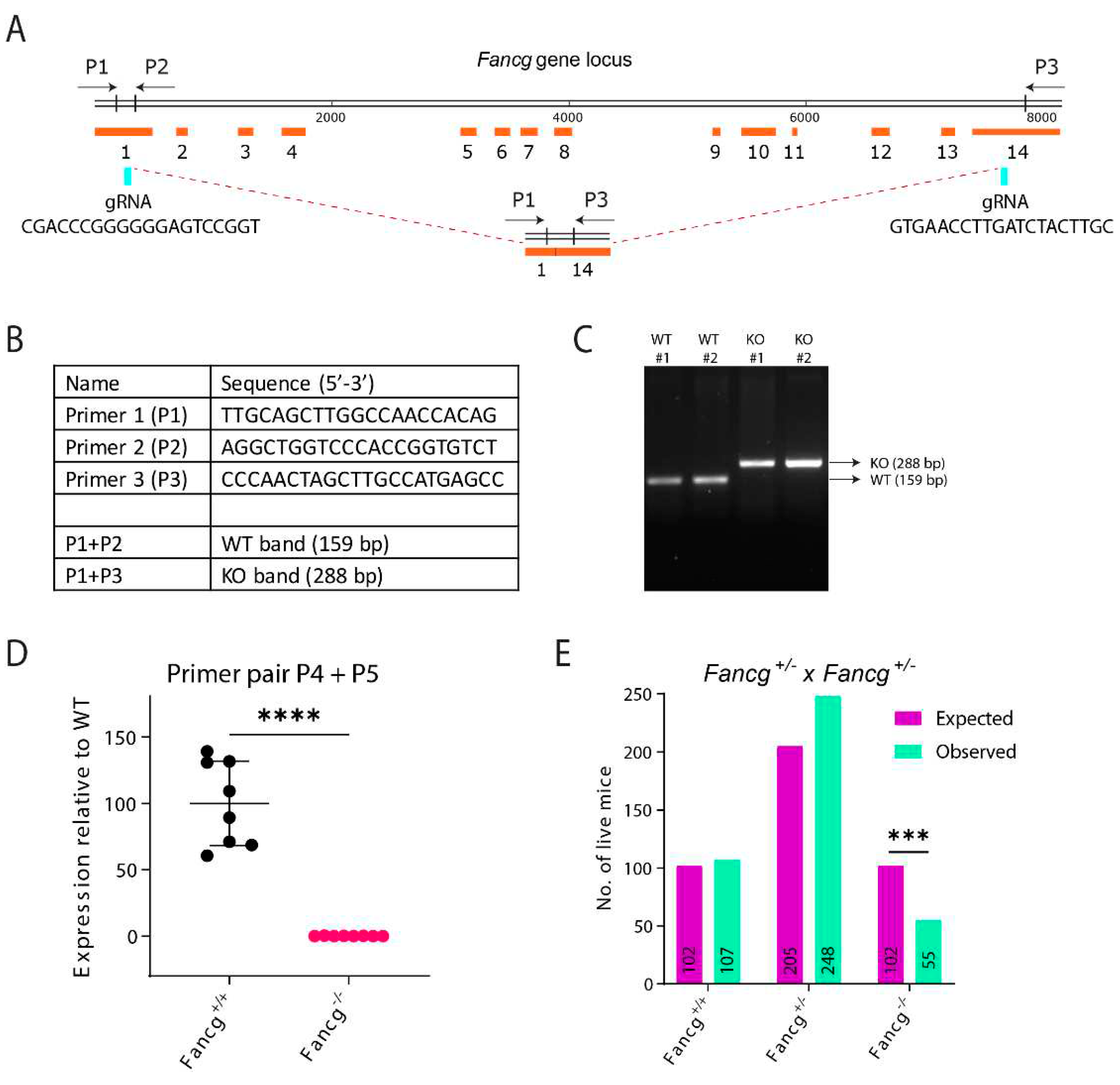
Establishing a Genetically and Immunologically Defined C57BL/6J Fancg-KO Mouse Model
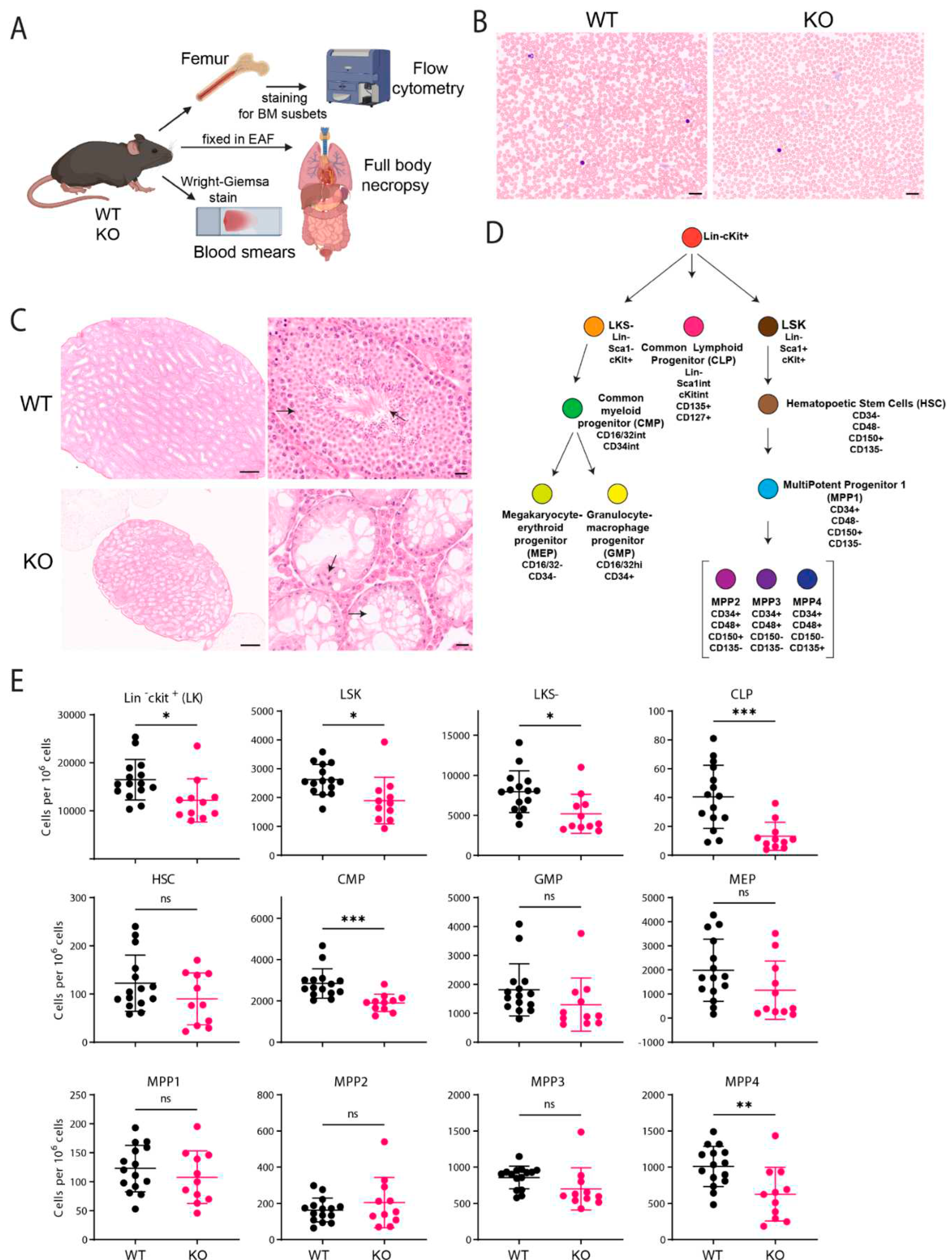
C57BL/6J Fancg-KO Mice Display Stem Cell Defects
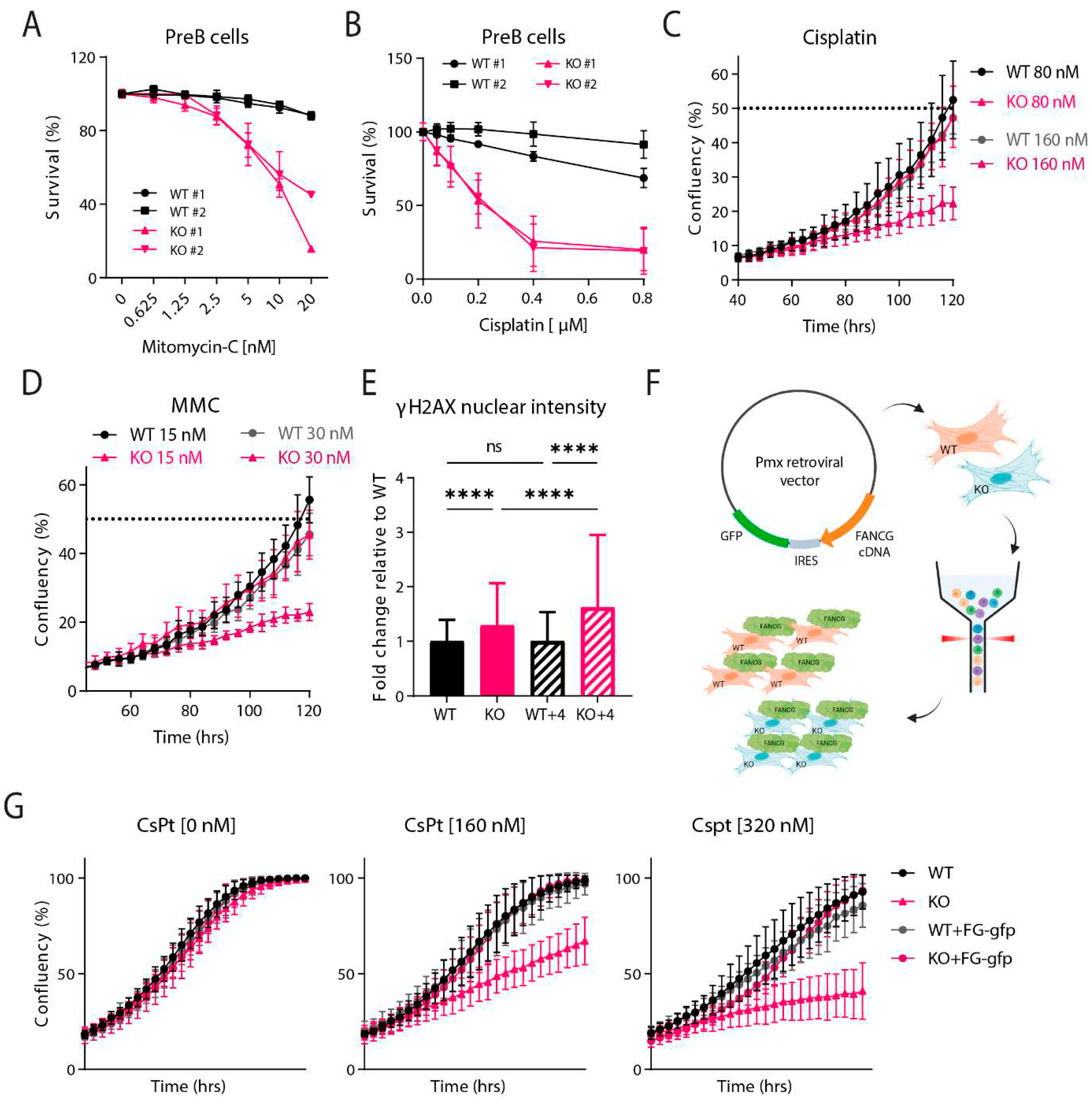
Fancg-KO Cells Are Hypersensitive to Crosslinking Agents
Discussion
Materials and Methods
Generation of Fancg-/- Mouse
Generation of Primary Cell-Lines and Cell Culture
PCR Genotyping
qRT-PCR
Immunofluorescence of Bone Marrow (BM) Cells
Histopathology
PreB Survival Assay
IncuCyte Proliferation Assay
γ-H2AX Immunofluorescence
Fancg-Flag Reconstitution
Statistical Analysis
Author Contributions
Funding
Institutional Board Review Statement
Acknowledgements
Conflicts of Interest
References
- T. Yamashita and T. Nakahata, “Current Knowledge on the Pathophysiology of Fanconi Anemia: From Genes to Phenotypes,” Int. J. Hematol., vol. 74, no. 1, pp. 33–41, Jul. 2001. [CrossRef]
- R. Ceccaldi, P. Sarangi, and A. D. D’Andrea, “The Fanconi anaemia pathway: new players and new functions,” Nat. Rev. Mol. Cell Biol., vol. 17, no. 6, pp. 337–349, Jun. 2016. [CrossRef]
- R. Errazquin et al., “Development of a mouse model for spontaneous oral squamous cell carcinoma in Fanconi anemia,” Oral Oncol., vol. 134, p. 106184, Nov. 2022. [CrossRef]
- G. Nalepa and D. W. Clapp, “Fanconi anaemia and cancer: an intricate relationship,” Nat. Rev. Cancer, vol. 18, no. 3, pp. 168–185, Mar. 2018. [CrossRef]
- R. Che, J. Zhang, M. Nepal, B. Han, and P. Fei, “Multifaceted Fanconi Anemia Signaling,” Trends Genet., vol. 34, no. 3, pp. 171–183, Mar. 2018. [CrossRef]
- A. F. Alpi and K. J. Patel, “Monoubiquitylation in the Fanconi anemia DNA damage response pathway,” DNA Repair, vol. 8, no. 4, pp. 430–435, Apr. 2009. [CrossRef]
- M. C. Kottemann and A. Smogorzewska, “Fanconi anaemia and the repair of Watson and Crick DNA crosslinks,” Nature, vol. 493, no. 7432, Art. no. 7432, Jan. 2013. [CrossRef]
- J. P. de Winter et al., “The Fanconi anaemia group G gene FANCG is identical with XRCC9,” Nat. Genet., vol. 20, no. 3, pp. 281–283, Nov. 1998. [CrossRef]
- N. Liu et al., “The human XRCC9 gene corrects chromosomal instability and mutagen sensitivities in CHO UV40 cells,” Proc. Natl. Acad. Sci., vol. 94, no. 17, pp. 9232–9237, Aug. 1997. [CrossRef]
- K. Saar et al., “Localisation of a Fanconi anaemia gene to chromosome 9p,” Eur. J. Hum. Genet., vol. 6, no. 5, Art. no. 5, Sep. 1998. [CrossRef]
- H. J. Van De Vrugt et al., “Characterization, expression and complex formation of the murine Fanconi anaemia gene product Fancg,” Genes Cells, vol. 7, no. 3, pp. 333–342, 2002. [CrossRef]
- I. Demuth et al., “Spectrum of mutations in the Fanconi anaemia group G gene, FANCG/XRCC9,” Eur. J. Hum. Genet. EJHG, vol. 8, no. 11, pp. 861–868, Nov. 2000. [CrossRef]
- I. Garcia-Higuera, Y. Kuang, D. Näf, J. Wasik, and A. D. D’Andrea, “Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex,” Mol. Cell. Biol., vol. 19, no. 7, pp. 4866–4873, Jul. 1999. [CrossRef]
- Q. Waisfisz et al., “A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA,” Proc. Natl. Acad. Sci. U. S. A., vol. 96, no. 18, pp. 10320–10325, Aug. 1999. [CrossRef]
- I. Garcia-Higuera, Y. Kuang, J. Denham, and A. D. D’Andrea, “The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex,” Blood, vol. 96, no. 9, pp. 3224–3230, Nov. 2000.
- J. P. de Winter et al., “The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG,” Hum. Mol. Genet., vol. 9, no. 18, pp. 2665–2674, Nov. 2000. [CrossRef]
- K. Yamamoto et al., “Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells,” Mol. Cell. Biol., vol. 23, no. 15, pp. 5421–5430, Aug. 2003. [CrossRef]
- K. Kersten, K. E. de Visser, M. H. van Miltenburg, and J. Jonkers, “Genetically engineered mouse models in oncology research and cancer medicine,” EMBO Mol. Med., vol. 9, no. 2, pp. 137–153, Feb. 2017. [CrossRef]
- W. Hill, D. R. Caswell, and C. Swanton, “Capturing cancer evolution using genetically engineered mouse models (GEMMs),” Trends Cell Biol., vol. 31, no. 12, pp. 1007–1018, Dec. 2021. [CrossRef]
- S. T. Bakker, J. P. de Winter, and H. te Riele, “Learning from a paradox: recent insights into Fanconi anaemia through studying mouse models,” Dis. Model. Mech., vol. 6, no. 1, pp. 40–47, Jan. 2013. [CrossRef]
- E. L. Dubois et al., “A Fanci knockout mouse model reveals common and distinct functions for FANCI and FANCD2,” Nucleic Acids Res., vol. 47, no. 14, pp. 7532–7547, Aug. 2019. [CrossRef]
- N. C. Cheng et al., “Mice with a targeted disruption of the Fanconi anemia homolog Fanca,” Hum. Mol. Genet., vol. 9, no. 12, pp. 1805–1811, Jul. 2000. [CrossRef]
- J. D. Sander and J. K. Joung, “CRISPR-Cas systems for editing, regulating and targeting genomes,” Nat. Biotechnol., vol. 32, no. 4, pp. 347–355, Apr. 2014. [CrossRef]
- F. Zhang, Y. Wen, and X. Guo, “CRISPR/Cas9 for genome editing: progress, implications and challenges,” Hum. Mol. Genet., vol. 23, no. R1, pp. R40–R46, Sep. 2014. [CrossRef]
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, and E. Charpentier, “A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity,” Science, vol. 337, no. 6096, pp. 816–821, Aug. 2012. [CrossRef]
- L. Cong et al., “Multiplex genome engineering using CRISPR/Cas systems,” Science, vol. 339, no. 6121, pp. 819–823, Feb. 2013. [CrossRef]
- W. Jiang, D. Bikard, D. Cox, F. Zhang, and L. A. Marraffini, “RNA-guided editing of bacterial genomes using CRISPR-Cas systems,” Nat. Biotechnol., vol. 31, no. 3, Art. no. 3, Mar. 2013. [CrossRef]
- B. Shen et al., “Generation of gene-modified mice via Cas9/RNA-mediated gene targeting,” Cell Res., vol. 23, no. 5, pp. 720–723, May 2013. [CrossRef]
- H. Wang et al., “One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering,” Cell, vol. 153, no. 4, pp. 910–918, May 2013. [CrossRef]
- M. Koomen et al., “Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice,” Hum. Mol. Genet., vol. 11, no. 3, pp. 273–281, Feb. 2002. [CrossRef]
- Y. Yang et al., “Targeted disruption of the murine Fanconi anemia gene,Fancg/Xrcc9,” Blood, vol. 98, no. 12, pp. 3435–3440, Dec. 2001. [CrossRef]
- D. I. Kutler et al., “A 20-year perspective on the International Fanconi Anemia Registry (IFAR),” Blood, vol. 101, no. 4, pp. 1249–1256, Feb. 2003. [CrossRef]
- C. G. Mathew, “Fanconi anaemia genes and susceptibility to cancer,” Oncogene, vol. 25, no. 43, Art. no. 43, Sep. 2006. [CrossRef]
- E. W. Sayers et al., “Database resources of the national center for biotechnology information,” Nucleic Acids Res., vol. 50, no. D1, pp. D20–D26, Jan. 2022. [CrossRef]
- A. Wilson et al., “Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair,” Cell, vol. 135, no. 6, pp. 1118–1129, Dec. 2008. [CrossRef]
- A. C. Pulliam-Leath et al., “Genetic disruption of both Fancc and Fancg in mice recapitulates the hematopoietic manifestations of Fanconi anemia,” Blood, vol. 116, no. 16, pp. 2915–2920, Oct. 2010. [CrossRef]
- E. M. Pietras et al., “Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions,” Cell Stem Cell, vol. 17, no. 1, pp. 35–46, Jul. 2015. [CrossRef]
- M. Chen et al., “Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia,” Nat. Genet., vol. 12, no. 4, pp. 448–451, Apr. 1996. [CrossRef]
- J. I. Garaycoechea et al., “Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells,” Nature, vol. 553, no. 7687, Art. no. 7687, Jan. 2018. [CrossRef]
- M. Wang, F. A. Dingler, and K. J. Patel, “Genotoxic aldehydes in the hematopoietic system,” Blood, vol. 139, no. 14, pp. 2119–2129, Apr. 2022. [CrossRef]
- A. J. Deans and S. C. West, “DNA interstrand crosslink repair and cancer,” Nat. Rev. Cancer, vol. 11, no. 7, Art. no. 7, Jul. 2011. [CrossRef]
- I. M. Ward and J. Chen, “Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress,” J. Biol. Chem., vol. 276, no. 51, pp. 47759–47762, Dec. 2001. [CrossRef]
- A. Jarysta et al., “Abnormal migration behavior linked to Rac1 signaling contributes to primordial germ cell exhaustion in Fanconi anemia pathway-deficient Fancg-/- embryos,” Hum. Mol. Genet., vol. 31, no. 1, pp. 97–110, Dec. 2021. [CrossRef]
- V. Barroca et al., “Impaired functionality and homing of Fancg-deficient hematopoietic stem cells,” Hum. Mol. Genet., vol. 21, no. 1, pp. 121–135, Jan. 2012. [CrossRef]
- Y. Li et al., “Mesenchymal stem/progenitor cells promote the reconstitution of exogenous hematopoietic stem cells in Fancg−/− mice in vivo,” Blood, vol. 113, no. 10, pp. 2342–2351, Mar. 2009. [CrossRef]
- M. A. Whitney et al., “Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene,” Blood, vol. 88, no. 1, pp. 49–58, Jul. 1996.
- M. A. Whitney, P. Jakobs, M. Kaback, R. E. Moses, and M. Grompe, “The Ashkenazi Jewish Fanconi anemia mutation: incidence among patients and carrier frequency in the at-risk population,” Hum. Mutat., vol. 3, no. 4, pp. 339–341, 1994. [CrossRef]
- Y. Zhou et al., “An abnormal bone marrow microenvironment contributes to hematopoietic dysfunction in Fanconi anemia,” Haematologica, vol. 102, no. 6, pp. 1017–1027, Jun. 2017. [CrossRef]
- K. Parmar et al., “Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1,” Stem Cells Dayt. Ohio, vol. 28, no. 7, pp. 1186–1195, Jul. 2010. [CrossRef]
- J. M. Kim et al., “Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype,” Dev. Cell, vol. 16, no. 2, pp. 314–320, Feb. 2009. [CrossRef]
- A. Veillette, “SLAM-Family Receptors: Immune Regulators with or without SAP-Family Adaptors,” Cold Spring Harb. Perspect. Biol., vol. 2, no. 3, p. a002469, Mar. 2010. [CrossRef]
- C. E. J. Pritchard, L. J. Kroese, and I. J. Huijbers, “Direct Generation of Conditional Alleles Using CRISPR/Cas9 in Mouse Zygotes,” in Site-Specific Recombinases: Methods and Protocols, N. Eroshenko, Ed., in Methods in Molecular Biology. New York, NY: Springer, 2017, pp. 21–35. [CrossRef]
- N. Wit et al., “Roles of PCNA ubiquitination and TLS polymerases κ and η in the bypass of methyl methanesulfonate-induced DNA damage,” Nucleic Acids Res., vol. 43, no. 1, pp. 282–294, Jan. 2015. [CrossRef]
- A. M. G. Dirac and R. Bernards, “Reversal of Senescence in Mouse Fibroblasts through Lentiviral Suppression of p53*210,” J. Biol. Chem., vol. 278, no. 14, pp. 11731–11734, Apr. 2003. [CrossRef]
- X. Wang, A. Spandidos, H. Wang, and B. Seed, “PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update,” Nucleic Acids Res., vol. 40, no. Database issue, pp. D1144–D1149, Jan. 2012. [CrossRef]
| Name | Sequence (5’-3’) |
|---|---|
| pmxFG_Internal primer A fwd | GAGGGATGTCCTTCTGACTGC |
| pmxFG_Internal primer A rev | GAGAACCTTGTCTCTGAGCCACCC |
| pmxFG_Internal primer B fwd | CTGCCGTGTTGCCCAGTCTGGGTC |
| pmxFG_Internal primer B rev | GGCTTCGCGGTTAGGGGGATGGAT |
| pmxFG_Internal primer C fwd | TCTTCCACTGTATTTAGAAACCTG |
| pmxFG_Internal primer C rev | TTATACACGTGGCTTTTGGCCGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





