Submitted:
05 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Insect rearing
2.2. Feeding behavior assay
2.3. Mating behavior assay
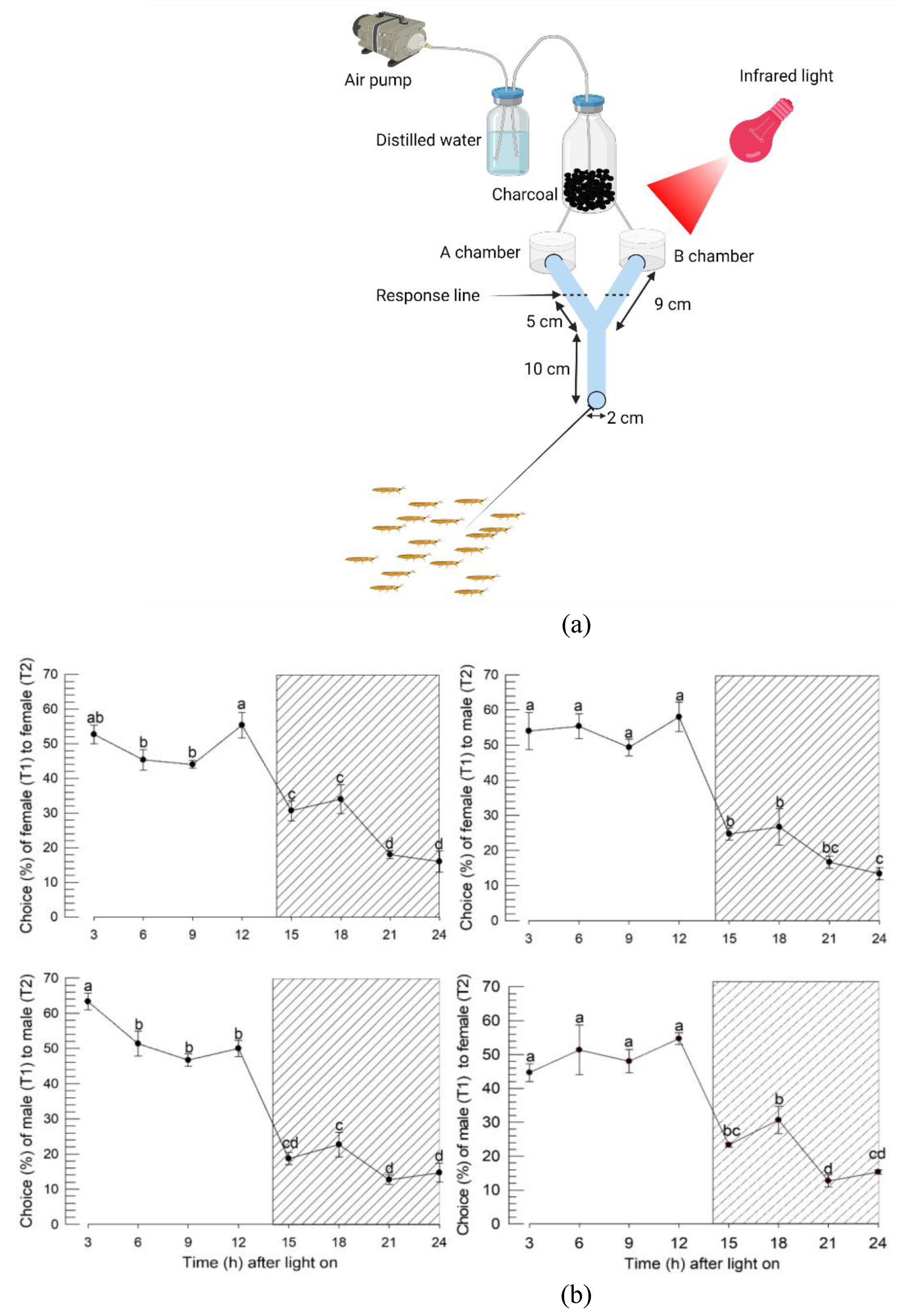
2.4. Y-tube olfactometer assay
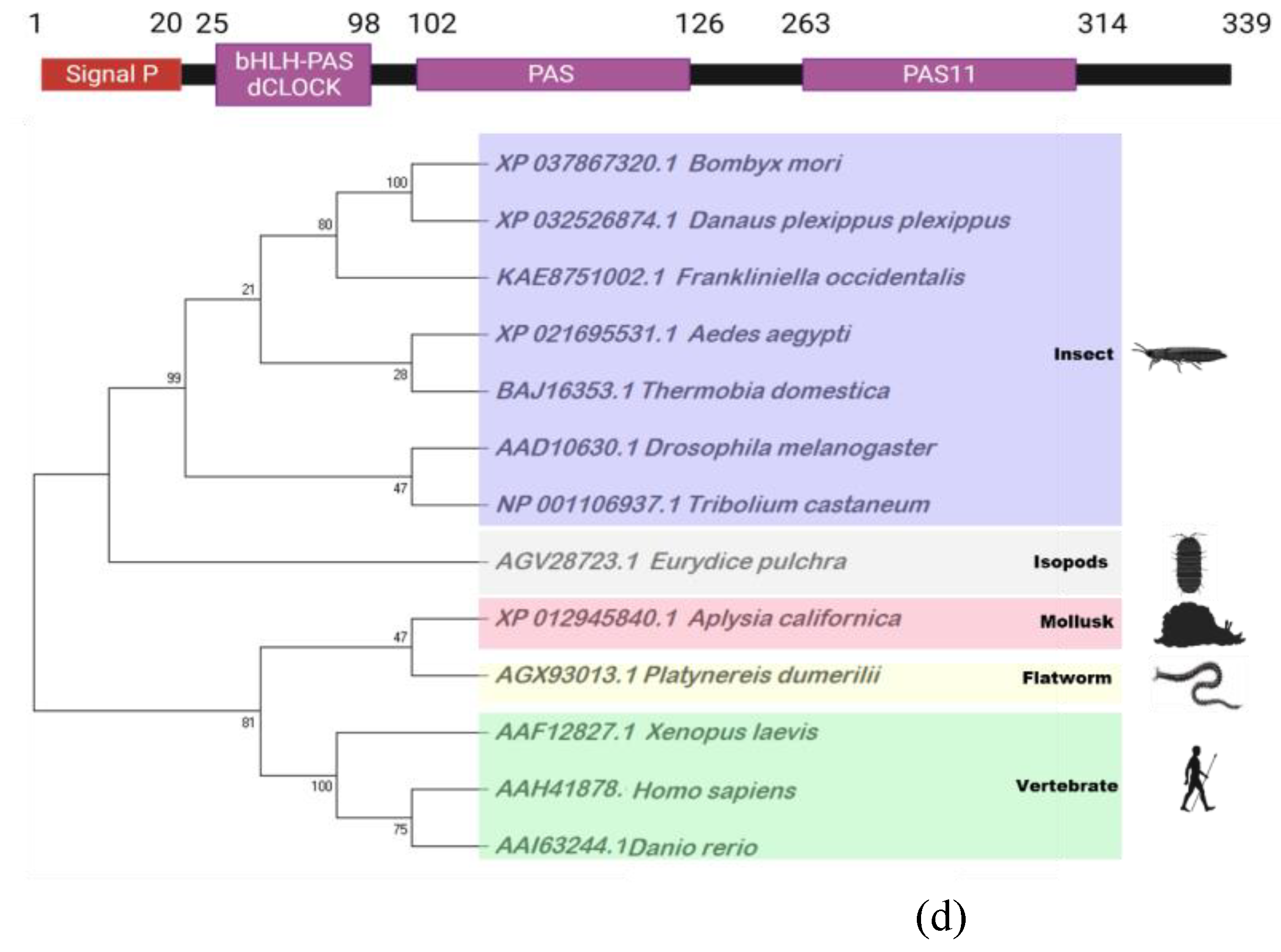
2.5. Bioinformatic analysis
2.6. RNA extraction and cDNA preparation
2.7. RT-PCR and RT-qPCR
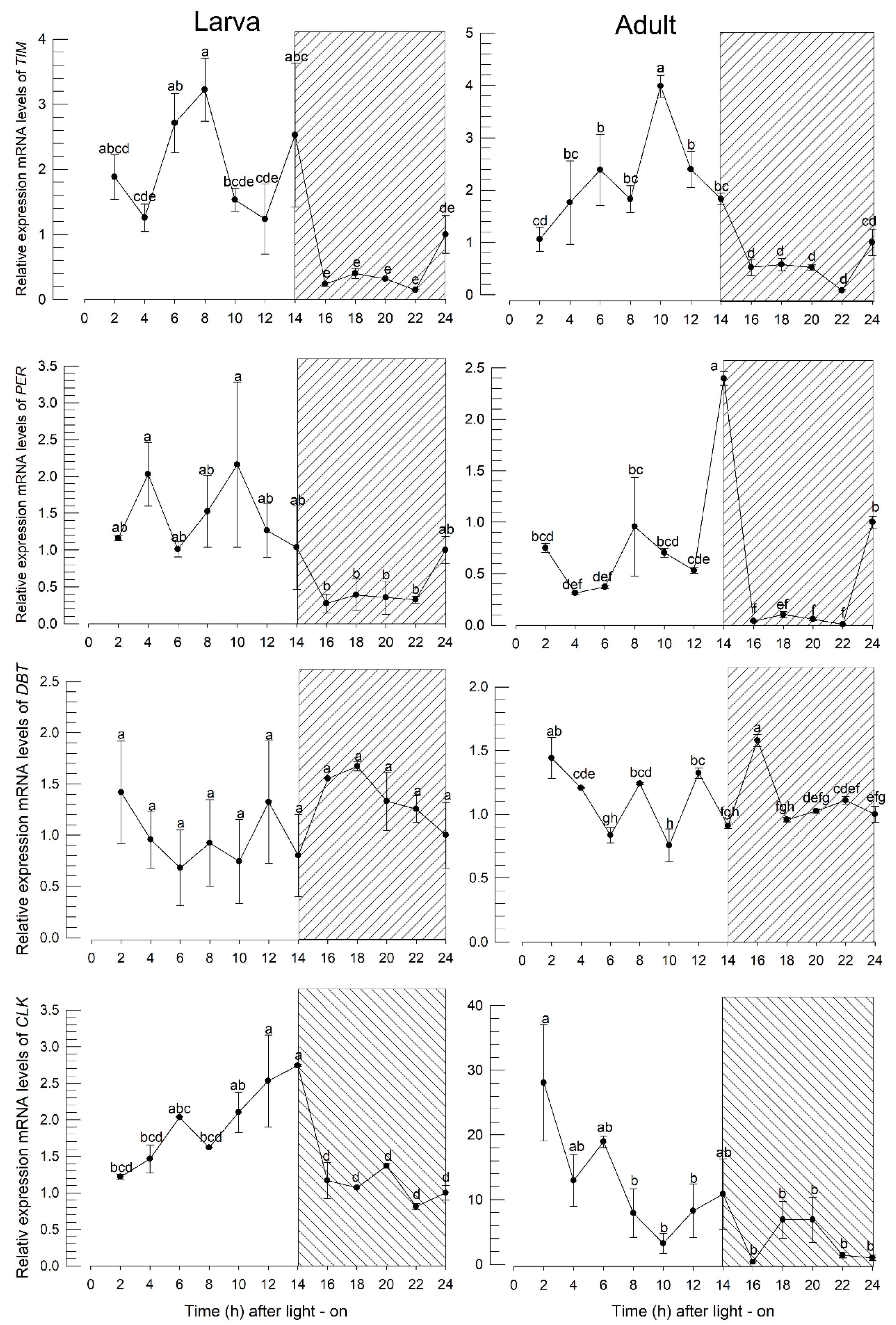
2.8. Measuring expression levels of circadian genes
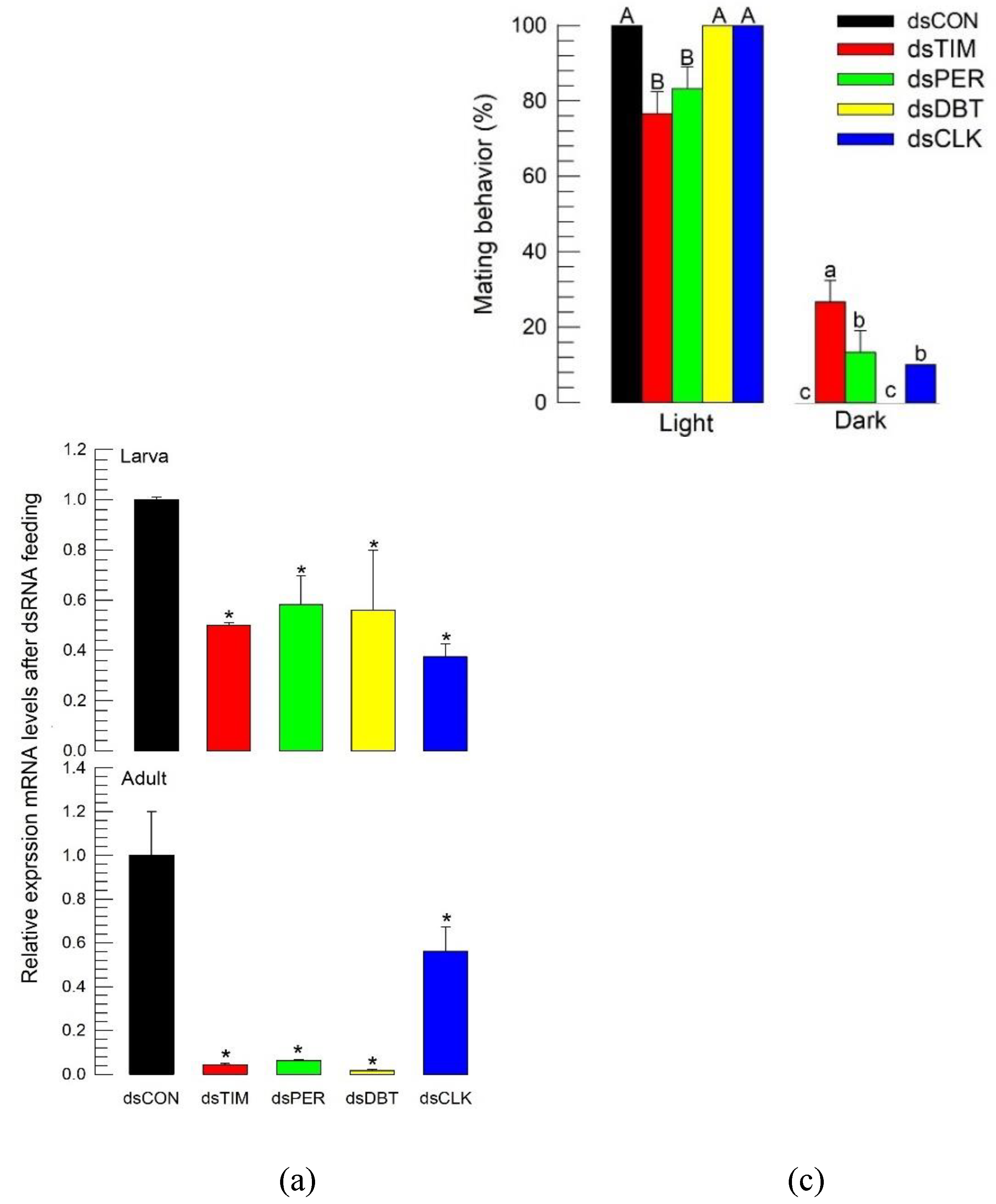
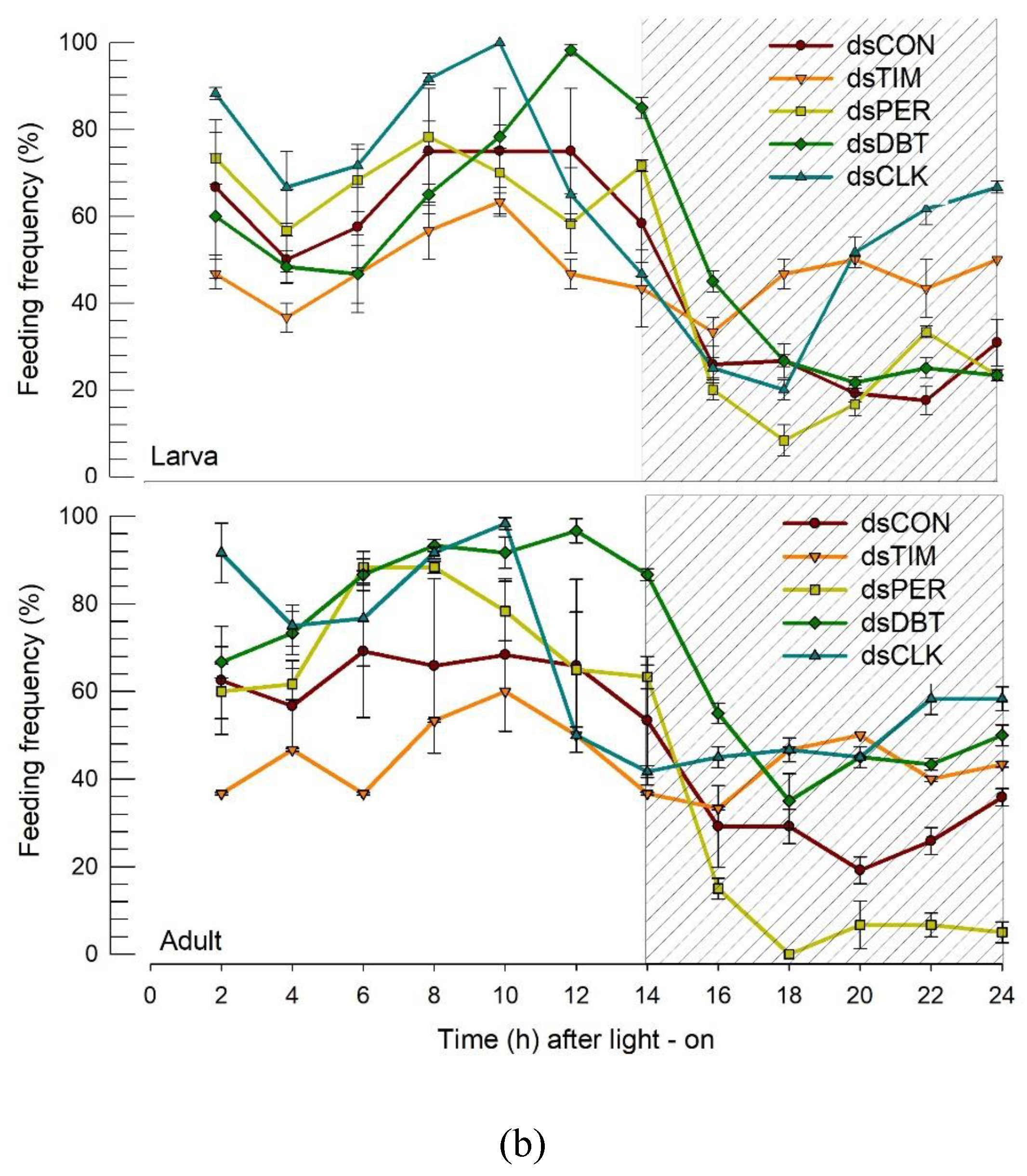
2.9. RNA interference (RNAi) treatment and subsequent behavior assays
2.10. Statistical analysis
3. Results
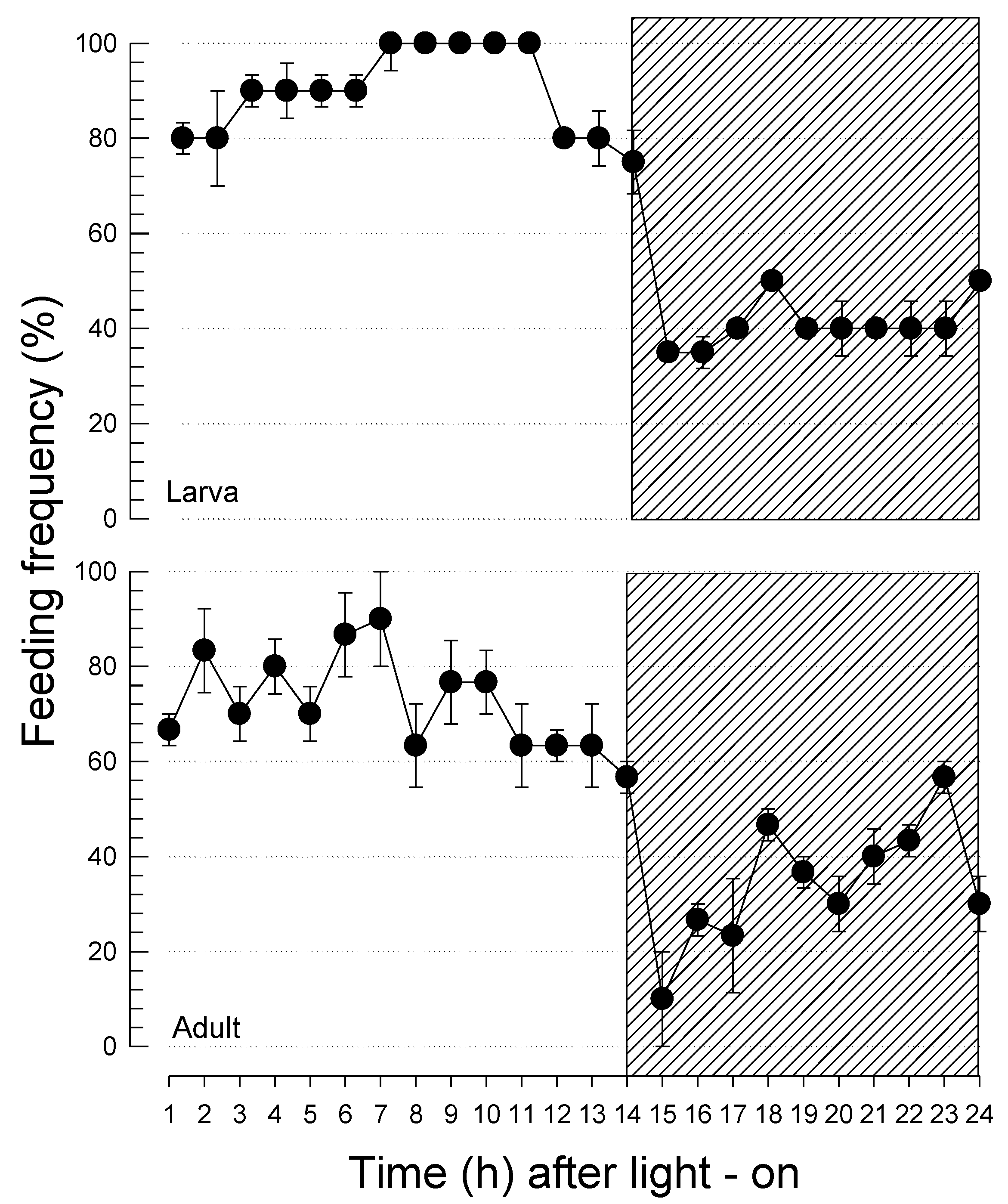
3.1. Diurnal feeding behavior of F. occidentalis
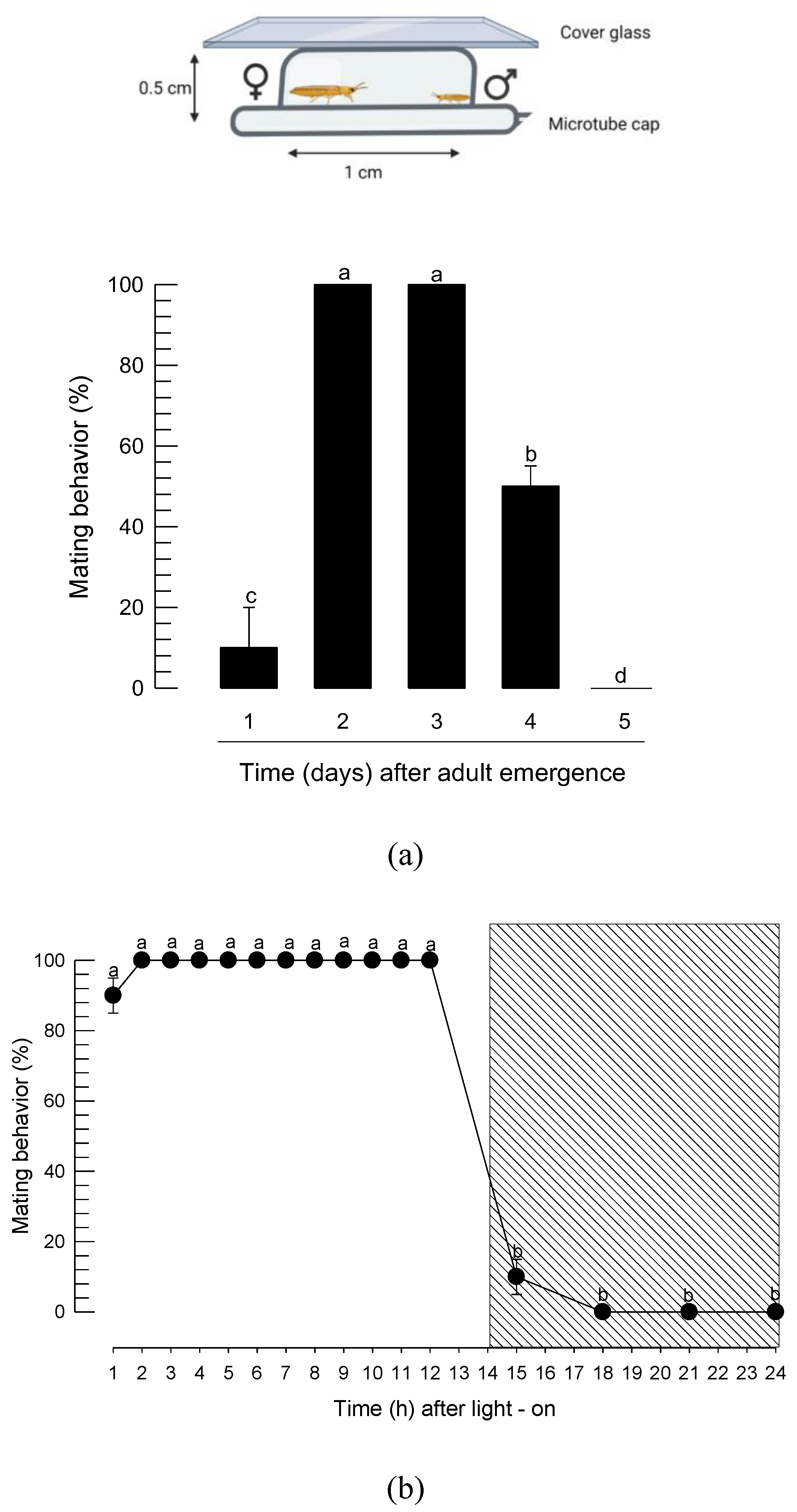
3.2. Diurnal mating behavior of F. occidentalis
3.3. Diurnal calling behavior of F. occidentalis
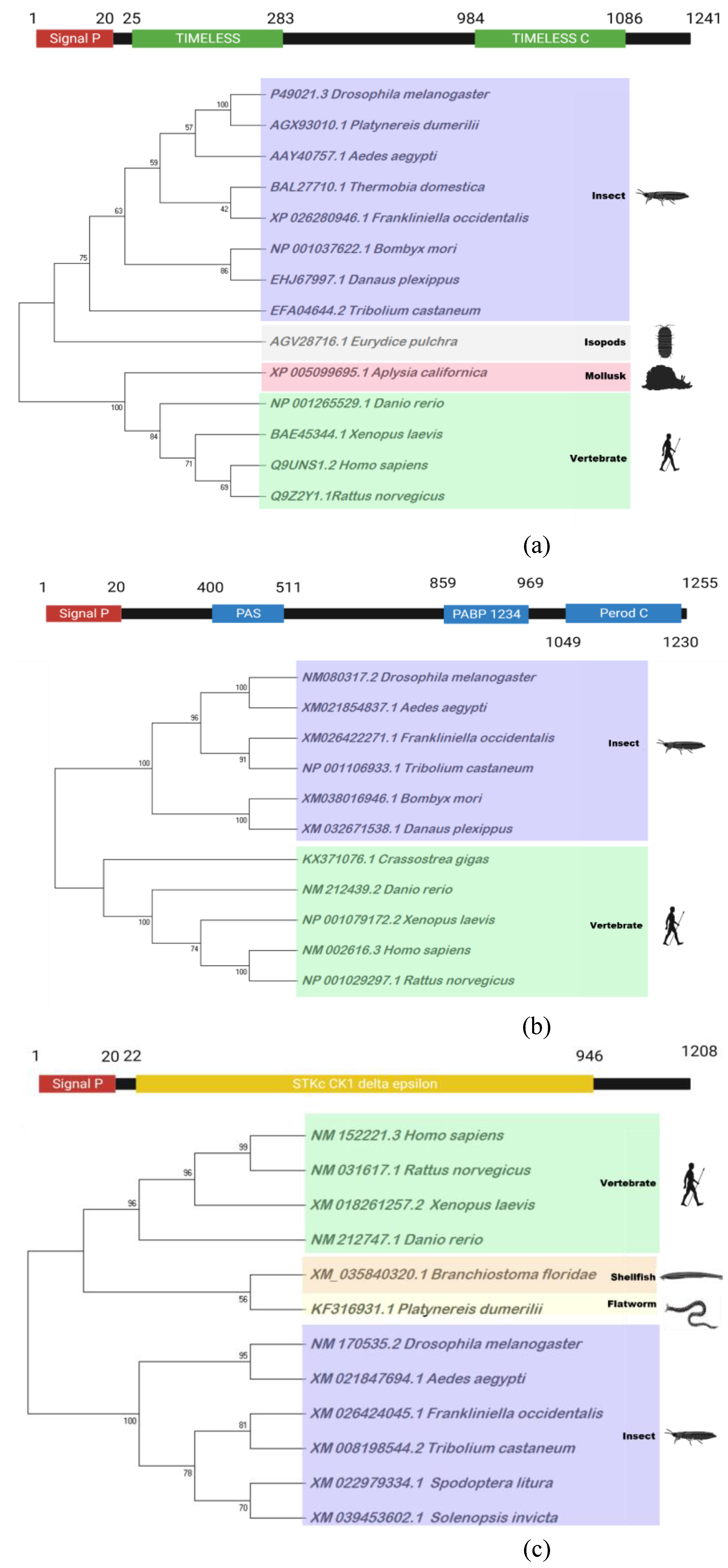
3.4. Prediction of four genes associated with circadian clock and their expression profiles
3.5. RNAi treatments of circadian genes disrupt the circadian rhythmicity of the feeding and mating behaviors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Woo, K.S.; Ahn, S.B.; Lee, S.H.; Kwon, H.M. First record of Frankliniella occidentalis and its distribution and host plants in Korea. Korean J. Appl. Entomol. 1994, 33, 127. [Google Scholar]
- Lee, G.S.; Lee, J.H.; Kang, S.H.; Woo, K.S. Thrips species (Thysanoptera: Thripidae) in winter season and their vernal activities on Jeju island, Korea. J. Asia Pac. Entomol. 2001, 4, 115–122. [Google Scholar] [CrossRef]
- Lim, U.T.; Kim, E.; Mainali, B.P. Flower model traps reduced thrips infestations on a pepper crop in field. J. Asia Pac. Entomol. 2013, 16, 143–145. [Google Scholar] [CrossRef]
- Kim, C.Y.; Choi, D.Y.; Kang, J.H.; Ahmed, S.; Kil, E.J.; Kwon, G.M.; Lee, G.S.; Kim, Y. Thrips infesting hot pepper cultured in greenhouses and variation in gene sequences encoded in TSWV. Korean J. Appl. Entomol. 2021, 60, 381–401. [Google Scholar]
- Shuqi, H.; Ying, L.; Lei, Q.; Zhihua, L.; Chao, X.; Lu, Y.; Furong, G. The influence of elevated CO2 concentration on the fitness traits of Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae). Environ. Entomol. 2017, 46, 722–728. [Google Scholar] [CrossRef]
- Bhuyain, M.M.H.; Lim, U.T. Relative susceptibility to pesticides and environmental conditions of Frankliniella intonsa and F. occidentalis (Thysanoptera: Thripidae), an underlying reason for their asymmetrical occurrence. PLoS ONE 2020, 15, e0237876. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wu, S.; Xing, Z.; Xu, R.; Cai, W.; Lei, Z. Behavioral responses of western flower thrips (Frankliniella occidentalis) to visual and olfactory cues at short distances. Insects 2020, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Stukenberg, N.; Pietruska, M.; Waldherr, A.; Meyhöfer, R. Wavelength-specific behavior of the western flower thrips (Frankliniella occidentalis): evidence for a blue-green chromatic mechanism. Insects 2020, 11, 423. [Google Scholar] [CrossRef] [PubMed]
- Brødsgaard, H.F. Effect of photoperiod on the bionomics of Frankliniella occidentalis (Pergande) (Thysanoptera, Thripidae). J. Appl. Entomol. 1994, 117, 498–507. [Google Scholar] [CrossRef]
- Crespo-Flores, S.L.; Barber, A.F. The Drosophila circadian clock circuit is a nonhierarchical network of peptidergic oscillators. Curr. Opin. Insect Sci. 2022, 52, 100944. [Google Scholar] [CrossRef]
- Lin, C.; Todo, T. The cryptochromes. Genome Biol. 2005, 6, 220. [Google Scholar] [CrossRef]
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Tokuoka, A.; Itoh, T.Q.; Hori, S.; Uryu, O.; Danbara, Y.; Nose, M.; Bando, T.; Tanimura, T.; Tomioka, K. cryptochrome genes form an oscillatory loop independent of the per/tim loop in the circadian clockwork of the cricket Gryllus bimaculatus. Zool. Lett. 2017, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Tomioka, K.; Matsumoto, A. The circadian system in insects: cellular, molecular, and functional organization. Adv. Insect Physiol. 2019, 56, 73–115. [Google Scholar]
- Emery, P.; Stanewsky, R.; Hall, J.C.; Rosbash, M. Drosophila cryptochromes: a unique circadian-rhythm photoreceptor. Nature 2000, 404, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Parikh, V.; Itsukaichi, T.; Bae, K.; Edery, I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science 1996, 271, 1740–1744. [Google Scholar] [CrossRef] [PubMed]
- Ceriani, M.F.; Darlington, T.K.; Staknis, D.; Mas, P.; Petti, A.A.; Weitz, C.J.; Kay, S.A. Light-dependent sequentation of TIMELESS by CRYPTOCHROME. Science 1999, 285, 553–556. [Google Scholar] [CrossRef]
- Zhu, H.; Sauman, I.; Yuan, Q.; Casselman, A.; Emery-Le, M.; Emery, P.; Reppert, S.M. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008, 6, e4. [Google Scholar] [CrossRef]
- Rotenberg, D.; Baumann, A.A.; Ben-Mahmoud, S.; Christiaens, O.; Dermauw, W.; Ioannidis, P.; Jacobs, C.G.C.; Vargas Jentzsch, I.M.; Oliver, J.E.; Poelchau, M.F.; et al. Genome-enabled insights into the biology of thrips as crop pests. BMC Biol. 2020, 18, 142. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data analysis using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vatanparast, M.; Ahmed, S.; Herrero, S.; Kim, Y. A non-venomous sPLA2 of a lepidopteran insect: its physiological functions in development and immunity. Dev. Comp. Immunol. 2018, 89, 83–92. [Google Scholar] [CrossRef] [PubMed]
- SAS Institute Inc. SAS/STAT user’s guide, Release 6.03, Ed. Cary, NC. 1989.
- Hughes, M.E.; Hogenesch, J.B. Kornacker, K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 2010, 25, 372–380. [Google Scholar] [CrossRef]
- Aliakbarpour, H.; Rawi, C.S. Diurnal activity of four species of thrips (Thysanoptera: Thripidae) and efficiencies of three nondestructive sampling techniques for thrips in mango inflorescences. J. Econ. Entomol. 2010, 103, 631–640. [Google Scholar] [CrossRef]
- Millar, A.J.; Kay, S.A. The genetics of phototransduction and circadian rhythms in Arabidopsis. BioEssays 1997, 19, 209–214. [Google Scholar] [CrossRef]
- Ishiura, M.; Kutsuna, S.; Aoki, S.; Iwasaki, H.; Andersson, C.R.; Tanabe, A.; Golden, S.S.; Johnson, C.H.; Kondo, T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 1998, 281, 1519–1523. [Google Scholar] [CrossRef]
- Dunlap, J.C. Genetics and molecular analysis of circadian rhythms. Annu. Rev. Genet. 1996, 30, 579–601. [Google Scholar] [CrossRef] [PubMed]
- Rosbash, M.; Allada, R.; Dembinska, M.E.; Guo, W.Q.; Le, M.; Marrus, S.B.; Qian, Z.; Rutila, J.E.; Yaglom, J.; Zeng, H. A Drosophila circadian clock. Cold Spring Harbor, Cold Spring Harbor Laboratory Press, NY, pp. 265–278. 1996.
- Vitaterna, M.H.; King, D.P.; Chang, A.M.; Kornhauser, J.M.; Lowrey, P.L.; McDonald, J.D.; Dove, W.F.; Pinto, L.H.; Turek, F.W.; Takahashi, J.S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 1994, 264, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef]
- Reddy, P.; Zehring, W.A.; Wheeler, D.A.; Pirrotta, V.; Hadfield, C.; Hall, J.C.; Rosbash, M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 1984, 38, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Vosshall, L.B.; Price, J.L.; Sehgal, A.; Saez, L.; Young, M.W. Specific block in nuclear localization of period protein by a second clock mutation, timeless. Science 1994, 263, 1606–1609. [Google Scholar] [CrossRef]
- Saez, L.; Young, M.W. Regulation of nuclear entry of the Drosophila clock proteins PERIOD and TIMELESS. Neuron. 1996, 17, 911–920. [Google Scholar] [CrossRef]
- Rutila, J.E.; Suri, V.; Le, M.; So, W.V.; Rosbash, M.; Hall, J.C. CYCLE is a second bHLH-PAS protein essential for circadian transcription of Drosophila period and timeless. Cell 1998, 93, 805–814. [Google Scholar] [CrossRef]
- Allada, R.; White, N.E.; So, W.V.; Hall, J.C.; Rosbash, M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 1998, 93, 791–804. [Google Scholar] [CrossRef]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, C.F.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Darlington, T.K.; Wager-Smith, K.; Ceriani, M.F.; Staknis, D.; Gekakis. N.; Steeves, T.D.L.; Weitz, C.J.; Takahashi, J.S.; Kay, S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 1998, 280, 1599–1603. [Google Scholar] [CrossRef] [PubMed]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Akinyemi, A.O.; Kirk, W.D.J. Experienced males recognise and avoid mating with non-virgin females in the western flower thrips. PLoS ONE 2019, 14, e0224115. [Google Scholar] [CrossRef] [PubMed]
- de Kogel, W.J.; van Deventer, P. Intraspecific attraction in the western flower thrips, Frankliniella occidentalis; indications for a male sex pheromone. Entomol. Exp. Appl. 2003, 107, 87–89. [Google Scholar] [CrossRef]
- Khan, F.; Kim, K.; Sung, J.; Lim, H.; Kim, S.G.; Choi, M.Y.; Kim, Y. A novel physiological function of pheromone biosynthesis-activating neuropeptide in production of aggregation pheromone. Sci. Rep. 2023, 13, 5551. [Google Scholar] [CrossRef]
- Foley, L.E.; Emery, P. Drosophila cryptochrome: variations in blue. J. Biol. Rhythms 2020, 35, 16–27. [Google Scholar] [CrossRef]
- Moriyama,Y. ; Takeuchi, K.; Shinohara, T.; Miyagawa, K.; Matsuka, M.; Yoshii, T.; Tomioka, K. Timeless plays an important role in compound eye-dependent photic entrainment of the circadian rhythm in the cricket Gryllus bimaculatus. Zool. Sci. 2022, 39, 397–405. [Google Scholar]
- Liu, T.; Mahesh, G.; Yu, W.; Hardin, P.E. CLOCK stabilizes CYCLE to initiate clock function in Drosophila. Proc. Natl. Acad. Sci. USA, 2017, 114, 10972–10977. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, M.S.; Kirk, W.D.J. The effect of photoperiod on walking, feeding, and oviposition in the western flower thrips. Entomol. Exp. Appl. 2004, 2004. 111, 209–214. [Google Scholar] [CrossRef]
- Andongma, A.A.; Greig, C.; Dyson, P.J.; Flynn, N.; Whitten, M.M.A. Optimization of dietary RNA interference delivery to western flower thrips Frankliniella occidentalis and onion thrips Thrips tabaci. Arch. Insect Biochem. Physiol. 2020, 103, e21645. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.; Dyson, P. Gene silencing in non-model insects: overcoming hurdles using symbiotic bacteria for trauma-free sustainable delivery of RNA interference. BioEssays 2017, 39, 1600247. [Google Scholar] [CrossRef]
- Zotti, M.; Smagghe, G. RNAi technology for insect management and protection of beneficial insects from diseases: lessons, challenges, and risk assessments. Neotrop. Entomol. 2015, 44, 197–213. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



