Submitted:
27 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Microbiological testing of Legionella spp.
2.3. DNA extraction from Legionella isolates
2.4. Whole Genome Sequencing
2.5. Data Analysis
3. Results
3.1. Prevalence of L. pneumophila in residential buildings
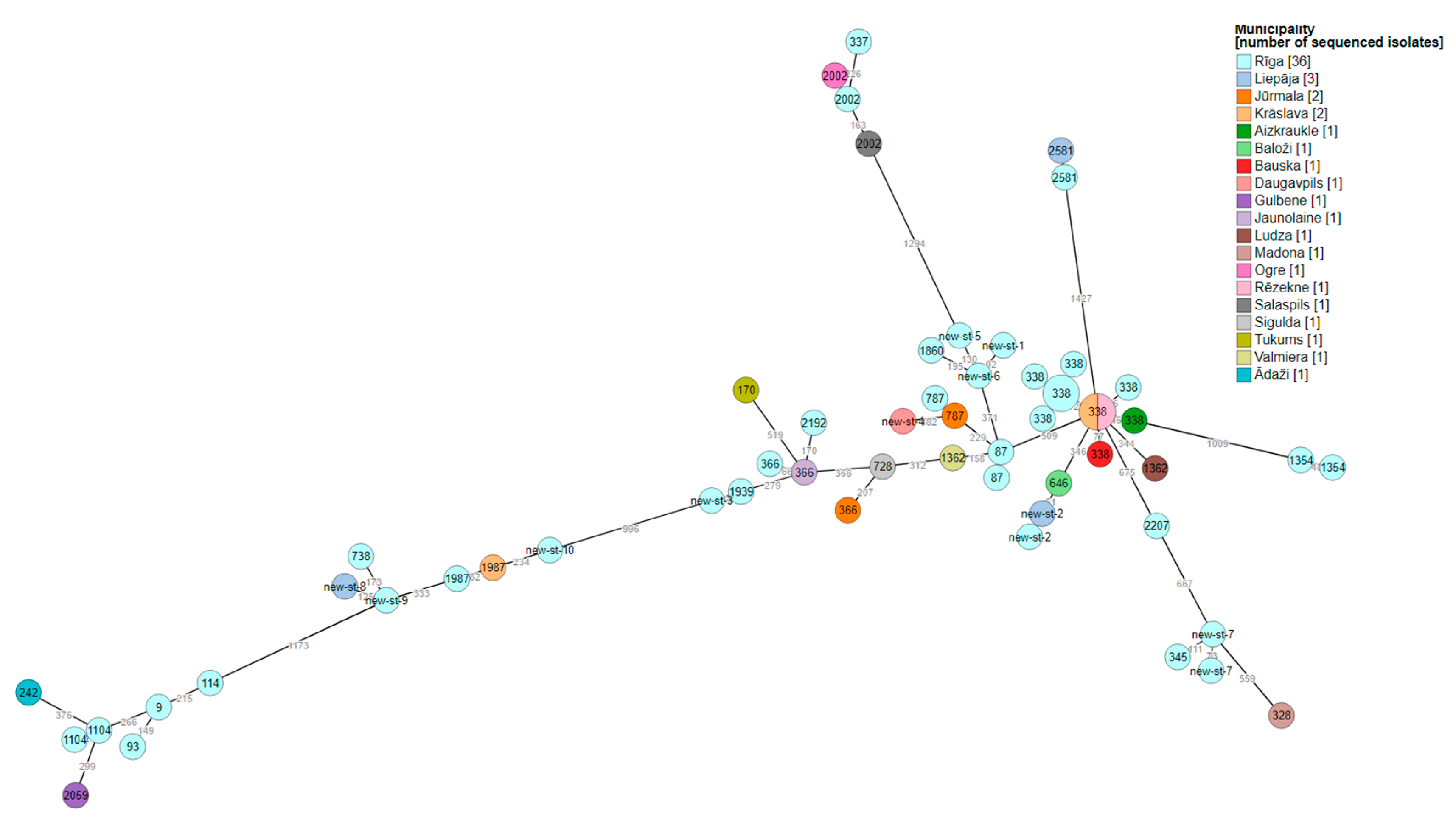
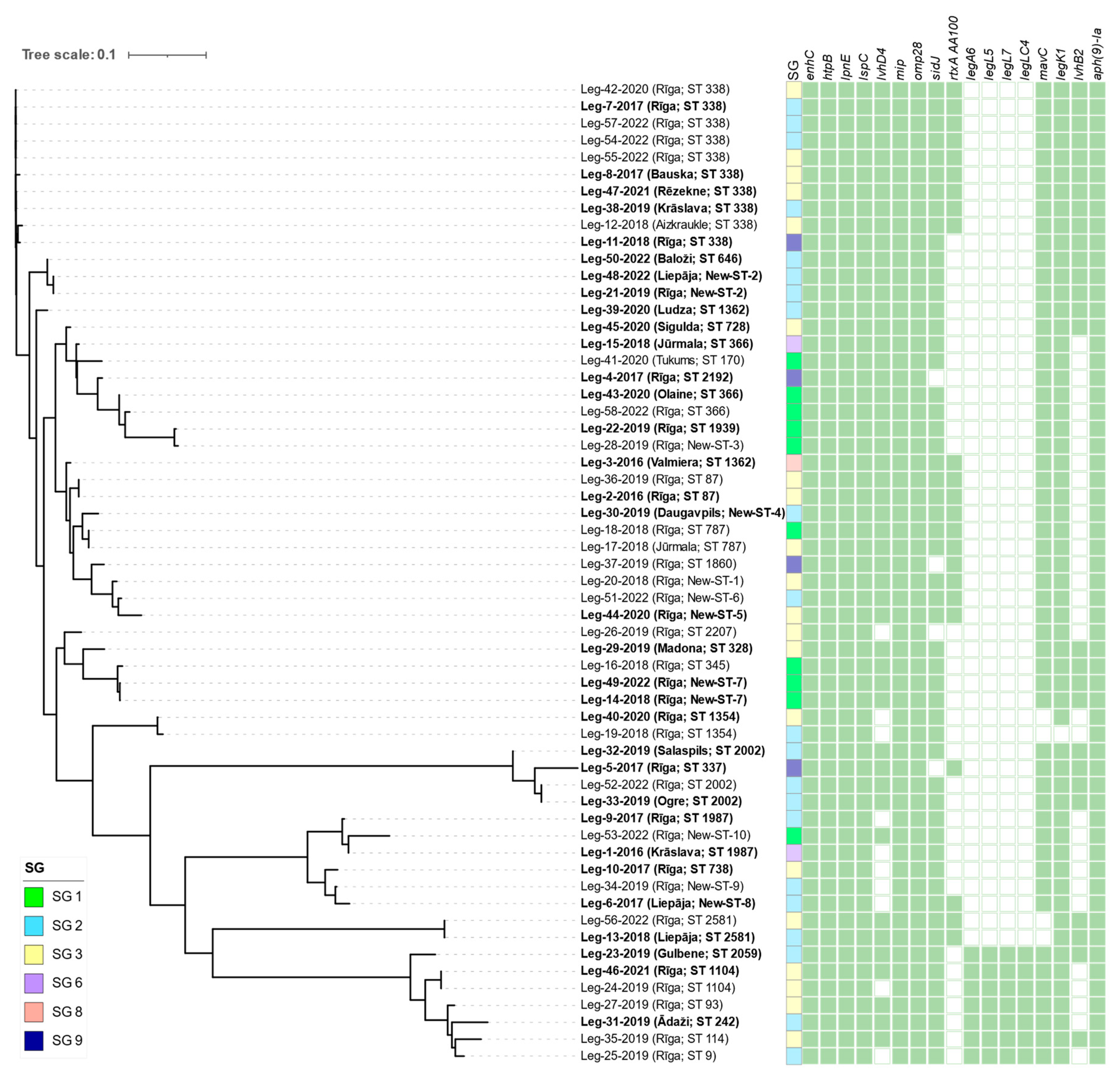
3.2. Whole-genome sequencing of L. pneumophila
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mondino, S.; Schmidt, S.; Rolando, M.; Escoll, P.; Gomez-Valero, L.; Buchrieser, C. Legionnaires’ Disease: State of the Art Knowledge of Pathogenesis Mechanisms of Legionella. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 439–466. [Google Scholar] [CrossRef] [PubMed]
- Legionnaires’ Disease-Annual Epidemiological Report for 2020 Available online: https://www.ecdc.europa.eu/en/publications-data/legionnaires-disease-annual-epidemiological-report-2020.
- Byrne, B.G.; McColm, S.; McElmurry, S.P.; Kilgore, P.E.; Sobeck, J.; Sadler, R.; Love, N.G.; Swanson, M.S. Prevalence of Infection-Competent Serogroup 6 Legionella Pneumophila within Premise Plumbing in Southeast Michigan. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.T.; Kamali, A.; Vugia, D.J. Legionella Epidemiologic and Environmental Risks. Current Epidemiology Reports 2019, 6, 310–320. [Google Scholar] [CrossRef]
- Kessler, M.A.; Osman, F.; Marx, J.; Pop-Vicas, A.; Safdar, N. Hospital-Acquired Legionella Pneumonia Outbreak at an Academic Medical Center: Lessons Learned. American Journal of Infection Control 2021. [Google Scholar] [CrossRef]
- De Filippis, P.; Mozzetti, C.; Messina, A.; D'Alò, G.L. Prevalence of Legionella in Retirement Homes and Group Homes Water Distribution Systems. Science of The Total Environment 2018, 643, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Mentula, S.; Kääriäinen, S.; Jaakola, S.; Niittynen, M.; Airaksinen, P.; Koivula, I.; Lehtola, M.; Mauranen, E.; Mononen, I.; Savolainen, R.; et al. Tap Water as the Source of a Legionnaires’ Disease Outbreak Spread to Several Residential Buildings and One Hospital, Finland, 2020 to 2021. Eurosurveillance 2023, 28. [Google Scholar] [CrossRef]
- Nisar, M.A.; Ross, K.E.; Brown, M.H.; Bentham, R.; Whiley, H. Water Stagnation and Flow Obstruction Reduces the Quality of Potable Water and Increases the Risk of Legionelloses. Frontiers in Environmental Science 2020, 8. [Google Scholar] [CrossRef]
- Singh, R.; Chauhan, D.; Fogarty, A.; Rasheduzzaman, M.; Gurian, P.L. Practitioners’ Perspective on the Prevalent Water Quality Management Practices for Legionella Control in Large Buildings in the United States. Water 2022, 14, 663. [Google Scholar] [CrossRef]
- Kruse, E.-B.; Wehner, A.; Wisplinghoff, H. Prevalence and Distribution of Legionella Spp in Potable Water Systems in Germany, Risk Factors Associated with Contamination, and Effectiveness of Thermal Disinfection. American Journal of Infection Control 2016, 44, 470–474. [Google Scholar] [CrossRef]
- Ge, Z.; Yuan, P.; Chen, L.; Chen, J.; Shen, D.; She, Z.; Lu, Y. New Global Insights on the Regulation of the Biphasic Life Cycle and Virulence via ClpP-Dependent Proteolysis in Legionella Pneumophila. Molecular & Cellular Proteomics 2022, 21, 100233. [Google Scholar] [CrossRef]
- Chauhan, D.; Shames, S.R. Pathogenicity and Virulence Of Legionella: Intracellular Replication and Host Response. Virulence 2021, 12, 1122–1144. [Google Scholar] [CrossRef] [PubMed]
- Gattuso, G.; Rizzo, R.; Lavoro, A.; Spoto, V.; Porciello, G.; Montagnese, C.; Cinà, D.; Cosentino, A.; Lombardo, C.; Mezzatesta, M.L.; et al. Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies. Antibiotics 2022, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Abu Khweek, A.; Amer, A.O. Factors Mediating Environmental Biofilm Formation by Legionella Pneumophila. Frontiers in Cellular and Infection Microbiology 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, L.; Karagöz, M.S.; Mayer, B.E.; Steinert, M. Protein Sociology of ProA, Mip and Other Secreted Virulence Factors at the Legionella Pneumophila Surface. Frontiers in Cellular and Infection Microbiology 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-L.; Li, D.; Zhan, X.-Y. Concept about the Virulence Factor of Legionella. Microorganisms 2022, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization ISO 19458:2006 Water Quality—Sampling for Microbiological Analysis. Available online: https://www.iso.org/standard/33845.html. (accessed on 15 November 2022).
- International Organization for Standardization ISO 11731:2017 Water Quality—Enumeration of Legionella. Available online: https://www.iso.org/standard/61782.html. (accessed on 15 November 2022).
- Valciņa, O.; Pūle, D.; Ķibilds, J.; Lazdāne, A.; Trofimova, J.; Makarova, S.; Konvisers, G.; Ķimse, L.; Krūmiņa, A.; Bērziņš, A. Prevalence and Genetic Diversity of Legionella Spp. In Hotel Water-Supply Systems in Latvia. Microorganisms 2023, 11, 596. [Google Scholar] [CrossRef]
- Prjibelski, A.D.; Puglia, G.D.; Antipov, D.; Bushmanova, E.; Giordano, D.; Mikheenko, A.; Vitale, D.; Lapidus, A. Extending RnaSPAdes Functionality for Hybrid Transcriptome Assembly. BMC Bioinformatics 2020, 21. [Google Scholar] [CrossRef]
- Gaia, V.; Fry, N.K.; Afshar, B.; Luck, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus Sequence-Based Scheme for Epidemiological Typing of Clinical and Environmental Isolates of Legionella Pneumophila. Journal of Clinical Microbiology 2005, 43, 2047–2052. [Google Scholar] [CrossRef]
- Mentasti, M.; Underwood, A.; Lück, C.; Kozak-Muiznieks, N.A.; Harrison, T.G.; Fry, N.K. Extension of the Legionella Pneumophila Sequence-Based Typing Scheme to Include Strains Carrying a Variant of the N-Acylneuraminate Cytidylyltransferase Gene. Clinical Microbiology and Infection 2014, 20, 435–441. [Google Scholar] [CrossRef]
- Moran-Gilad, J.; Prior, K.; Yakunin, E.; Harrison, T.G.; Underwood, A.; Lazarovitch, T.; Valinsky, L.; Lück, C.; Krux, F.; Agmon, V.; et al. Design and Application of a Core Genome Multilocus Sequence Typing Scheme for Investigation of Legionnaires’ Disease Incidents. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Research 2021, 50, D912–D917. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. Journal of Computational Biology 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Samrakandi, M.M.; Cirillo, S.L.G.; Ridenour, D.A.; Bermudez, L.E.; Cirillo, J.D. Genetic and Phenotypic Differences between Legionella Pneumophila Strains. Journal of Clinical Microbiology 2002, 40, 1352–1362. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.; Birney, E. Automated Generation of Heuristics for Biological Sequence Comparison. BMC Bioinformatics 2005, 6, 31. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. Journal of Antimicrobial Chemotherapy 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Conover, G.M.; Isberg, R.R. Legionella Pneumophila EnhC Is Required for Efficient Replication in Tumour Necrosis Factor α-Stimulated Macrophages. Cellular Microbiology 2008, 10, 1906–1923. [Google Scholar] [CrossRef]
- Hoffman, P.C.; Garduno, R.A. Surface-Associated Heat Shock Proteins Of Legionella Pneumophila and Helicobacter Pylori: Roles in Pathogenesis and Immunity. Infectious Diseases in Obstetrics and Gynecology 1999, 7, 58–63. [Google Scholar] [CrossRef]
- Helbig, J.H.; König, B.; Knospe, H.; Bubert, B.; Yu, C.; Lück, C.P.; Riboldi-Tunnicliffe, A.; Hilgenfeld, R.; Jacobs, E.; Hacker, J.; et al. The PPIase Active Site of Legionella Pneumophila Mip Protein Is Involved in the Infection of Eukaryotic Host Cells. Biological Chemistry 2003, 384. [Google Scholar] [CrossRef]
- Bellinger-Kawahara, C.; Horwitz, M.A. Complement Component C3 Fixes Selectively to the Major Outer Membrane Protein (MOMP) of Legionella Pneumophila and Mediates Phagocytosis of Liposome-MOMP Complexes by Human Monocytes. The Journal of Experimental Medicine 1990, 172, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Gan, N.; Nakayasu, E.S.; Hollenbeck, P.J.; Luo, Z.-Q. Legionella Pneumophila Inhibits Immune Signalling via MavC-Mediated Transglutaminase-Induced Ubiquitination of UBE2N. Nature Microbiology 2018, 4, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Losick, V.P.; Haenssler, E.; Moy, M.-Y.; Isberg, R.R. LnaB: A Legionella Pneumophila Activator of NF-ΚB. Cellular Microbiology 2010, 12, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Bhogaraju, S.; Bonn, F.; Mukherjee, R.; Adams, M.; Pfleiderer, M.M.; Galej, W.P.; Matkovic, V.; Lopez-Mosqueda, J.; Kalayil, S.; Shin, D.; et al. Inhibition of Bacterial Ubiquitin Ligases by SidJ–Calmodulin Catalysed Glutamylation. Nature 2019, 572, 382–386. [Google Scholar] [CrossRef]
- Bandyopadhyay, P.; Lang, E.A.S.; Rasaputra, K.S.; Steinman, H.M. Implication of the VirD4 Coupling Protein of the Lvh Type 4 Secretion System in Virulence Phenotypes of Legionella Pneumophila. Journal of Bacteriology 2013, 195, 3468–3475. [Google Scholar] [CrossRef]
- Newton, H.J.; Browning, G.F.; Dao, J.; McAlister, A.D.; Sloan, J.; Cianciotto, N.P.; Hartland, E.L. Sel1 Repeat Protein LpnE Is a Legionella Pneumophila Virulence Determinant That Influences Vacuolar Trafficking. 2007, 75, 5575–5585. [Google Scholar] [CrossRef]
- Rossier, O.; Starkenburg, S.R.; Cianciotto, N.P. Legionella Pneumophila Type II Protein Secretion Promotes Virulence in the A/J Mouse Model of Legionnaires’ Disease Pneumonia. Infection and Immunity 2003, 72, 310–321. [Google Scholar] [CrossRef]
- Cirillo, S.L.G.; Bermudez, L.E.; El-Etr, S.H.; Duhamel, G.E.; Cirillo, J.D. Legionella Pneumophila Entry Gene RtxA Is Involved in Virulence. Infection and Immunity 2001, 69, 508–517. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Research 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Research 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Dilger, T.; Melzl, H.; Gessner, A. Legionella Contamination in Warm Water Systems: A Species-Level Survey. International Journal of Hygiene and Environmental Health 2018, 221, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Felice, A.; Franchi, M.; De Martin, S.; Vitacolonna, N.; Iacumin, L.; Civilini, M. Environmental Surveillance and Spatio-Temporal Analysis of Legionella Spp. In a Region of Northeastern Italy (2002–2017). PLoS ONE 2019, 14, e0218687. [Google Scholar] [CrossRef] [PubMed]
- Gleason, J.A.; Conner, L.E.; Ross, K.M. Associations of Household Factors, Hot Water Temperature, and Chlorine Residual with Legionella Occurrence in Single-Family Homes in New Jersey. Science of The Total Environment 2023, 870, 161984. [Google Scholar] [CrossRef] [PubMed]
- Valciņa, O.; Pūle, D.; Mališevs, A.; Trofimova, J.; Makarova, S.; Konvisers, G.; Bērziņš, A.; Krūmiņa, A. Co-Occurrence of Free-Living Amoeba and Legionella in Drinking Water Supply Systems. Medicina 2019, 55, 492. [Google Scholar] [CrossRef] [PubMed]
- Cabinet of Ministers of Latvia MK Nr. 906, 2010.28.09. Rules of Sanitary Maintenance of the Residential House. Available online: https://likumi.lv/ta/id/218830-dzivojamas-majas-sanitaras-apkopes-noteikumi (accessed on 11 May 2023).
- Valciņa, O.; Pūle, D.; Lucenko, I.; Krastiņa, D.; Šteingolde, Ž.; Krūmiņa, A.; Bērziņš, A. Legionella Pneumophila Seropositivity-Associated Factors in Latvian Blood Donors. International Journal of Environmental Research and Public Health 2015, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Bešić, A.; Karakaš, S.; Obradović, Z.; Mušović, A.; Hrapović, E. Travel-Related Epidemiological Studies of Legionellosis in Federation of Bosnia and Herzegovina. Health and Technology 2021, 11, 971–979. [Google Scholar] [CrossRef]
- Raphael, B.H.; Huynh, T.; Brown, E.; Smith, J.C.; Ruberto, I.; Getsinger, L.; White, S.; Winchell, J.M. Culture of Clinical Specimens Reveals Extensive Diversity of Legionella Pneumophila Strains in Arizona. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, S.; Cai, X.; Mu, D.; Zhang, X.; Kang, J.; Zhao, L.; Chen, Y. Sequence-Based Typing of Clinical and Environmental Legionella Pneumophila Isolates in Shenyang, China. Enfermedades infecciosas y microbiologia clinica (English ed.) 2021, 39, 383–389. [Google Scholar] [CrossRef]
- Keše, D.; Obreza, A.; Rojko, T.; Kišek, T.C. Legionella Pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020. Diagnostics 2021, 11, 1201. [Google Scholar] [CrossRef]
- Lévesque, S.; Lalancette, C.; Bernard, K.; Pacheco, A.L.; Dion, R.; Longtin, J.; Tremblay, C. Molecular Typing of Legionella Pneumophila Isolates in the Province of Quebec from 2005 to 2015. PLoS ONE 2016, 11, e0163818. [Google Scholar] [CrossRef]
- Best, A.; Abu Kwaik, Y. Evolution of the Arsenal of Legionella Pneumophila Effectors to Modulate Protist Hosts. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; O’Connor, T.J. Beyond Paralogs: The Multiple Layers of Redundancy in Bacterial Pathogenesis. Frontiers in Cellular and Infection Microbiology 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Ridenour, D.A.; Cirillo, S.L.G.; Feng, S.; Samrakandi, M.M.; Cirillo, J.D. Identification of a Gene That Affects the Efficiency of Host Cell Infection by Legionella Pneumophila in a Temperature-Dependent Fashion. Infection and Immunity 2003, 71, 6256–6263. [Google Scholar] [CrossRef] [PubMed]
- Sawczyn-Domańska, A. Detection of Legionella Spp. And Occurrence of Virulence Genes: Lvh, RtxA and EnhC in Water Samples from Artificial Water Systems. Annals of Agricultural and Environmental Medicine 2021, 28, 617–620. [Google Scholar] [CrossRef]
- Zeng, L.Z.; Liao, H.Y.; Luo, L.Z.; He, S.S.; Qin, T.; Zhou, H.J.; Li, H.X.; Chen, D.L.; Chen, J.P. An Investigation on the Molecular Characteristics and Intracellular Growth Ability among Environmental and Clinical Isolates of Legionella Pneumophila in Sichuan Province, China. Biomedical and Environmental Sciences 2019, 32, 520–530. [Google Scholar] [CrossRef]
- D’Auria, G.; Jiménez, N.; Peris-Bondia, F.; Pelaz, C.; Latorre, A.; Moya, A. Virulence Factor Rtx in Legionella Pneumophila, Evidence Suggesting It Is a Modular Multifunctional Protein. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
| Gene | Virulence factor | Roles | |
| Bacterial surface structures | enhC | EnhC | Enhanced entry, trafficking of Legionella-containing vacuole [31] |
| htpB | Hsp60 | Attachment, modulation of invasion [32] | |
| mip | MIP | Penetration of the lung epithelial barrier [33] | |
| omp28 | MOMP | Mediating phagocytosis [34] | |
| T4SS effectors | mavC | MavC | Inhibiting host immunity [35] |
| legK1 | LegK1 | Activation of NF-kB [36] | |
| sidJ | SidJ | Calmodulin-activated glutamylase for SidE [37] | |
| lvhD4 | VirD4 | Coupling protein, reversing virulence defects [38] | |
| lpnE | LpnE | Entry into macrophages and epithelial cells; manipulate host cell trafficking [39] | |
| T2SS effectors | lsp | Lsp | Transport proteins from the periplasm to the extracellular space [40] |
| T1SS effectors | rtxA | RtxA | Ensures adherence and entry into host and enhances replication and cytotoxicity and pore forming [41] |
| Buildings with known LD cases | Buildings without known LD cases | Total | ||||
| Samples tested/ positive samples (%) | Average water temperature, °C | Samples tested/ positive samples (%) | Average water temperature, °C | Samples tested/ positive samples (%) | Average water temperature, °C | |
| Cold water | 120/22 (18.3%) | 12.9 ± 0.4 | 44/19 (43.2%) | 15.6 ± 0.7 | 164/41 (25%) | 13.5 ± 0.3 |
| Hot water | 242/107 (44.2%) | 52.1 ± 0.4 | 86/49 (57.0%) | 45.8 ± 1.1 | 328/156 (47.6%) | 50.7 ± 0.4 |
| Total | 362/129 (35.6%) | -- | 130/68 (52.3%) | -- | 492/197 (40.0%) | -- |
| Serogroup | No. of L. pneumophila isolates (%) | ||
|---|---|---|---|
| Buildings with known LD cases | Buildings without known LD cases | Total | |
| SG 1 | 15 (11.6%) | 4 (6.0%) | 19 (9.7%) |
| SG 2 | 69 (53.5%) | 39 (58.2%) | 108 (55.1%) |
| SG 3 | 35 (27.1%) | 20 (29.9%) | 55 (28.1%) |
| SG 6 | 3 (2.3%) | 1 (1.5%) | 4 (2.0%) |
| SG 8 | 1 (0.8%) | -- | 1 (0.5%) |
| SG 9 | 2 (1.6%) | 2 (3.0%) | 4 (2.0%) |
| SG 3, SG 2 | 3 (2.3%) | -- | 3 (1.5%) |
| SG 3, SG 1 | -- | 1 (1.5%) | 1 (0.5%) |
| SG 3, SG 9 | 1 (0.8%) | -- | 1 (0.5%) |
| Serogroup |
Levels of L. pneumophila colonization (min – max (average)), CFU/L |
||
| Buildings with known LD cases | Buildings without known LD cases | p-value | |
| SG 1 | 50 – 4.0×103 (8.4×102) | 4.0×102 – 5.5×103 (2.6×103) |
0.056 |
| SG 2 | 50 – 1.3×103 (1.5×103) |
50 – 6.4×103 (2.1×103) |
0.158 |
| SG 3 | 50 - 1.7×104 (2.8×103) |
1.0×102 – 3.9×103 (8.2×102) |
0.033 |
| Total | 50 - 1.7×104 (2.0×103) |
50 – 6.4×103 (1.7×103) |
0.574 |
| Isolate Id | Year of sampling |
Municipality | SG | Linked with LD cases | Allelic profile | Sequence type | Number of observed virulence genes |
|---|---|---|---|---|---|---|---|
| Leg-1-2016 | 2016 | Krāslava | 6 | Yes | 7,6,17,3,50,11,9 | 1987 | 377 |
| Leg-2-2016 | 2016 | Rīga | 3 | Yes | 2,10,3,28,9,4,13 | 87 | 372 |
| Leg-3-2016 | 2016 | Valmiera | 8 | Yes | 2,10,3,28,9,4,207 | 1362 | 372 |
| Leg-4-2017 | 2017 | Rīga | 9 | Yes | 2,10,24,3,9,4,6 | 2192 | 370 |
| Leg-5-2017 | 2017 | Rīga | 9 | Yes | 10,22,7,28,16,18,6 | 337 | 358 |
| Leg-6-2017 | 2017 | Liepāja | 2 | Yes | 7,10,17,6,9,11,9 | New-ST-8 | 378 |
| Leg-7-2017 | 2017 | Rīga | 2 | Yes | 2,10,15,28,9,4,13 | 338 | 375 |
| Leg-8-2017 | 2017 | Bauska | 3 | Yes | 2,10,15,28,9,4,13 | 338 | 378 |
| Leg-9-2017 | 2017 | Rīga | 2 | Yes | 7,6,17,3,50,11,9 | 1987 | 378 |
| Leg-10-2017 | 2017 | Rīga | 3 | Yes | 7,10,17,28,17,11,9 | 738 | 376 |
| Leg-11-2018 | 2018 | Rīga | 9 | Yes | 2,10,15,28,9,4,13 | 338 | 378 |
| Leg-12-2018 | 2018 | Aizkraukle | 3 | No | 2,10,15,28,9,4,13 | 338 | 378 |
| Leg-13-2018 | 2018 | Liepāja | 2 | Yes | 2,32,20,38,34,35,219 | 2581 | 344 |
| Leg-14-2018 | 2018 | Rīga | 1 | Yes | 6,10,19,3,98,4,novel neuA allele | New-ST-7 | 374 |
| Leg-15-2018 | 2018 | Jūrmala | 6 | Yes | 2,10,3,3,9,4,6 | 366 | 374 |
| Leg-16-2018 | 2018 | Rīga | 1 | No | 6,10,19,3,19,4,11 | 345 | 374 |
| Leg-17-2018 | 2018 | Jūrmala | 3 | No | 2,10,1,3,9,4,3 | 787 | 372 |
| Leg-18-2018 | 2018 | Rīga | 1 | No | 2,10,1,3,9,4,3 | 787 | 372 |
| Leg-19-2018 | 2018 | Rīga | 2 | No | 2,10,24,28,4,4,207 | 1354 | 312 |
| Leg-20-2018 | 2018 | Rīga | 3 | No | 2,10,3,3,50,4,3 | New-ST-1 | 372 |
| Leg-21-2019 | 2019 | Rīga | 2 | Yes | 2,10,21,28,9,4,6 | New-ST-2 | 374 |
| Leg-22-2019 | 2019 | Rīga | 1 | Yes | 2,10,1,3,9,4,6 | 1939 | 374 |
| Leg-23-2019 | 2019 | Gulbene | 2 | Yes | 3,4,1,6,35,9,220 | 2059 | 415 |
| Leg-24-2019 | 2019 | Rīga | 3 | No | 3,13,1,28,14,9,13 | 1104 | 397 |
| Leg-25-2019 | 2019 | Rīga | 2 | No | 3,10,1,3,14,9,11 | 9 | 396 |
| Leg-26-2019 | 2019 | Rīga | 3 | No | 2,10,19,28,19,4,3 | 2207 | 360 |
| Leg-27-2019 | 2019 | Rīga | 3 | No | 3,10,1,28,14,9,13 | 93 | 415 |
| Leg-28-2019 | 2019 | Rīga | 1 | No | 2,10,17,3,9,4,6 | New-ST-3 | 373 |
| Leg-29-2019 | 2019 | Madona | 3 | Yes | 6,10,19,28,19,4,9 | 328 | 375 |
| Leg-30-2019 | 2019 | Daugavpils | 2 | Yes | 2,10,17,3,9,4,9 | New-ST-4 | 369 |
| Leg-31-2019 | 2019 | Ādaži | 2 | Yes | 3,10,1,28,1,9,3 | 242 | 415 |
| Leg-32-2019 | 2019 | Salaspils | 2 | Yes | 10,22,7,28,16,18,8 | 2002 | 363 |
| Leg-33-2019 | 2019 | Ogre | 2 | Yes | 10,22,7,28,16,18,8 | 2002 | 357 |
| Leg-34-2019 | 2019 | Rīga | 2 | No | 7,10,17,6,17,11,9 | New-ST-9 | 377 |
| Leg-35-2019 | 2019 | Rīga | 3 | No | 3,6,1,6,14,11,9 | 114 | 415 |
| Leg-36-2019 | 2019 | Rīga | 3 | No | 2,10,3,28,9,4,13 | 87 | 373 |
| Leg-37-2019 | 2019 | Rīga | 9 | No | 2,10,3,3,9,4,207 | 1860 | 371 |
| Leg-38-2019 | 2019 | Krāslava | 2 | Yes | 2,10,15,28,9,4,13 | 338 | 378 |
| Leg-39-2020 | 2020 | Ludza | 2 | Yes | 2,10,3,28,9,4,207 | 1362 | 376 |
| Leg-40-2020 | 2020 | Rīga | 3 | Yes | 2,10,24,28,4,4,207 | 1354 | 328 |
| Leg-41-2020 | 2020 | Tukums | 1 | Yes | 2,10,3,10,9,4,11 | 170 | 375 |
| Leg-42-2020 | 2020 | Rīga | 3 | No | 2,10,15,28,9,4,13 | 338 | 371 |
| Leg-43-2020 | 2020 | Olaine | 1 | Yes | 2,10,3,3,9,4,6 | 366 | 379 |
| Leg-44-2020 | 2020 | Rīga | 3 | Yes | 2,22,3,28,50,4,3 | New-ST-5 | 371 |
| Leg-45-2020 | 2020 | Sigulda | 3 | Yes | 2,10,3,28,9,4,3 | 728 | 373 |
| Leg-46-2021 | 2021 | Rīga | 3 | Yes | 3,13,1,28,14,9,13 | 1104 | 412 |
| Leg-47-2021 | 2021 | Rēzekne | 3 | Yes | 2,10,15,28,9,4,13 | 338 | 378 |
| Leg-48-2022 | 2022 | Liepāja | 2 | Yes | 2,10,21,28,9,4,6 | New-ST-2 | 372 |
| Leg-49-2022 | 2022 | Rīga | 1 | Yes | 6,10,19,3,17,4,11 | New-ST-7 | 374 |
| Leg-50-2022 | 2022 | Baloži | 2 | Yes | 2,10,21,28,9,4,13 | 646 | 375 |
| Leg-51-2022 | 2022 | Rīga | 2 | No | 2,10,3,3,50,4,6 | New-ST-6 | 372 |
| Leg-52-2022 | 2022 | Rīga | 2 | No | 10,22,7,28,16,18,8 | 2002 | 360 |
| Leg-53-2022 | 2022 | Rīga | 1 | No | 7,6,17,3,50,4,9 | New-ST-10 | 390 |
| Leg-54-2022 | 2022 | Rīga | 2 | No | 2,10,15,28,9,4,13 | 338 | 375 |
| Leg-55-2022 | 2022 | Rīga | 3 | No | 2,10,15,28,9,4,13 | 338 | 375 |
| Leg-56-2022 | 2022 | Rīga | 3 | No | 2,32,20,38,34,35,219 | 2581 | 344 |
| Leg-57-2022 | 2022 | Rīga | 2 | No | 2,10,15,28,9,4,13 | 338 | 375 |
| Leg-58-2022 | 2022 | Rīga | 1 | No | 2,10,3,3,9,4,6 | 366 | 375 |
| Virulence gene | No. of positive / No. of sequenced isolates | Relative frequency, % | ||||||||
| Overall | Isolates from buildings linked to LD cases N=34 |
Isolates from buildings not linked to LD cases N=24 |
SG 1 isolates N=10 |
SG 2 isolates N=21 |
SG 3 isolates N=20 |
SG 6 isolates N=2 |
SG 8 isolates N=1 |
SG 9 isolates N=4 |
||
| enhC | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| htpB | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| omp28 | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| mip | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| mavC | 54/58 | 93 | 94 | 92 | 100 | 90 | 90 | 100 | 100 | 100 |
| legK1 | 57/58 | 98 | 100 | 96 | 100 | 95 | 100 | 100 | 100 | 100 |
| sidJ | 54/58 | 93 | 94 | 92 | 100 | 100 | 95 | 100 | 100 | 25 |
| lvhD4 | 48/58 | 83 | 85 | 79 | 100 | 76 | 80 | 50 | 100 | 100 |
| lpnE | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| lspC | 58/58 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| rtxA AA100 | 23/58 | 40 | 32 | 50 | 10 | 38 | 55 | 0 | 100 | 50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





