Submitted:
24 May 2023
Posted:
25 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Salt stress reactions of the selected F2:3 progenies and the parental genotypes
2.2. Assessing agronomic characters under salt-stress
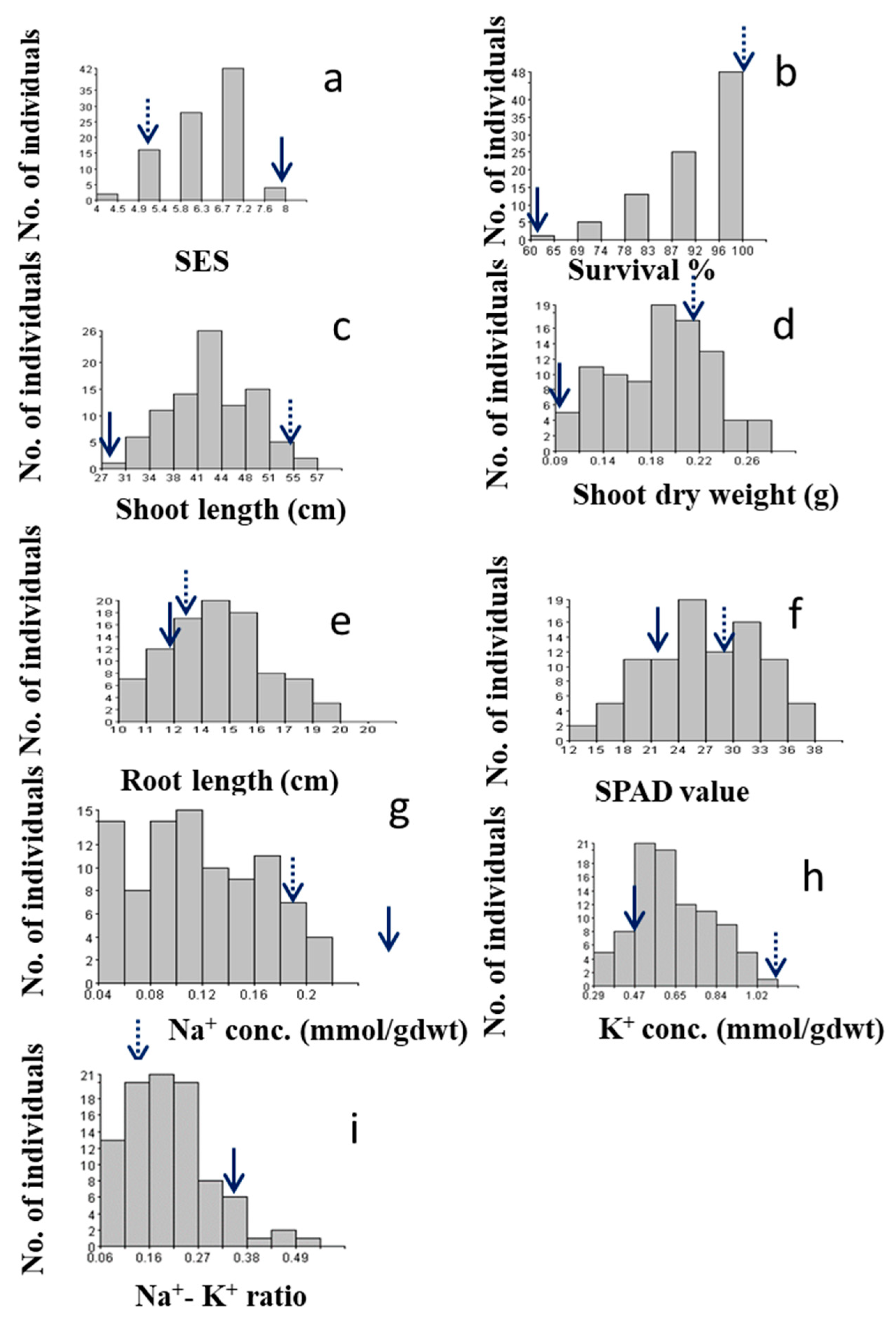
2.2.1. SES score
2.2.2. Survival rate
2.2.3. Shoot length
2.2.4. Shoot dry weight
2.2.5. Root length
2.3. Characterizing physiological parameters under stress
2.3.1. SPAD value
2.3.2. Na+ concentration
2.3.3. K+ concentration
2.3.4. Na+/K+ ratio
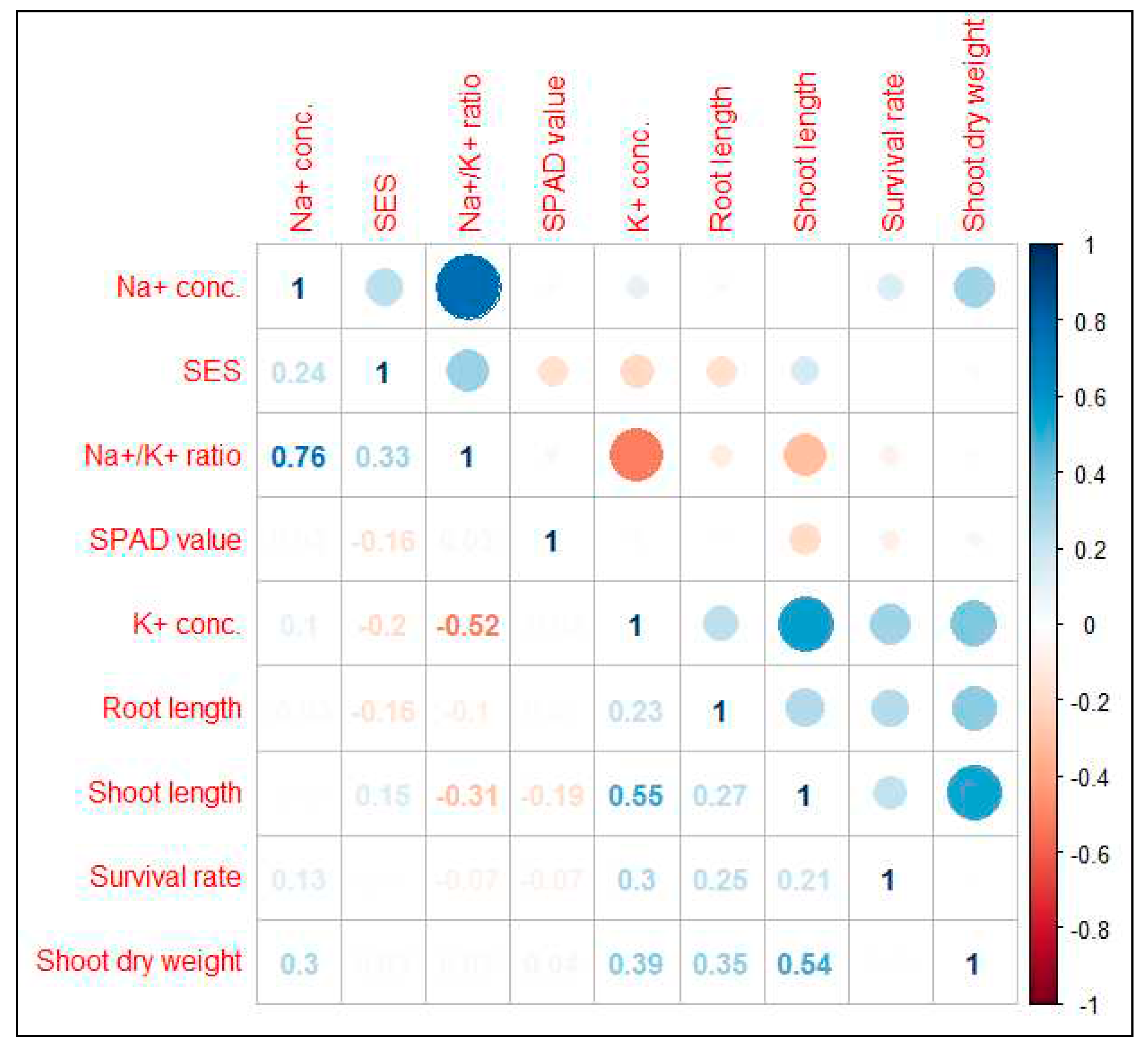
2.4. Trait correlation analysis between different characters
2.5. Determining the contribution of component agronomic and physiological traits (independent variables) to overall phenotypic performance (SES score: dependent variable) through path analysis
2.6. SNP marker polymorphism and construction of genetic linkage map
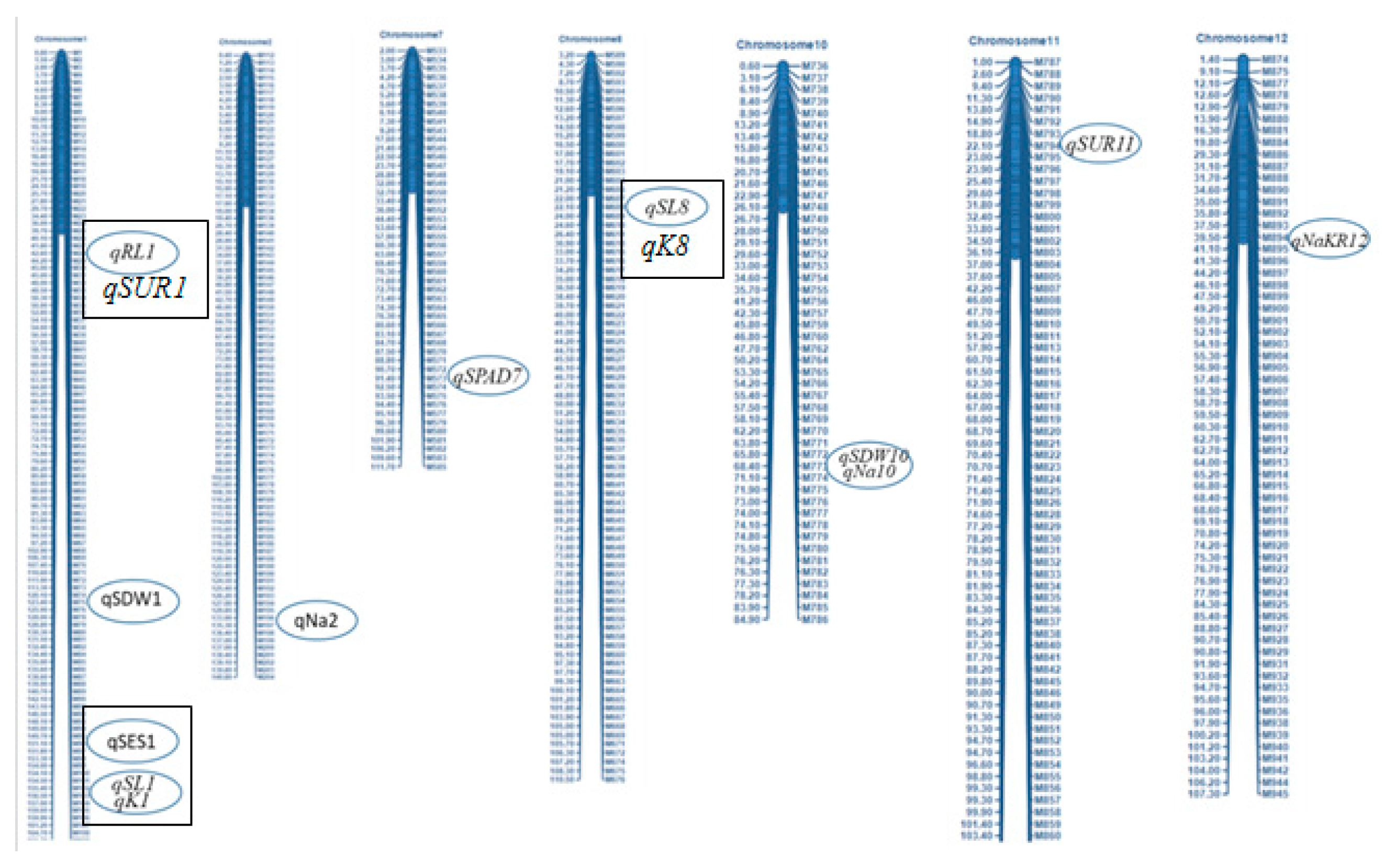
2.7. Salinity tolerance QTLs controlling agronomic traits
2.8. QTL regions governing physiological characters
2.9. Probable functional genes detection in the different QTL regions
2.10. Epistatic interaction
2.11. Stable QTLs for different agronomic and physiological traits responsible for salinity tolerance
| Trait | Population-1 | Favorable allele contributing parent | Trait | Population-2 | Favorable allele contributing parent | Common and stable QTLs in different genetic background | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| QTL detected in pop. 1 | Chr. | QTL Position (cM) |
QTL detected in pop. 2 | Chr. | QTL Position (cM) | |||||
| SES | qSES1 | 1 | 151.8 | Akundi | SES | qSES1 | 1 | 151.8 | Akundi | qSES1 |
| Shoot length | qSL1 | 1 | 156 | BR28 | Shoot length | qSL1 | 1 | 156 | BR49 | qSL1 |
| Root length | qRL1 | 1 | 42 | BR28 | Root length | qRL1 | 1 | 48.8 | Akundi | qRL1 |
| Root length | qRL1 | 1 | 42 | BR28 | Survival (%) |
qSUR1 | 1 | 50 | BR49 | qSUR1 |
| Shoot length | qSL8 | 8 | 22 | BR28 | K+ Conc. | qK8 | 8 | 18.8 | Akundi | qSL8 |
| Shoot length | qSL8 | 8 | 22 | BR28 | K+ Conc. | qK8 | 8 | 18.8 | Akundi | qK8 |
| K+ conc. | qK1 | 1 | 156.5 | BR28 | Shoot length | qSL1 | 1 | 156 | BR49 | qK1 |
3. Discussion
3.1. Assessing the salt-stress responses in the selected F2:3 progenies with their parents and determining cause and effect relationship via path analysis
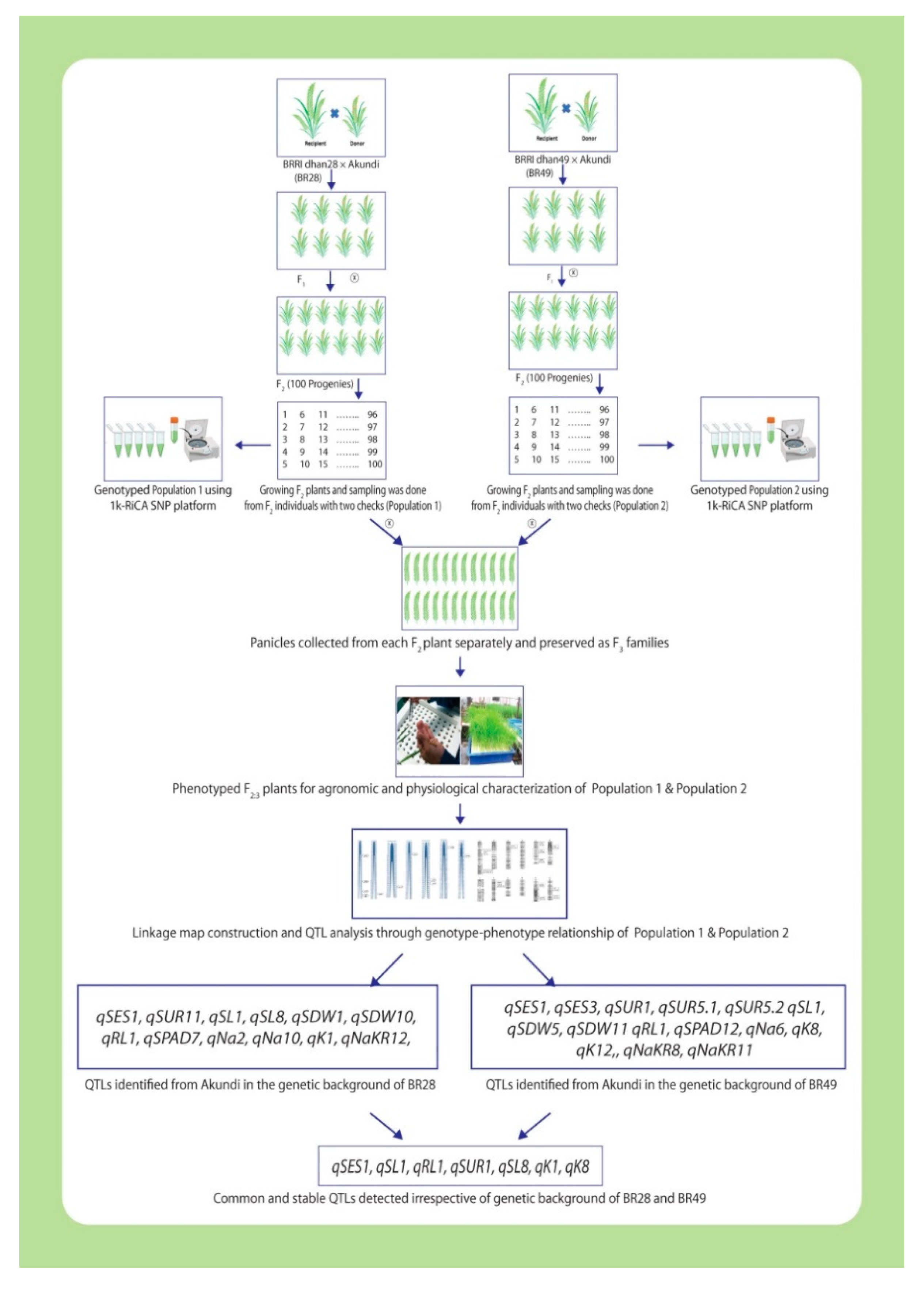
3.2. Marker segregation and important salt-tolerant QTL regions
3.3. Comparing the QTLs revealed in the present study with previously reported QTLs
4. Materials and Methods
4.1. Parent selection
4.2. Growing conditions
4.3. Characterizing agronomic traits
4.4. Determining physiological traits response
4.5. SPAD reading
4.6. Analyzing trait associations (correlation)
4.7. Path coefficient analysis
4.8. SNP genotyping and genetic linkage map construction
4.9. QTL dissection
4.10. Epistatic QTLs identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, M.S.; Ali, M.A.; Khan, M.A.I.; Maniruzzaman, S. (2020). Prevalence and Transmission of Fusarium moniliforme: A Seed Borne Pathogen of Rice. Bangladesh Rice J. 24, 11–19. [CrossRef]
- Hossain, M. S., Ivy, N. A., Maniruzzaman, S., Raihan, M. S., Ruma, A. S., Akter, A., ... & Kabir, K. F. (2023). Genetic Variability of Floral and Agronomic Characteristics that Influence Outcrossing Rate Percentage of Cytoplasmic Male Sterile Rice. Asian Journal of Advances in Agricultural Research, 22(1), 46-57. [CrossRef]
- Maniruzzaman, S.; Hossain, M.; Ahmed, H. U., Hossain, M., Jahan, G. S., Kundu, P. K., & Haque, M. M. (2023). Development of Rice Varieties for Stress-Prone Tidal Ecosystem of Bangladesh. Middle-East Journal of Scientific Research, 31(1), 22-31. [CrossRef]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; & Yang, J. (2016) Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genomics 43:529–532. [CrossRef]
- Maniruzzaman, S., Rahman, M. A., Hasan, M., Rasul, M. G., Molla, A. H., Khatun, H., & Akter, S. (2022). Genetic Mapping to Detect Stringent QTLs Using 1k-RiCA SNP Genotyping Platform from the New Landrace Associated with Salt Tolerance at the Seedling Stage in Rice. Plants, 11(11), 1409. [CrossRef]
- Islam, F., Wang, J., Farooq, M. A., Yang, C., Jan, M., Mwamba, T. M., ... & Zhou, W. (2019). Rice responses and tolerance to salt stress: deciphering the physiological and molecular mechanisms of salinity adaptation. In Advances in rice research for abiotic stress tolerance (pp. 791-819). Woodhead Publishing. [CrossRef]
- -Tamimi, N., Brien, C., Oakey, H., Berger, B., Saade, S., Ho, Y. S., ... & Negrão, S. (2016). Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nature communications, 7(1), 1-11. [CrossRef]
- Rasel, M., Tahjib-Ul-Arif, M., Hossain, M. A., Hassan, L., Farzana, S., & Brestic, M. (2021). Screening of salt-tolerant rice landraces by seedling stage phenotyping and dissecting biochemical determinants of tolerance mechanism. Journal of Plant Growth Regulation, 40(5), 1853-1868. [CrossRef]
- Rahman, M. A., Thomson, M. J., De Ocampo, M., Egdane, J. A., Salam, M. A., & Ismail, A. M. (2019). Assessing trait contribution and mapping novel QTL for salinity tolerance using the Bangladeshi rice landrace Capsule. Rice, 12(1), 1-18. [CrossRef]
- Hossain, H., Rahman, M. A., Alam, M. S., & Singh, R. K. (2015). Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. Journal of Agronomy and Crop Science, 201(1), 17-31. [CrossRef]
- Nayyeripasand, L., Garoosi, G. A., & Ahmadikhah, A. (2021). Genome-wide association study (GWAS) to identify salt-tolerance QTLs carrying novel candidate genes in rice during early vegetative stage. Rice, 14(1), 1-21. [CrossRef]
- Rahman, M. A., Thomson, M. J., Shah-E-Alam, M., de Ocampo, M., Egdane, J., & Ismail, A. M. (2016). Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization. Annals of botany, 117(6), 1083-1097. [CrossRef]
- de Souza Freitas, W. E., de Oliveira, A. B., Mesquita, R. O., de Carvalho, H. H., Prisco, J. T., & Gomes-Filho, E. (2019). Sulfur-induced salinity tolerance in lettuce is due to a better P and K uptake, lower Na/K ratio and an efficient antioxidative defense system. Scientia Horticulturae, 257, 108764. [CrossRef]
- Singh, R.K., Kota, S.; & Flowers, T.J. (2021). Salt tolerance in rice: seedling and reproductive stage QTL mapping come of age. Theor Appl Genet 134, 3495–3533. [CrossRef]
- Mackay, T. F., & Huang, W. (2018). Charting the genotype–phenotype map: lessons from the Drosophila melanogaster Genetic Reference Panel. Wiley Interdisciplinary Reviews: Developmental Biology, 7(1), e289. [CrossRef]
- Rahman M.A., Khatun H., Sarker M.R.A., Hossain H., Quddus M.R., Iftekharuddaula K.M.; & Kabir, M. S. (2021). Enhancing abiotic stress tolerance to develop climate-smart rice using holistic breeding approach. Cereal Grains - Volume 2. [CrossRef]
- BRRI.(2020). Adhunik dhaner chash (23th edition), Bangladesh Rice Research Institute, Gazipur-1701, Bangladesh.
- Yoshida, S. Forno, D. A., & Cock, J. H. (1976). Laboratory manual for physiological studies of rice. Laboratory manual for physiological studies of rice.
- IRRI (2014). Standard evaluation system for rice (SES), 5th edn. Los Banos, International Rice Research Institute, p 57.
- Flowers, T. J., & Yeo, A. R. (1981). Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytologist, 88(2), 363-373. 10.1111/j.1469-8137.1981.tb01731.x.
- Moradi, F., & Ismail, A. M. (2007). Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Annals of botany, 99(6), 1161-1173. 10.1093/aob/mcm052.
- RStudio Team (2020). RStudio: Integrated Development for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/.
- Wright, S. (1922). Coefficients of inbreeding and relationship. The American Naturalist, 56(645), 330-338. [CrossRef]
- Dewey, D. R., & Lu, K. (1959). A correlation and path-coefficient analysis of components of crested wheatgrass seed production 1. Agronomy journal, 51(9), 515-518. [CrossRef]
- Arbelaez, J.D., Dwiyanti, M.S., & Tandayu, E. (2019) 1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice. Rice 12, 55. [CrossRef]
- Chen, M.; Presting, G.; Barbazuk, W.B.; Goicoechea, J.L.; Blackmon, B.; Fang, G.; Kim, H.; Frisch, D.; Yu, Y.; Sun, S.; et al. (2002) An integrated physical and genetic map of the Rice genome. Plant Cell 14:537–545. [CrossRef]
- Joehanes, R., & Nelson, J. C. (2008). QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics, 24(23), 2788-2789. [CrossRef]
- Meng, L., Li, H., Zhang, L., & Wang, J. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop Journal. 3(3), 269-283. [CrossRef]
- Li, H., Ye, G., & Wang, J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361-374. [CrossRef]
- Zhang, L., Li, H., Li, Z., & Wang, J. (2008). Interactions between markers can be caused by the dominance effect of quantitative trait loci. Genetics 180(2), 1177-1190. [CrossRef]
- Churchill, G.A.; & Doerge, R.W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. [CrossRef]
- Li, H., Ribaut, J.-M., Li, Z., & Wang, J. (2008a). Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet. 116(2), 243-260. [CrossRef]
- Kosambi, D.D. (1944). The estimation of map distance from recombination values. Ann. Eugen. 12, 172–175. [CrossRef]
- Rahman, M. A., Bimpong, I. K., Bizimana, J. B., Pascual, E. D., Arceta, M., Swamy, B. P., ... & Singh, R. K. (2017). Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice, 10(1), 1-17. [CrossRef]
- Nakhla, W. R., Sun, W., Fan, K., Yang, K., Zhang, C., & Yu, S. (2021). Identification of QTLs for salt tolerance at the germination and seedling stages in rice. Plants, 10(3), 428. [CrossRef]
- Debsharma, S. K., Syed, M., Ali, M., Maniruzzaman, S., Roy, P. R., Brestic, M., ... & Hossain, A. (2023). Harnessing on Genetic Variability and Diversity of Rice (Oryza sativa L.) Genotypes Based on Quantitative and Qualitative Traits for Desirable Crossing Materials. Genes, 14(1), 10. [CrossRef]
- Hossain, M.; Ivy, N.; Raihan, M.; Kayesh, E.; & Maniruzzaman, S. (2020). Genetic Variability, Correlation and Path Analysis of Floral, Yield and its Component Traits of Maintainer Lines of Rice (Oryza sativa L.). Bangladesh Rice Journal. 24, 1–9. [CrossRef]
- Rajasekar, R., Jeyaprakash, P., Manonmani, K., Nithila, S., & Thirumurugan, T. (2021). Trait relationship and path analysis under sodicity in Nagina 22 rice mutants. Electronic Journal of Plant Breeding, 12(3), 963-968.
- Thomson, M. J., de Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., ... & Ismail, A. M. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice, 3(2), 148-160. [CrossRef]
- Manohara, K. K., Morajkar, S., Shanbhag, Y., Phadte, P., & Singh, N. K. (2021). Haplotype analysis of Saltol QTL region in diverse landraces, wild rice and introgression lines of rice (Oryza sativa L.). Plant Genetic Resources, 19(4), 289-298. [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; & Chao, D.Y. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37: 1141–1146. [CrossRef]
- Kaur, N.; Prashanth, K.V.H.; Bhatti, M.S.; & Pati, P.K. (2022). OsSalT gene cloned from rice provides evidence of its role in salinity and drought stress tolerance. Plant Science, 320, 111306. [CrossRef]
- Claes, B.; Dekeyser, R.; Villaroel, R.; Van den Bulcke, M.; Bauw, G. M.V.; Montagu, & Caplan, A. (1990). Characterization of rice gene showing organ specific expression in response to salt stress and drought. Plant Cell 2: 19-27. [CrossRef]
- Jahan, N. , Zhang, Y. U., Lv, Y., Song, M., Zhao, C., Hu, H., ... & Guo, L. (2020). QTL analysis for rice salinity tolerance and fine mapping of a candidate locus qSL7 for shoot length under salt stress. Plant Growth Regulation, 90(2), 307-319. [CrossRef]
- O’Connor, K. , Hayes, B., Hardner, C., Nock, C., Baten, A., Alam, M., ... & Topp, B. (2020). Genome-wide association studies for yield component traits in a macadamia breeding population. BMC genomics, 21(1), 1-12. [CrossRef]

| Variable name | Correlation | % Survival | Shoot | Root length | SPAD value | Na+ concentration | K+ concentration | Na+/K+ Ratio | Total effect | |
|---|---|---|---|---|---|---|---|---|---|---|
| length | dry weight | |||||||||
| % Survival | 0.008 | 0.035 | 0.093 | 0.001 | -0.042 | 0.006 | 0.021 | -0.096 | -0.011 | 0.008 |
| Shoot length | 0.147 | 0.007 | 0.440 | -0.046 | -0.044 | 0.016 | -0.001 | -0.173 | -0.052 | 0.147 |
| Shoot dry weight | 0.018 | 0.001 | 0.235 | -0.086 | -0.058 | -0.004 | 0.049 | -0.121 | 0.003 | 0.018 |
| Root length | -0.161 | 0.009 | 0.118 | -0.030 | -0.164 | -0.001 | -0.004 | -0.072 | -0.017 | -0.161 |
| SPAD value | -0.162 | -0.003 | -0.083 | -0.004 | -0.003 | -0.086 | 0.005 | 0.005 | 0.006 | -0.162 |
| Na+ conc. | 0.239 | 0.004 | -0.002 | -0.025 | 0.004 | -0.003 | 0.164 | -0.031 | 0.127 | 0.239 |
| K+ conc. | -0.201 | 0.011 | 0.243 | -0.033 | -0.038 | 0.001 | 0.016 | -0.314 | -0.087 | -0.201 |
| NaK Ratio | 0.328 | -0.002 | -0.137 | -0.001 | 0.016 | -0.003 | 0.125 | 0.163 | 0.168 | 0.328 |
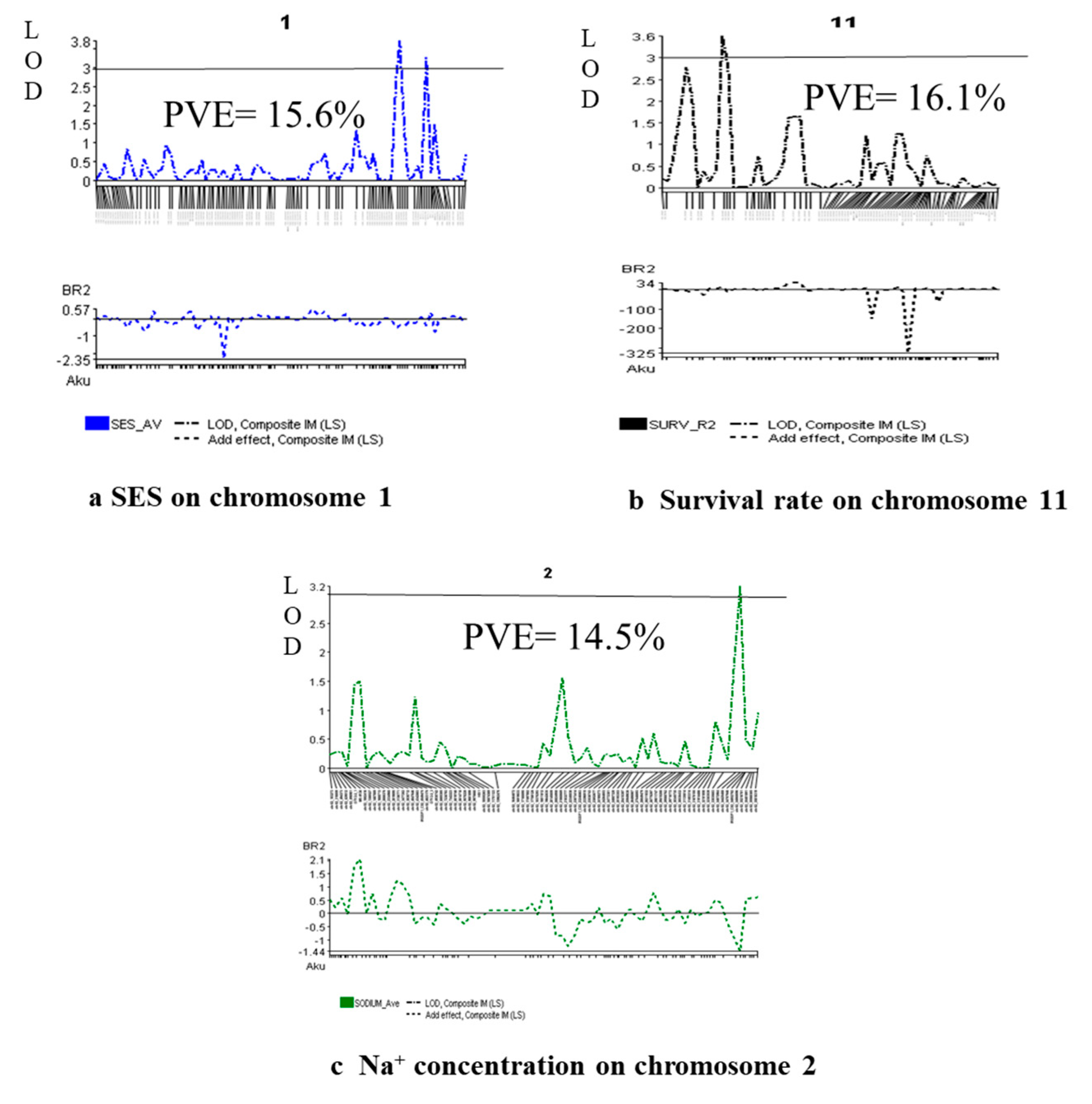
| Characters | QTL identified | Chr. | QTL peak marker | QTL position (cM) | Additive effect | LOD | PVE (%) | QTL detection method | Favorable allele contributing parent | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QGene | ICIM | QGene | ICIM | QGene | ICIM | |||||||
| SES | qSES1 | 1 | chr01_38632196 | 151.8 | 150.8 | -0.61 | 3.4 | 3.0 | 15.6 | 2.59 | IM, CIM | Akundi |
| Survival rate (%) | qSUR11 | 11 | chr11_5615885 | 21 | 21 | 13.73 | 3.5 | 4.0 | 16.1 | 0.85 | IM, CIM | BR28 |
| Shoot length | qSL1 | 1 | QSES1-2_2 | 156 | - | 4.86 | 7.3 | - | 30.7 | - | IM, CIM | BR28 |
| qSL8 | 8 | GM4_4 | 22 | - | 4.29 | 3.2 | - | 16.3 | - | IM, CIM | BR28 | |
| Shoot dry weight | qSDW1 | 1 | chr01_231396842 | 123.4 | 137.8 | -0.25 | 3.6 | 6.0 | 16.4 | 13.44 | IM, CIM | Akundi |
| qSDW10 | 10 | chr10_17397576 | 68.6 | 64.6 | -0.2 | 3.6 | 5.0 | 17.1 | 17.31 | IM, CIM | Akundi | |
| Root length | qRL1 | 1 | chr01_1045259 | 42 | - | 1.93 | 3.5 | - | 26 | - | SMA | BR28 |
| SPAD value | qSPAD7 | 7 | AG3_1 | 91.4 | - | -2.32 | 3.0 | - | 12.1 | - | SMR | Akundi |
| Na+ conc. | qNa2 | 2 | chr02_34072964 | 134.4 | - | -1.43 | 3.1 | - | 14.5 | - | IM, CIM | Akundi |
| qNa10 | 10 | chr10_17397576 | 68.8 | 68.6 | -0.74 | 3.3 | 5.0 | 15.5 | 21.20 | IM, CIM | Akundi | |
| K+ conc. | qK1 | 1 | QSES1-2_2 | 156.5 | 156.8 | 0.102 | 4.28 | 4.0 | 19.3 | 18.75 | SMR | BR28 |
| Na+/K+ ratio | qNaKR12 | 12 | chr12_10051752 | 39.4 | - | -0.04 | 3.0 | - | 12.1 | - | CIM | Akundi |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





