Submitted:
10 May 2023
Posted:
11 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
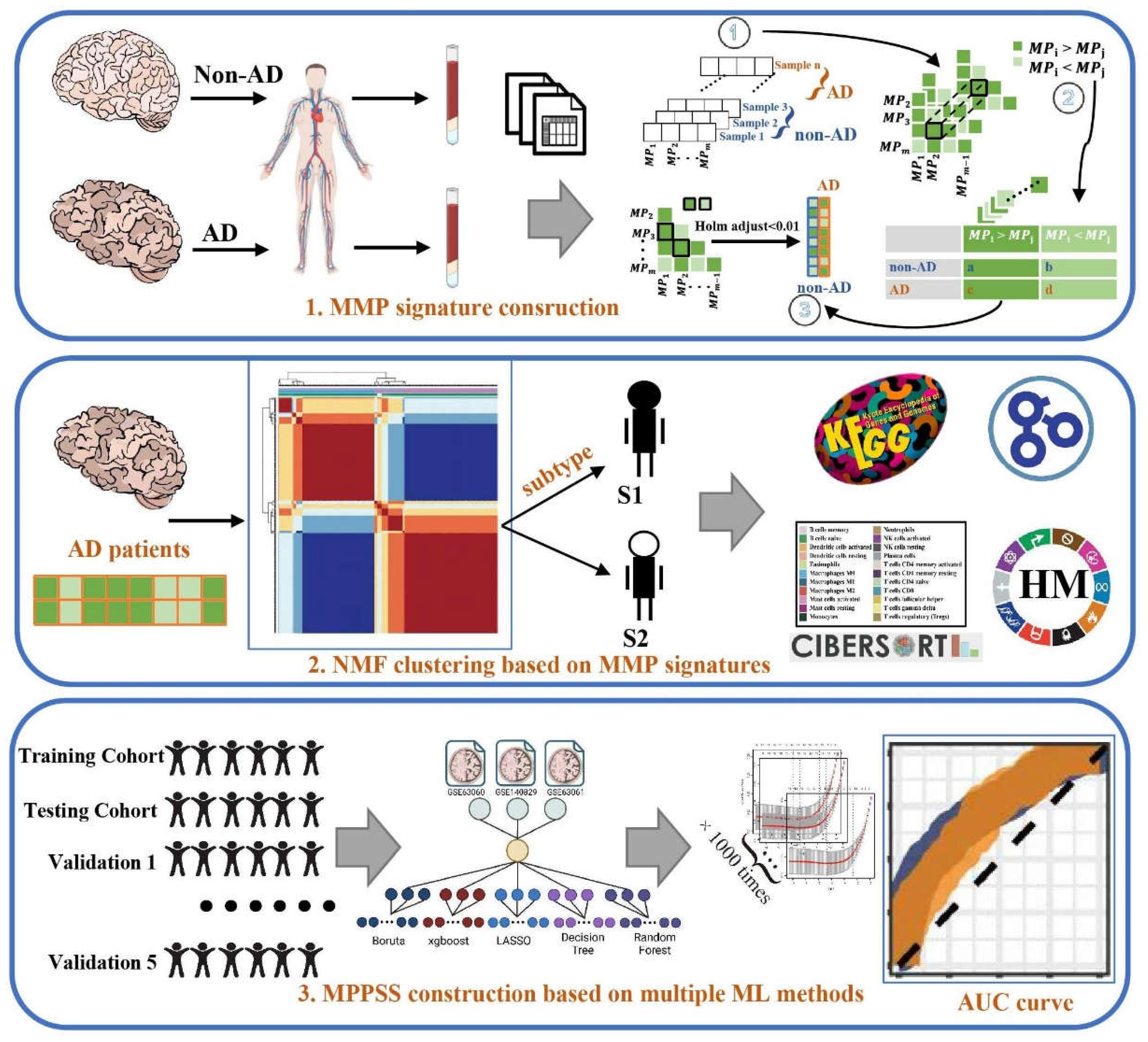
2.1. Data acquisition and preprocess
2.2. Construction of MPP signatures
2.2.1. Within-sample analysis
2.2.2. Cross-sample analysis
| Type | ||
| Non-AD | a | b |
| AD | c | d |
2.3. Unsupervised clustering to characterize AD patient subgroups
2.4. Establishment of AD diagnostic MPPSS by using multiple machine learning approaches
2.5. Immune infiltration analysis by CIBERSORT algorithm
2.6. Gene differential expression analysis and functional annotation analysis between the AD and non-AD groups, as well as within the two AD subgroups.
3. Results
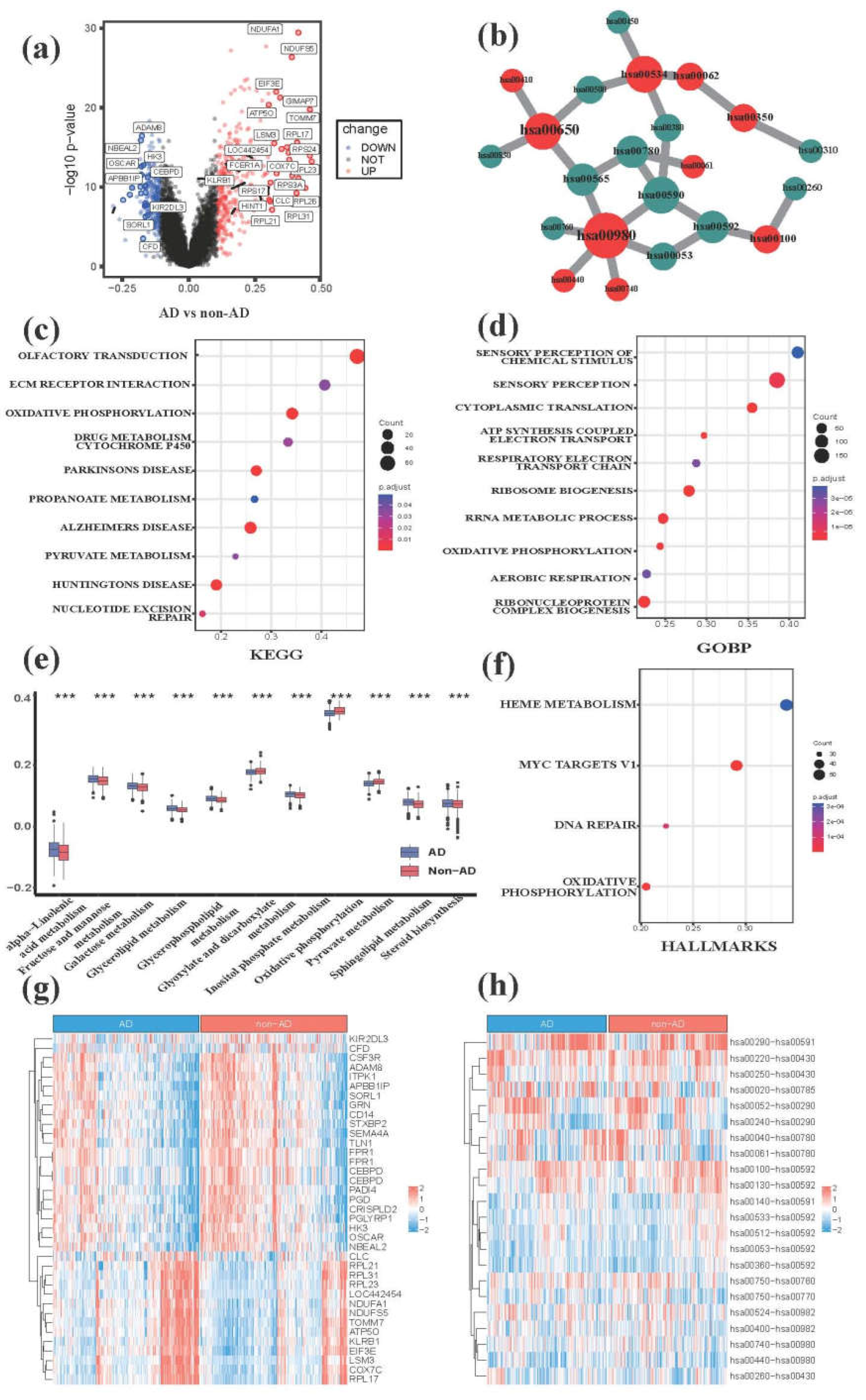
3.1. Comparative transcriptome analysis characterizes metabolic hallmarks of peripheral blood in AD
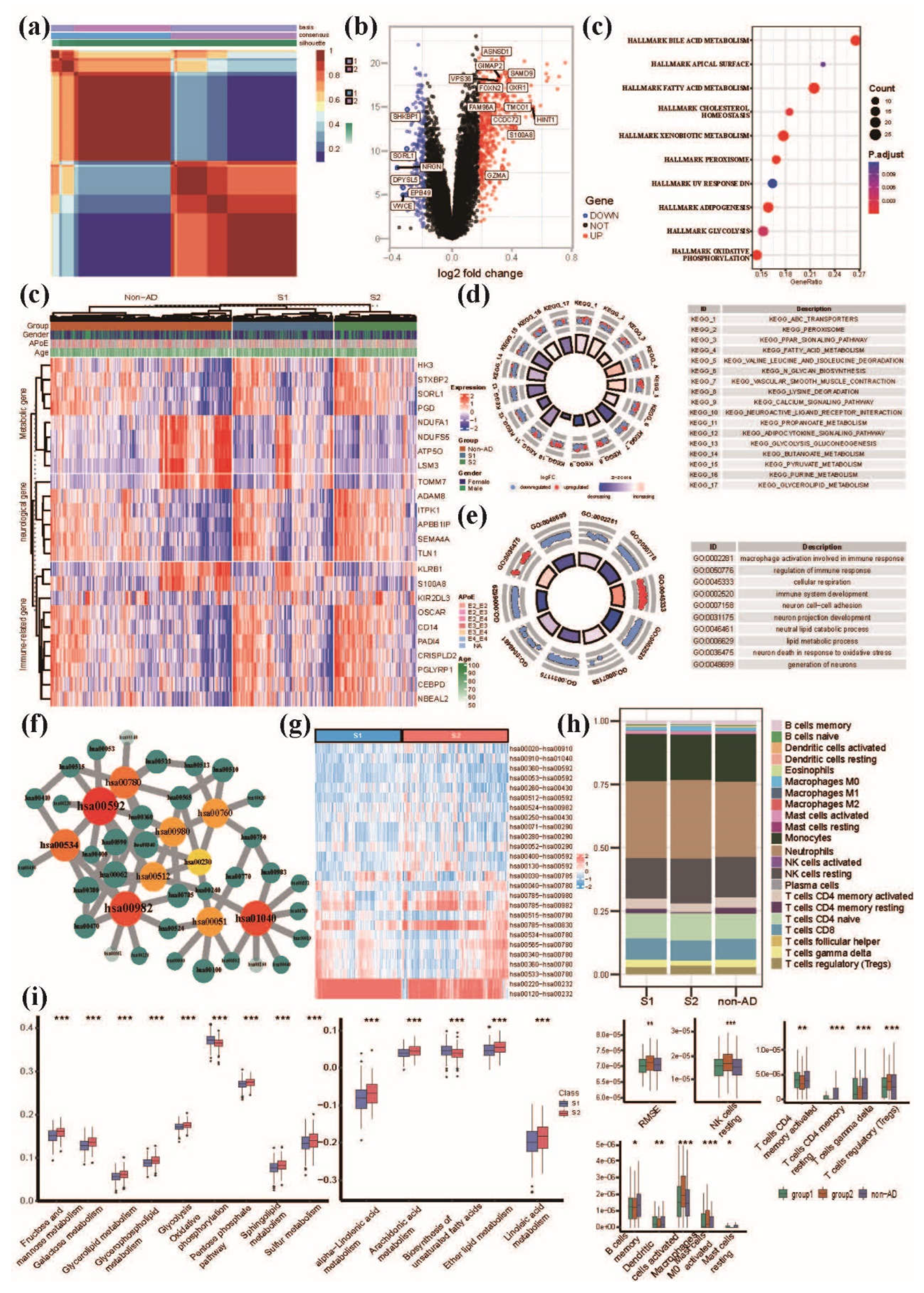
3.2. NMF clustering analysis of AD patients based on peripheral blood MMP signatures reveals distinct pattern of lipid, glucose and energy metabolism
3.3. Comprehensive evaluation of immune cell infiltration characteristics in AD subgroups and non-AD control group
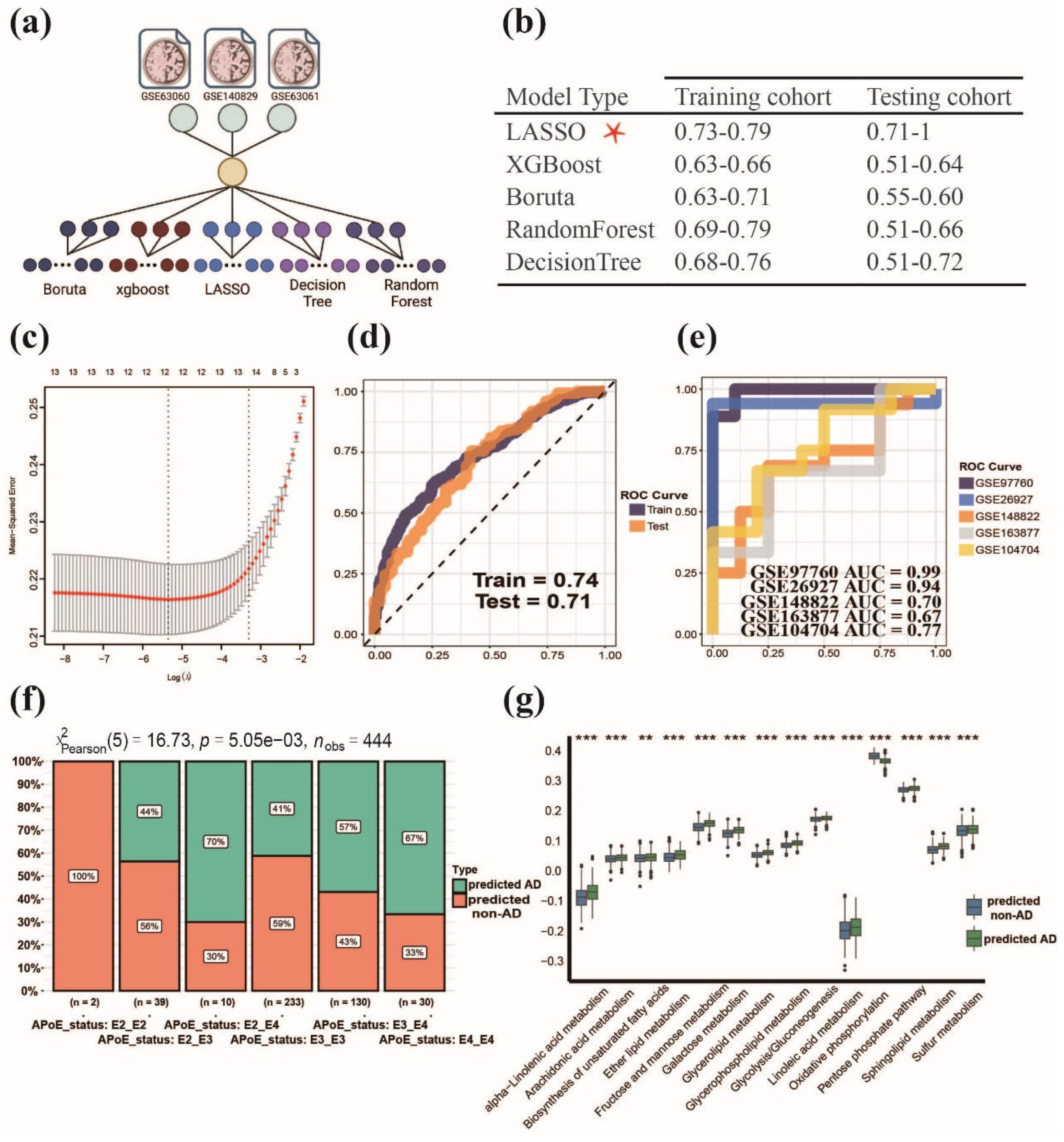
3.4. Establishment of MPPSS for distinguishing AD patients from non-AD patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer's, A. 2012 Alzheimer's disease facts and figures. Alzheimers Dement 2012, 8, 131–168. [Google Scholar] [CrossRef]
- Ayodele, T.; Rogaeva, E.; Kurup, J.T.; Beecham, G.; Reitz, C. Early-Onset Alzheimer’s Disease: What Is Missing in Research? Current Neurology and Neuroscience Reports 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yeo, S.H.; Park, J.M.; Choi, J.Y.; Lee, T.H.; Park, S.Y.; Ock, M.S.; Eo, J.; Kim, H.S.; Cha, H.J. Genetic markers for diagnosis and pathogenesis of Alzheimer's disease. Gene 2014, 545, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.S.; DeKosky, S.T.; Dickson, D.; Dubois, B.; Feldman, H.H.; Fox, N.C.; Gamst, A.; Holtzman, D.M.; Jagust, W.J.; Petersen, R.C.; et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011, 7, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Villain, N.; Frisoni, G.B.; Rabinovici, G.D.; Sabbagh, M.; Cappa, S.; Bejanin, A.; Bombois, S.; Epelbaum, S.; Teichmann, M.; et al. Clinical diagnosis of Alzheimer's disease: recommendations of the International Working Group. Lancet Neurol 2021, 20, 484–496. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, G.; Shapiro, F.; Vanderstichele, H.; Vanmechelen, E.; Engelborghs, S.; De Deyn, P.P.; Coart, E.; Hansson, O.; Minthon, L.; Zetterberg, H.; et al. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol 2010, 67, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Kim, S.J.; Hong, S.; Kim, Y. Diagnosis of Alzheimer's disease utilizing amyloid and tau as fluid biomarkers. Exp Mol Med 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Olsson, B.; Lautner, R.; Andreasson, U.; Öhrfelt, A.; Portelius, E.; Bjerke, M.; Hölttä, M.; Rosén, C.; Olsson, C.; Strobel, G. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. The Lancet Neurology 2016, 15, 673–684. [Google Scholar] [CrossRef]
- Teunissen, C.E.; Verberk, I.M.; Thijssen, E.H.; Vermunt, L.; Hansson, O.; Zetterberg, H.; van der Flier, W.M.; Mielke, M.M.; Del Campo, M. Blood-based biomarkers for Alzheimer's disease: towards clinical implementation. The Lancet Neurology 2022, 21, 66–77. [Google Scholar] [CrossRef]
- Iwata, N.; Tsubuki, S.; Takaki, Y.; Shirotani, K.; Lu, B.; Gerard, N.P.; Gerard, C.; Hama, E.; Lee, H.-J.; Saido, T.C. Metabolic Regulation of Brain Aβ by Neprilysin. Science 2001, 292, 1550–1552. [Google Scholar] [CrossRef]
- Kapogiannis, D.; Mattson, M.P. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer's disease. Lancet Neurol 2011, 10, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Poddar, M.K.; Banerjee, S.; Chakraborty, A.; Dutta, D. Metabolic disorder in Alzheimer’s disease. Metabolic Brain Disease 2021, 36, 781–813. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kim, S.H.; Bishayee, K. Dysfunctional Glucose Metabolism in Alzheimer's Disease Onset and Potential Pharmacological Interventions. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer's diseases. Neurobiol Dis 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef]
- Martins, I.J.; Hone, E.; Foster, J.K.; Sünram-Lea, S.I.; Gnjec, A.; Fuller, S.J.; Nolan, D.; Gandy, S.E.; Martins, R.N. Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer's disease and cardiovascular disease. Molecular Psychiatry 2006, 11, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, S.H.; Bakhuraysah, M.M.; Cram, D.S.; Petratos, S. The Beta-amyloid protein of Alzheimer's disease: communication breakdown by modifying the neuronal cytoskeleton. Int J Alzheimers Dis 2013, 2013, 910502. [Google Scholar] [CrossRef]
- Munoz, D.G.; Feldman, H. Causes of Alzheimer's disease. CMAJ 2000, 162, 65–72. [Google Scholar]
- Yan, X.; Hu, Y.; Wang, B.; Wang, S.; Zhang, X. Metabolic Dysregulation Contributes to the Progression of Alzheimer's Disease. Front Neurosci 2020, 14, 530219. [Google Scholar] [CrossRef]
- Small, G.W.; Kepe, V.; Barrio, J.R. Seeing is believing: neuroimaging adds to our understanding of cerebral pathology. Curr Opin Psychiatry 2006, 19, 564–569. [Google Scholar] [CrossRef]
- Arias, C.; Montiel, T.; Quiroz-Baez, R.; Massieu, L. beta-Amyloid neurotoxicity is exacerbated during glycolysis inhibition and mitochondrial impairment in the rat hippocampus in vivo and in isolated nerve terminals: implications for Alzheimer's disease. Exp Neurol 2002, 176, 163–174. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, X.; Leung, K.S.; Wong, M.H.; Tsui, S.K.; Cheng, L. meGPS: a multi-omics signature for hepatocellular carcinoma detection integrating methylome and transcriptome data. Bioinformatics 2022, 38, 3513–3522. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014, 8 Suppl 4, S11. [Google Scholar] [CrossRef]
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Browne, M.W. Cross-validation methods. Journal of mathematical psychology 2000, 44, 108–132. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T. Xgboost: extreme gradient boosting. R package version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. Journal of statistical software 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Therneau, T.M.; Atkinson, E.J. An introduction to recursive partitioning using the RPART routines; Technical report Mayo Foundation: 1997.
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Zou, H. The adaptive lasso and its oracle properties. Journal of the American statistical association 2006, 101, 1418–1429. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Cancer Systems Biology: Methods and Protocols 2018, 243–259. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 2015, 43, e47–e47. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Kim, C.; Park, S.; Joo, J.; Choi, B.; Yang, D.; Jun, K.; Eom, J.; Lee, S.-J.; Chung, S.J.; et al. Characterization of altered molecular mechanisms in Parkinson’s disease through cell type–resolved multiomics analyses. Science Advances 2023, 9, eabo2467. [Google Scholar] [CrossRef]
- Wu, Q.; Kong, W.; Wang, S. Peripheral Blood Biomarkers CXCL12 and TNFRSF13C Associate with Cerebrospinal Fluid Biomarkers and Infiltrating Immune Cells in Alzheimer Disease. J Mol Neurosci 2021, 71, 1485–1494. [Google Scholar] [CrossRef]
- Holtzman, D.M.; Morris, J.C.; Goate, A.M. Alzheimer's disease: the challenge of the second century. Sci Transl Med 2011, 3, 77sr71. [Google Scholar] [CrossRef]
- Kang, S.; Lee, Y.H.; Lee, J.E. Metabolism-Centric Overview of the Pathogenesis of Alzheimer's Disease. Yonsei Med J 2017, 58, 479–488. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenetic basis for therapeutic optimization in Alzheimer's disease. Mol Diagn Ther 2007, 11, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Hooijmans, C.R.; Kiliaan, A.J. Fatty acids, lipid metabolism and Alzheimer pathology. European Journal of Pharmacology 2008, 585, 176–196. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Henderson, C.J.; Scott, C.L.; Wolf, C.R. Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J 2009, 417, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Rezen, T.; Rozman, D. Regulation of hepatic cytochromes p450 by lipids and cholesterol. Curr Drug Metab 2011, 12, 173–185. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, L.R.B.; Fleming, I. Role of cytochrome P450-derived, polyunsaturated fatty acid mediators in diabetes and the metabolic syndrome. Prostaglandins & Other Lipid Mediators 2020, 148, 106407. [Google Scholar] [CrossRef]
- Kao, Y.-C.; Ho, P.-C.; Tu, Y.-K.; Jou, I.-M.; Tsai, K.-J. Lipids and Alzheimer’s Disease. International Journal of Molecular Sciences 2020, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Montine, T.J.; Neely, M.D.; Quinn, J.F.; Beal, M.F.; Markesbery, W.R.; Roberts, L.J.; Morrow, J.D. Lipid peroxidation in aging brain and Alzheimer’s disease1, 2 1Guest Editors: Mark A. Smith and George Perry 2This article is part of a series of reviews on “Causes and Consequences of Oxidative Stress in Alzheimer’s Disease.” The full list of papers may be found on the homepage of the journal. Free Radical Biology and Medicine 2002, 33, 620–626. [Google Scholar] [CrossRef]
- Bhatt, S.; Puli, L.; Patil, C.R. Role of reactive oxygen species in the progression of Alzheimer’s disease. Drug Discovery Today 2021, 26, 794–803. [Google Scholar] [CrossRef]
- Suzuki, N.; Sawada, K.; Takahashi, I.; Matsuda, M.; Fukui, S.; Tokuyasu, H.; Shimizu, H.; Yokoyama, J.; Akaike, A.; Nakaji, S. Association between Polyunsaturated Fatty Acid and Reactive Oxygen Species Production of Neutrophils in the General Population. Nutrients 2020, 12. [Google Scholar] [CrossRef]
- Snowden, S.G.; Ebshiana, A.A.; Hye, A.; An, Y.; Pletnikova, O.; O'Brien, R.; Troncoso, J.; Legido-Quigley, C.; Thambisetty, M. Association between fatty acid metabolism in the brain and Alzheimer disease neuropathology and cognitive performance: A nontargeted metabolomic study. PLoS Med 2017, 14, e1002266. [Google Scholar] [CrossRef]
- Campbell, K.; Vowinckel, J.; Keller, M.A.; Ralser, M. Methionine Metabolism Alters Oxidative Stress Resistance via the Pentose Phosphate Pathway. Antioxid Redox Signal 2016, 24, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Yellen, G. Fueling thought: Management of glycolysis and oxidative phosphorylation in neuronal metabolism. J Cell Biol 2018, 217, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Molecular Neurodegeneration 2020, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.-I.; Bormann, M.K.; Shen, M.; Kim, D.; Forester, B.; Park, Y.; So, J.; Seo, H.; Sonntag, K.-C.; Cohen, B.M. Brain cells derived from Alzheimer’s disease patients have multiple specific innate abnormalities in energy metabolism. Molecular Psychiatry 2021, 26, 5702–5714. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, B.E.; Stone, M.L.; Zhu, X.; Perry, G.; Smith, M.A. Heme deficiency in Alzheimer's disease: a possible connection to porphyria. J Biomed Biotechnol 2006, 2006, 24038. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.J.; Gomez-Pinilla, F.; Nagel, M.; Nakagawa, T.; Rodriguez-Iturbe, B.; Sanchez-Lozada, L.G.; Tolan, D.R.; Lanaspa, M.A. Cerebral Fructose Metabolism as a Potential Mechanism Driving Alzheimer's Disease. Front Aging Neurosci 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Germain, R.N. T-cell development and the CD4–CD8 lineage decision. Nature Reviews Immunology 2002, 2, 309–322. [Google Scholar] [CrossRef]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast cell: a multi-functional master cell. Frontiers in immunology 2016, 620. [Google Scholar] [CrossRef]
- Kvetnoĭ, I.M.; Kvetnaia, T.V.; Riadnova, I.; Fursov, B.B.; Ernandes-Jago, H.; Blesa, J.R. [Expression of beta-amyloid and tau-protein in mastocytes in Alzheimer disease]. Arkh Patol 2003, 65, 36–39. [Google Scholar]
- Maslinska, D.; Laure-Kamionowska, M.; Maslinski, K.T.; Gujski, M.; Maslinski, S. Distribution of tryptase-containing mast cells and metallothionein reactive astrocytes in human brains with amyloid deposits. Inflammation Research 2007, 56, S17–S18. [Google Scholar] [CrossRef]
- Dimitriadou, V.; Rouleau, A.; Tuong, M.D.T.; Newlands, G.J.F.; Miller, H.R.P.; Luffau, G.; Schwartz, J.C.; Garbarg, M. Functional relationships between sensory nerve fibers and mast cells of dura mater in normal and inflammatory conditions. Neuroscience 1997, 77, 829–839. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Group | No. of cases (%) |
|---|---|---|
| Samples | AD | 488 (50.05%) |
| Non-AD | 487 (49.95%) | |
| Age | ≤60 | 11 (1.13%) |
| >60&≤70 | 277 (28.41%0 | |
| >70&≤80 | 479 (49.13%) | |
| >80&≤90 | 206 (21.13%) | |
| >90 | 2 (0.21%) | |
| Gender | Male | 405 (41.54%) |
| Female | 570 (58.46%) | |
| Race | Western European | 385 (39.49%) |
| Other Caucasian | 42 (4.31%) | |
| British | 3 (0.31%) | |
| British Welsh | 2 (0.21%) | |
| British English | 69 (7.08%) | |
| British Scottish | 2 (0.21%) | |
| British Other Background | 1 (0.10%) | |
| Irish | 5 (0.51%) | |
| Indian | 2 (0.21%) | |
| White And Asian | 1 (0.10%) | |
| Any Other White Background | 7 (0.72%) | |
| Any Other Asian Background | 1 (0.10%) | |
| Unknown | 455 (46.67%) | |
| APoE status | apoe_E2_E2 | 2 (0.21%) |
| apoe_E2_E3 | 39 (4.00%) | |
| apoe_E2_E4 | 10 (1.03%) | |
| apoe_E3_E3 | 233 (23.90%) | |
| apoe_E3_E4 | 130 (13.33%) | |
| apoe_E4_E4 | 30 (3.08%) | |
| Unknown | 531 (45.54%) | |
| Subgroups | S1 | 295 (54.92%) |
| S2 | 193 (40.98%) |
| MPPS | coef | Pathway pairwise function |
|---|---|---|
| hsa00100-hsa00190 | 1.0285978 | Steroid hormone biosynthesis - oxidative phosphorylation |
| hsa00563-hsa00190 | 1.4211556 | GPI-anchor biosynthesis - oxidative phosphorylation |
| hsa00534-hsa00190 | 1.0289191 | Glycosaminoglycan biosynthesis-heparan sulfate/heparin - oxidative phosphorylation |
| hsa00900-hsa00190 | 1.1686399 | Terpenoid backbone biosynthesis - oxidative phosphorylation |
| hsa00310-hsa00534 | -0.6982631 | Lysine degradation - glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate |
| hsa00760-hsa00190 | 1.1373381 | Nicotinate and nicotinamide metabolism - oxidative phosphorylation |
| hsa00531-hsa00860 | 0.1188049 | Glycosaminoglycan degradation - porphyrin metabolism |
| hsa00513-hsa00620 | 1.3416181 | Various types of N-glycan biosynthesis - pyruvate metabolism |
| hsa01040-hsa00190 | 1.0216412 | Unsaturated fatty acid biosynthesis - oxidative phosphorylation |
| hsa00310-hsa00600 | -0.8491503 | Lysine degradation - sphingolipid metabolism |
| hsa00534-hsa00620 | 0.1976417 | Glycosaminoglycan biosynthesis-heparan sulfate/heparin - pyruvate metabolism |
| hsa00310-hsa00531 | -1.1770501 | Lysine degradation - glycosaminoglycan degradation |
| hsa00051-hsa00860 | 0.4476219 | Fructose and mannose metabolism - porphyrin metabolism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





