Submitted:
07 May 2023
Posted:
08 May 2023
You are already at the latest version
Abstract
Keywords:
Introduction
2. Materials and Methods
3. Results and Discussion
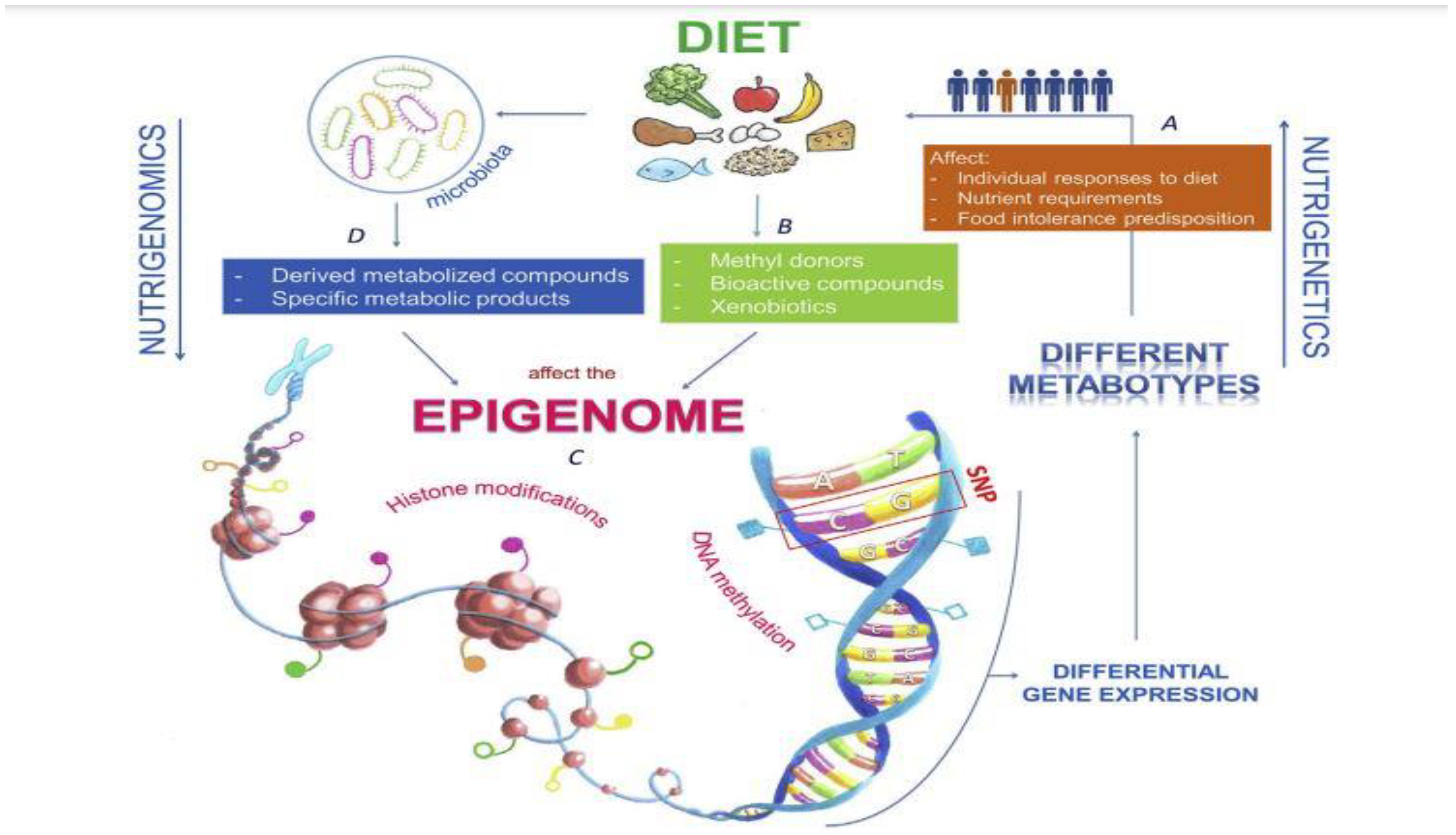
3.1. The effect of nutrition on human gut microbiome
3.2. The effect of nutrition on human genome expression
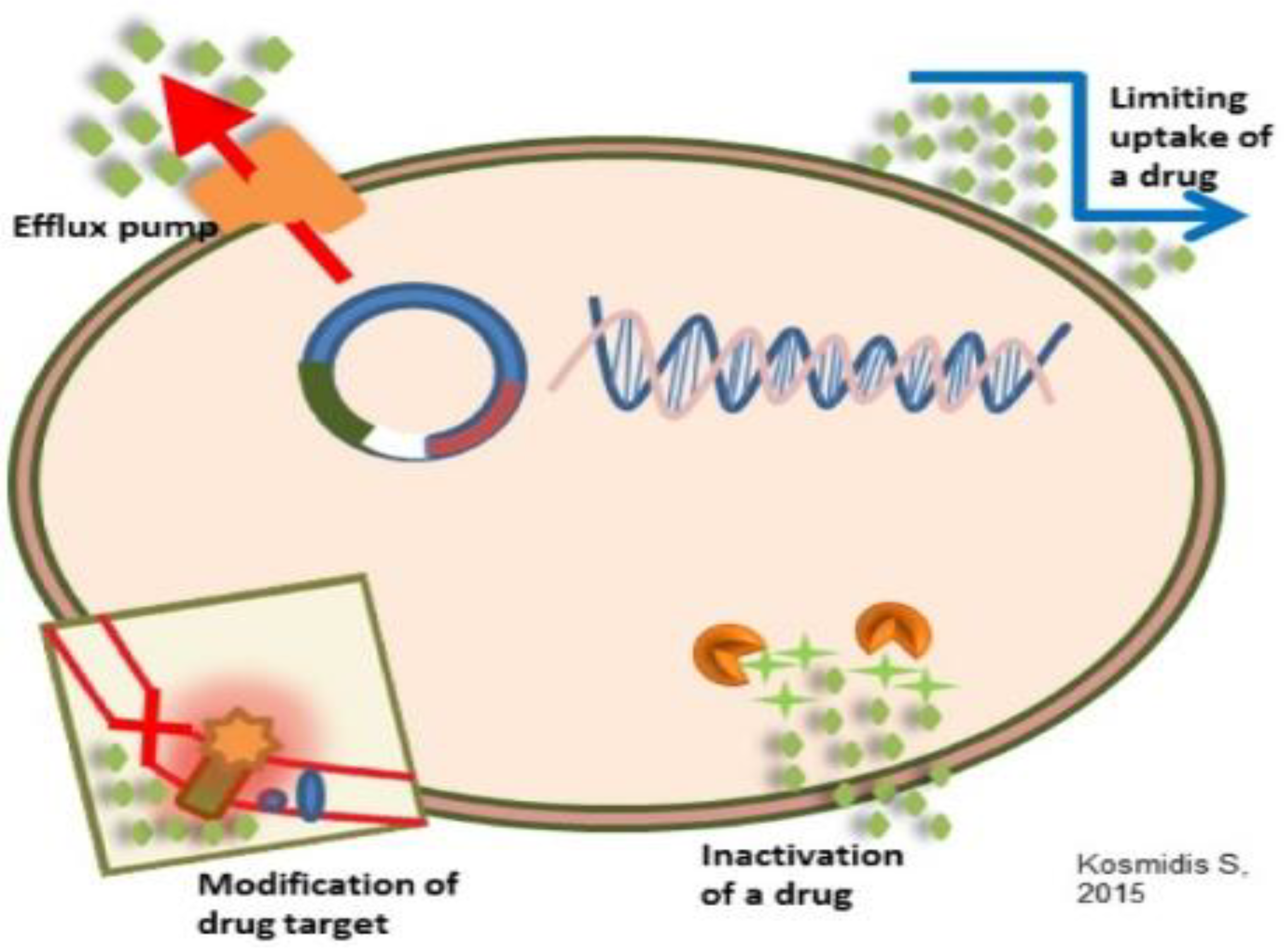
3.3. Antimicrobial resistance mechanisms
3.4. Antimicrobial effect on human gut microbiome
3.5. Phylogenetic Groups and Antimicrobial Resistance Genes from poultry
3.6. New antibiotics against microbial resistance
3.7. How genomics mitigates the public health impact of antimicrobial resistance
| Case 1: International surveillance— determination the population structure and epidemiology of carbapenem-resistant K. pneumoniae (CR-Kp) across Europe [61] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| The primary reservoirs and transmission dynamics of CR-Kp in Europe are still poorly understood. | For sequencing, hospital European laboratories have submitted consecutive clinical isolates of CR-Kp, along with a susceptible strain for comparison. | Carbapenemase acquisition was the primary cause of CR-Kp dissemination; nosocomial acquisition was the other main source of CR-Kp spread. | Provided a baseline for continuous CR-Kp monitoring. The importance of nosocomial spread was emphasized. |
| Case 2: Enhancing the national surveillance of antimicrobial resistance in the Philippines [55] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| National laboratory-based surveillance has revealed an increase in AMR incidences over the preceding ten years, but the understanding of the epidemiology and causes of AMR have remained limited. | The capacity of WGS was added to the current surveillance project. To provide baseline data and guide control strategies, retrospective sequencing of MDR GNB collected before the introduction was performed and examined with phenotypic and epidemiological data. | Through the discovery of the introduction and country-wide dissemination of a high-risk epidemic clone, E. coli ST410, drivers of carbapenem resistance at several healthcare system levels were found, including a localized outbreak of plasmid-driven CR-Kp impacting a single healthcare facility. | The implementation of efficient infection control methods was made possible by a thorough understanding of the epidemiology and causes of AMR. Data was provided to worldwide AMR surveillance initiatives, which improved global coverage. |
| Case3: Investigating an MRSA outbreak in a neonatal unit in the UK [62] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| Over a 6-month period, phenotypically comparable MRSA isolates were found in patients on a special baby care unit but could not be connected chronologically or geographically, implying that the entire breadth of the epidemic had not been recognized. | WGS was performed on all MRSA isolates received from a special baby unit patients over a 6-month period, independent of phenotypic features. MRSA isolates from the community with antibiograms comparable to the epidemic strain, as well as screening samples from elsewhere in the hospital, were also sequenced. | Phylogenetic research revealed that two previously excluded isolates were part of the epidemic, allowing temporal linkages between patients to be established. Beyond the newborn unit, a large transmission network was discovered. | WGS enabled the testing of a large number of isolates and the precise identification of related strains, allowing for comprehensive epidemic reconstruction. Combining WGS data with clinical and epidemiological data allowed for the identification of the source of the epidemic and the successful implementation of infection control measures. |
| Case 4: Investigating the direction of transmission in an A. baumannii outbreak in a UK hospital [63] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| The molecular typing of a cluster of A. baumannii isolates acquired at a UK hospital suggested a clonal epidemic, but the route of transmission between cases could not be established based on the available laboratory, clinical, and epidemiological data. | WGS analysis was performed on a group of isolates acquired from patients with similar molecular typing profiles and antibiograms in order to better understand direct transmission between patients. | The index case was identified using phylogenetic analysis, and the subsequent chain of transmission was determined. One patient/isolate was found to be unconnected, and the outbreak investigation was abandoned. | The directionality of transmission may be identified by WGS, allowing for a precise reconstruction of the outbreak. |
| Case 5: Contact tracing and detection of secondary cases of TB in the Netherlands [64] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| Secondary TB detection and screening are critical for TB control. The poor precision of molecular typing makes the accurate identification of case clusters and transmission networks difficult. | In 2016, clinical TB isolates in the Netherlands were examined using both molecular typing and WGS. The two techniques were evaluated in terms of discrimination and accuracy in identifying possibly related situations. | WGS proved more capable of determining the relatedness of isolates than molecular typing, grouping a lesser proportion of isolates as related (25% vs. 14%) and boosting the proportion verified as epidemiologically connected (57% vs. 31%). | WGS aided in the identification of transmission episodes, allowing for contact tracing and generating a broader knowledge of TB control. |
| Case 6: Identifying the drivers of AMR in atypical enteropathogenic E. coli (aEPEC) strains isolated from children < 5 years in four sub-Saharan African countries and three South Asian countries [65] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| The incidence, causes, and drivers of AMR in E. coli intestinal isolates from children in the community in many places throughout the world were unclear. | The phenotypic susceptibility of isolates and WGS were investigated and linked with antibiotic usage, disease state (symptomatic/asymptomatic), phylogenetic lineage, and geographic location. | AMR was shown to be prevalent, with 65% of isolates resistant to at least three antimicrobial medication classes. A wide spectrum of genetic pathways of AMR was discovered, with geographic location and antimicrobial usage pattern being the best predictors of AMR. | WGS was utilized to conduct a thorough examination of AMR across a vast geographical area, revealing information about AMR epidemiology, distribution, and causes. |
| Case 7: Investigation of colistin resistance detected in commensal E. coli in food stock animals in China [66] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| Routine surveillance had revealed a significant increase in the rates of colistin resistance in bacteria colonizing pigs in China, but the cause of this resistance remained unknown. | To validate the presence of plasmid-associated, transmissible colistin resistance, conjugation tests were performed. The WGS on plasmids was utilized to identify the relevant gene. | The plasmid-associated colistin resistance gene sequence was identified and named mcr-1. | The genetic foundation of a novel AMR mechanism has been identified and characterized, allowing for continuing surveillance and guiding inquiry and identification of this growing danger. |
| Case 8: Understanding of the epidemiology of MDR and XDR pathogens amenable to control by vaccination [67,68] | |||
| Justification | WGS/workflow | Main findings | Advantages of WGS |
| AMR is affecting the effectiveness of typhoid fever therapy. Resistance to azithromycin, the final effective oral drug, was discovered in Bangladesh and later in Pakistan, but the genetic basis and likelihood of spread remained unclear. | WGS was used to examine clinical isolates of azithromycin-resistant S. Typhi. The phylogenetic analysis allowed the strains to be contextualized among contemporaneous S. Typhi isolates in both contexts. | Phylogenetic analysis revealed that resistant isolates in Bangladesh and Pakistan arose from the separate acquisition of mutations in the same gene, showing the breadth of azithromycin selection pressure and the critical need for disease management by vaccination. | Two independent epidemics of azithromycin-resistant S. Typhi were identified and investigated using WGS. These findings aided in the development of innovative typhoid conjugate vaccines for infection control. |
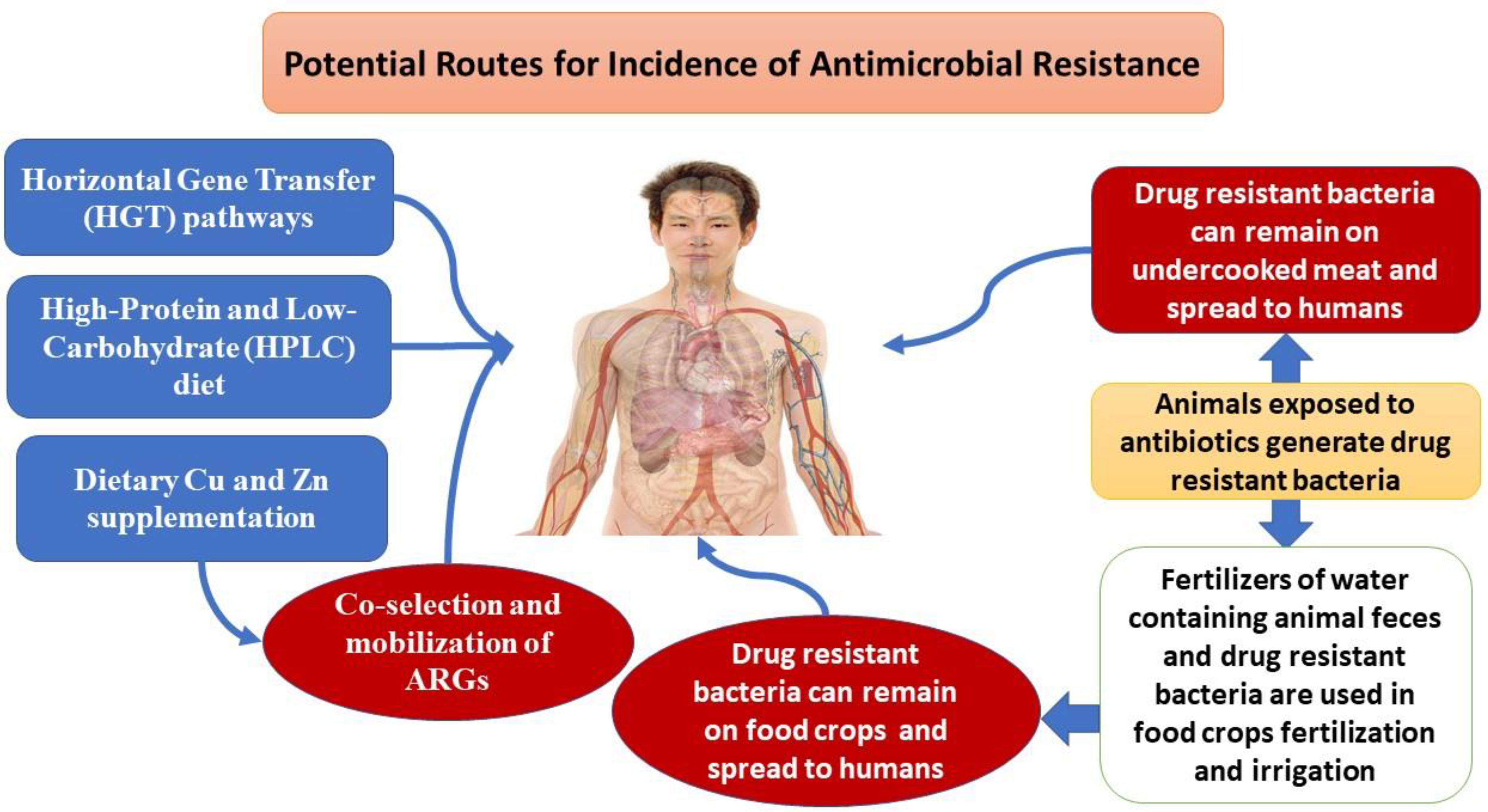
3.8. Potential effect of nutrigenomics on increased antimicrobial resistance against new antibiotics
| Procedure | Medium | Evaluation temperature (°C) |
Species | Antimicrobial Resistance Genes (ARGs) present | Stated antimicrobial resistance profiles | Recipient species | ARGs detected post-treatment from non-culturable samples |
Transformation demonstrated | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cooking—boiled (20 min), grilled (10 min), microwaved (5 min, 900 W), or autoclaved (20 min, 121 °C) | Chicken, beef, pork ?? |
Not Stated | E. faecalis | aac(6′)-Ie-aph(2″)-Ia | Aminoglycosides, except to streptomycin(predicted profile, not tested) | E. faecalis | YES | NO | [80] | |
| General heat treatments | Saline | 40, 50,60, 70, 80, 90, 100 | E. coli |
blaCTX-M-1, blaCMY-2, tetA, strA |
Cephalosporins, tetracycline, streptomycin | E. coli | YES | YES | 70 °C for 30 min | [81] |
| Milk pasteurizatio-n (sterilization) | Milk and elution buffer | 63.5, 121 |
S. aureus, S. sciuri |
blaZ, mecC, tetK | Penicillin, methicillin, tetracycline | S. aureus | YES | YES | 63.5 °C for 30 min | [82] |
| Non-food autoclaving | Distilled water and in presence of salt | 121, 135 | Plasmid (pUC18) | NS | Ampicillin | E. coli | - | YES | 121 °C for 15 min | [83] |
4. Conclusions
5. Recommendations
6. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Michalak, M.; Agellon, L.B. Importance of nutrients and nutrient metabolism on human health. Yale J. Biol. Med. 2018, 91, 95–103. [Google Scholar]
- Carlberg, C. Nutrigenomics in the context of evolution. Redox Biol. 2023, 62, 102656. [Google Scholar] [CrossRef]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361, bmj.k2173. [Google Scholar] [CrossRef]
- Mishra, U.N.; Jena, D.; Sahu, C.; Devi, R.; Kumar, R.; Jena, R.; Irondi, E.A.; Rout, S.; Tiwari, R.K.; Lal, M.K.; et al. Nutrigenomics: An Inimitable Interaction amid Genomics, Nutrition and Health. Innovative Food Science & Emerging Technologies 2022, 82, 103196. [Google Scholar]
- Neeha, V.S.; Kinth, P. Nutrigenomics Research: A Review. Journal of Food Science and Technology 2012, 50, 415–428. [Google Scholar] [CrossRef]
- Brennan, L.; de Roos, B. Nutrigenomics: Lessons Learned and Future Perspectives. The American Journal of Clinical Nutrition 2021, 113, 503–516. [Google Scholar] [CrossRef]
- Naushad, S.M.; Rama Devi, A.R. Role of Parental Folate Pathway Single Nucleotide Polymorphisms in Altering the Susceptibility to Neural Tube Defects in South India. Journal of Perinatal Medicine 2010, 38. [Google Scholar] [CrossRef]
- Vara Prasad, S.S.; Jeya Kumar, S.S.; Kumar, P.U.; Qadri, S.S.; Vajreswari, A. Dietary Fatty Acid Composition Alters 11β-Hydroxysteroid Dehydrogenase Type 1 Gene Expression in Rat Retroperitoneal White Adipose Tissue. Lipids in Health and Disease 2010, 9. [Google Scholar] [CrossRef]
- Valiakos, G.; Kapna, I. Colistin Resistant Mcr Genes Prevalence in Livestock Animals (Swine, Bovine, Poultry) from a Multinational Perspective. A Systematic Review. Veterinary Sciences 2021, 8, 265. [Google Scholar] [CrossRef]
- Di Francesco, A.; Salvatore, D.; Sakhria, S.; Bertelloni, F.; Catelli, E.; Ben Yahia, S.; Tlatli, A. Colistin Resistance Genes in Broiler Chickens in Tunisia. Animals 2023, 13, 1409. [Google Scholar] [CrossRef]
- Gilani, S.M.H.; Rashid, Z.; Galani, S.; Ilyas, S.; Sahar, S.; Zahoor-ul-Hassan; Al-Ghanim, K.; Zehra, S.; Azhar, A.; Al-Misned, F.; et al. Growth Performance, Intestinal Histomorphology, Gut Microflora and Ghrelin Gene Expression Analysis of Broiler by Supplementing Natural Growth Promoters: A Nutrigenomics Approach. Saudi Journal of Biological Sciences 2021, 28, 3438–3447. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, P.; Wu, Y.; Wang, X.; Lu, X.; Liu, C.; Li, N.; Sun, J.; Xiao, J.; Jesus, S.-G. The Formation of Antibiotic Resistance Genes in Bacterial Communities During Garlic Powder Processing. Frontiers in Nutrition 2021, 8. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host & Microbe 2016, 19, 731–743. [Google Scholar]
- Martín-Peláez, S.; Fito, M.; Castaner, O. Mediterranean Diet Effects on Type 2 Diabetes Prevention, Disease Progression, and Related Mechanisms. A Review. Nutrients 2020, 12, 2236. [Google Scholar] [CrossRef]
- Soldati, L.; Di Renzo, L.; Jirillo, E.; Ascierto, P.A.; Marincola, F.M.; De Lorenzo, A. The Influence of Diet on Anti-Cancer Immune Responsiveness. Journal of Translational Medicine 2018, 16. [Google Scholar] [CrossRef]
- Mansour, S.R.; Moustafa, M.A.A.; Saad, B.M.; Hamed, R.; Moustafa, A.-R.A. Impact of Diet on Human Gut Microbiome and Disease Risk. New Microbes and New Infections 2021, 41, 100845. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill–Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology 2009, 137, 1716–1724.e2. [Google Scholar] [CrossRef]
- Agans, R.; Gordon, A.; Kramer, D.L.; Perez-Burillo, S.; Rufián-Henares, J.A.; Paliy, O. Dietary Fatty Acids Sustain the Growth of the Human Gut Microbiota. Applied and Environmental Microbiology 2018, 84. [Google Scholar] [CrossRef]
- Doaei, S.; Gholamalizadeh, M.; Akbari, M.E.; Akbari, S.; Feradova, H.; Rahimzadeh, G.; Mosavi Jarrahi, A. Dietary Carbohydrate Promotes Cell Survival in Cancer Via the Up-Regulation of Fat Mass and Obesity-Associated Gene Expression Level. Malaysian Journal of Medical Sciences 2019, 26, 8–17. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. Journal of Lipid Research 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut Microbiome Development along the Colorectal Adenoma–Carcinoma Sequence. Nature Communications 2015, 6. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Boba, A.; Czemplik, M.; Kulma, A.; Wojtasik, W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients 2021, 13, 3673. [Google Scholar] [CrossRef]
- Paoloni-Giacobino, A.; Grimble, R.; Pichard, C. Genetics and Nutrition. Clinical Nutrition 2003, 22, 429–435. [Google Scholar] [CrossRef]
- Leonardson, A.S.; Zhu, J.; Chen, Y.; Wang, K.; Lamb, J.R.; Reitman, M.; Emilsson, V.; Schadt, E.E. The Effect of Food Intake on Gene Expression in Human Peripheral Blood. Human Molecular Genetics 2009, 19, 159–169. [Google Scholar] [CrossRef]
- Wolfrum, C.; Asilmaz, E.; Luca, E.; Friedman, J.M.; Stoffel, M. Foxa2 Regulates Lipid Metabolism and Ketogenesis in the Liver during Fasting and in Diabetes. Nature 2004, 432, 1027–1032. [Google Scholar] [CrossRef]
- Kobayashi, H.; Oishi, K.; Hanai, S.; Ishida, N. Effect of Feeding on Peripheral Circadian Rhythms and Behaviour in Mammals. Genes to Cells 2004, 9, 857–864. [Google Scholar] [CrossRef]
- Bordoni, L.; Gabbianelli, R. Primers on Nutrigenetics and Nutri(Epi)Genomics: Origins and Development of Precision Nutrition. Biochimie 2019, 160, 156–171. [Google Scholar] [CrossRef]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial Resistance, Mechanisms and Its Clinical Significance. Disease-a-Month 2020, 66, 100971. [Google Scholar] [CrossRef]
- C Reygaert, W. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiology 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of Antibiotics on the Human Microbiome and Consequences for Host Health. MicrobiologyOpen 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Stanton, C.; Lane, J.; Smith, G.; Ross, R. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients 2019, 11, 923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, D.-C. Facing a New Challenge. Chinese Medical Journal 2019, 132, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Gasparrini, A.J.; Wang, B.; Sun, X.; Kennedy, E.A.; Hernandez-Leyva, A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Dantas, G. Persistent Metagenomic Signatures of Early-Life Hospitalization and Antibiotic Treatment in the Infant Gut Microbiota and Resistome. Nature Microbiology 2019, 4, 2285–2297. [Google Scholar] [CrossRef]
- Gibson, M.K.; Wang, B.; Ahmadi, S.; Burnham, C.-A.D.; Tarr, P.I.; Warner, B.B.; Dantas, G. Developmental Dynamics of the Preterm Infant Gut Microbiota and Antibiotic Resistome. Nature Microbiology 2016, 1. [Google Scholar] [CrossRef]
- McDonald, L.C. Effects of Short- and Long-Course Antibiotics on the Lower Intestinal Microbiome as They Relate to Traveller’s Diarrhea. Journal of Travel Medicine 2017, 24, S35–S38. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Artacho, A.; Knecht, H.; Ferrús, M.L.; Friedrichs, A.; Ott, S.J.; Moya, A.; Latorre, A.; Gosalbes, M.J. Differential Effects of Antibiotic Therapy on the Structure and Function of Human Gut Microbiota. PLoS ONE 2013, 8, e80201. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Frontiers in Microbiology 2020, 11. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Yu, K.; Yu, M.; Zhang, C.; Su, Y.; Zhu, W. Alteration of Metabolomic Markers of Amino-Acid Metabolism in Piglets with in-Feed Antibiotics. Amino Acids 2017, 49, 771–781. [Google Scholar] [CrossRef]
- Yang, H.; Wei, S.-H.; Hobman, J.L.; Dodd, C.E.R. Antibiotic and Metal Resistance in Escherichia Coli Isolated from Pig Slaughterhouses in the United Kingdom. Antibiotics 2020, 9, 746. [Google Scholar] [CrossRef] [PubMed]
- Sacher-Pirklbauer, A.; Klein-Jöbstl, D.; Sofka, D.; Blanc-Potard, A.-B.; Hilbert, F. Phylogenetic Groups and Antimicrobial Resistance Genes in Escherichia Coli from Different Meat Species. Antibiotics 2021, 10, 1543. [Google Scholar] [CrossRef] [PubMed]
- Tohmaz, M.; Askari Badouei, M.; Kalateh Rahmani, H.; Hashemi Tabar, G. Antimicrobial Resistance, Virulence Associated Genes and Phylogenetic Background versus Plasmid Replicon Types: The Possible Associations in Avian Pathogenic Escherichia Coli (APEC). BMC Veterinary Research 2022, 18. [Google Scholar] [CrossRef] [PubMed]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline. [(accessed on 1 May 2021)];2019 Available online: http://apps.who.int/iris/bitstream/handle/10665/258965/WHO-EMP-IAU-2017.11-eng.pdf;jsessionid=9955EDC07F0D8D09ACAA2CAC34DB921F?sequence=1. (accessed on 1 May 2021).
- Bradford, P.A.; Miller, A.A.; O’Donnell, J.; Mueller, J.P. Zoliflodacin: An Oral Spiropyrimidinetrione Antibiotic for the Treatment of Neisseria Gonorrheae, Including Multi-Drug-Resistant Isolates. ACS Infectious Diseases 2020, 6, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Cho, J. Ridinilazole: A Novel Antimicrobial for Clostridium Difficile Infection. Annals of Gastroenterology 2018. [Google Scholar] [CrossRef] [PubMed]
- Tulkens, P.M.; Van Bambeke, F.; Zinner, S.H. Profile of a Novel Anionic Fluoroquinolone—Delafloxacin. Clinical Infectious Diseases 2019, 68, S213–S222. [Google Scholar] [CrossRef]
- Sartelli, M.; Chichom-Mefire, A.; Labricciosa, F.M.; Hardcastle, T.; Abu-Zidan, F.M.; Adesunkanmi, A.K.; Ansaloni, L.; Bala, M.; Balogh, Z.J.; Beltrán, M.A.; et al. The Management of Intra-Abdominal Infections from a Global Perspective: 2017 WSES Guidelines for Management of Intra-Abdominal Infections. World Journal of Emergency Surgery 2017, 12. [Google Scholar]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline Antibiotics and Resistance Mechanisms. Biological Chemistry 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Cheung, D.; Adam, H.; Zelenitsky, S.; Golden, A.; Schweizer, F.; Gorityala, B.; Lagacé-Wiens, P.R.S.; Walkty, A.; Gin, A.S.; et al. Review of Eravacycline, a Novel Fluorocycline Antibacterial Agent. Drugs 2016, 76, 567–588. [Google Scholar] [CrossRef] [PubMed]
- Endimiani, A.; Hujer, K.M.; Hujer, A.M.; Armstrong, E.S.; Choudhary, Y.; Aggen, J.B.; Bonomo, R.A. ACHN-490, a Neoglycoside with Potent In Vitro Activity against Multidrug-Resistant Klebsiella Pneumoniae Isolates. Antimicrobial Agents and Chemotherapy 2009, 53, 4504–4507. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.; Carey, M.E.; Boinett, C.J.; Higginson, E.; Veeraraghavan, B.; Baker, S. Exploiting Genomics to Mitigate the Public Health Impact of Antimicrobial Resistance. Genome Medicine 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Masim, M.A.L.; Gayeta, J.M.; Lagrada, M.L.; Macaranas, P.K.V.; Cohen, V.; Limas, M.T.; Espiritu, H.O.; Palarca, J.C.; Chilam, J.; et al. Integrating Whole-Genome Sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nature Communications 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Whole-Genome Sequencing as Part of National and International Surveillance Programmes for Antimicrobial Resistance: A Roadmap. BMJ Global Health 2020, 5, e002244. [CrossRef] [PubMed]
- Didelot, X.; Gardy, J.; Colijn, C. Bayesian Inference of Infectious Disease Transmission from Whole-Genome Sequence Data. Molecular Biology and Evolution 2014, 31, 1869–1879. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Identification of Salmonellafor Public Health Surveillance Using Whole Genome Sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef]
- Wainwright, C.; Altamirano, L.; Cheney, M.; Cheney, J.; Barber, S.; Price, D.; Moloney, S.; Kimberley, A.; Woolfield, N.; Cadzow, S.; et al. A Multicenter, Randomized, Double-Blind, Controlled Trial of Nebulized Epinephrine in Infants with Acute Bronchiolitis. New England Journal of Medicine 2003, 349, 27–35. [Google Scholar] [CrossRef]
- Blake, K.S.; Choi, J.; Dantas, G. Approaches for Characterizing and Tracking Hospital-Associated Multidrug-Resistant Bacteria. Cellular and Molecular Life Sciences 2021, 78, 2585–2606. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of Carbapenem-Resistant Klebsiella Pneumoniae in Europe Is Driven by Nosocomial Spread. Nature Microbiology 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Harris, S.R.; Cartwright, E.J.; Török, M.E.; Holden, M.T.; Brown, N.M.; Ogilvy-Stuart, A.L.; Ellington, M.J.; Quail, M.A.; Bentley, S.D.; Parkhill, J.; et al. Whole-Genome Sequencing for Analysis of an Outbreak of Meticillin-Resistant Staphylococcus Aureus: A Descriptive Study. The Lancet Infectious Diseases 2013, 13, 130–136. [Google Scholar] [CrossRef]
- Lewis, T.; Loman, N.J.; Bingle, L.; Jumaa, P.; Weinstock, G.M.; Mortiboy, D.; Pallen, M.J. High-Throughput Whole-Genome Sequencing to Dissect the Epidemiology of Acinetobacter Baumannii Isolates from a Hospital Outbreak. Journal of Hospital Infection 2010, 75, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Jajou, R.; Neeling, A. de; Hunen, R. van; Vries, G. de; Schimmel, H.; Mulder, A.; Anthony, R.; Hoek, W. van der; Soolingen, D. van Epidemiological Links between Tuberculosis Cases Identified Twice as Efficiently by Whole Genome Sequencing than Conventional Molecular Typing: A Population-Based Study. PLoS ONE 2018, 13, e0195413. [Google Scholar]
- Ingle, D.J.; Levine, M.M.; Kotloff, K.L.; Holt, K.E.; Robins-Browne, R.M. Dynamics of Antimicrobial Resistance in Intestinal Escherichia Coli from Children in Community Settings in South Asia and Sub-Saharan Africa. Nature Microbiology 2018, 3, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. The Lancet Infectious Diseases 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Hooda, Y.; Sajib, M.S.I.; Rahman, H.; Luby, S.P.; Bondy-Denomy, J.; Santosham, M.; Andrews, J.R.; Saha, S.K.; Saha, S. Molecular Mechanism of Azithromycin Resistance among Typhoidal Salmonella Strains in Bangladesh Identified through Passive Pediatric Surveillance. PLOS Neglected Tropical Diseases 2019, 13, e0007868. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Dehraj, I.F.; Carey, M.E.; Dyson, Z.A.; Garrett, D.; Seidman, J.C.; Kabir, F.; Saha, S.; Baker, S.; Qamar, F.N. A Race against Time: Reduced Azithromycin Susceptibility in Salmonella Enterica Serovar Typhi in Pakistan. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Jung, H.-J.; Littmann, E.R.; Seok, R.; Leiner, I.M.; Taur, Y.; Peled, J.; van den Brink, M.; Ling, L.; Chen, L.; Kreiswirth, B.N.; et al. Genome-Wide Screening for Enteric Colonization Factors in Carbapenem-Resistant ST258 Klebsiella Pneumoniae. mBio 2019, 10. [Google Scholar] [CrossRef]
- Taur, Y.; Xavier, J.B.; Lipuma, L.; Ubeda, C.; Goldberg, J.; Gobourne, A.; Lee, Y.J.; Dubin, K.A.; Socci, N.D.; Viale, A.; et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clinical Infectious Diseases 2012, 55, 905–914. [Google Scholar] [CrossRef]
- Bell, B.G.; Schellevis, F.; Stobberingh, E.; Goossens, H.; Pringle, M. A Systematic Review and Meta-Analysis of the Effects of Antibiotic Consumption on Antibiotic Resistance. BMC Infectious Diseases 2014, 14. [Google Scholar] [CrossRef]
- van Schaik, W. The Human Gut Resistome. Philosophical Transactions of the Royal Society B: Biological Sciences 2015, 370, 20140087. [Google Scholar] [CrossRef] [PubMed]
- Osthoff, M.; McGuinness, S.L.; Wagen, A.Z.; Eisen, D.P. Urinary Tract Infections Due to Extended-Spectrum Beta-Lactamase-Producing Gram-Negative Bacteria: Identification of Risk Factors and Outcome Predictors in an Australian Tertiary Referral Hospital. International Journal of Infectious Diseases 2015, 34, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Leal, H.F.; Azevedo, J.; Silva, G.E.O.; Amorim, A.M.L.; de Roma, L.R.C.; Arraes, A.C.P.; Gouveia, E.L.; Reis, M.G.; Mendes, A.V.; de Oliveira Silva, M.; et al. Bloodstream Infections Caused by Multidrug-Resistant Gram-Negative Bacteria: Epidemiological, Clinical and Microbiological Features. BMC Infectious Diseases 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- James, C.; Dixon, R.; Talbot, L.; James, S.J.; Williams, N.; Onarinde, B.A. Assessing the Impact of Heat Treatment of Food on Antimicrobial Resistance Genes and Their Potential Uptake by Other Bacteria—A Critical Review. Antibiotics 2021, 10, 1440. [Google Scholar] [CrossRef] [PubMed]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. International Journal of Environmental Research and Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, F.; Mercanoglu Taban, B. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Frontiers in Microbiology 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Rizzotti, L.; Felis, G.E.; Torriani, S. Horizontal Gene Transfer among Microorganisms in Food: Current Knowledge and Future Perspectives. Food Microbiology 2014, 42, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Kharazmi, M.; Bauer, T.; Hammes, W.P.; Hertel, C. Effect of Food Processing on the Fate of DNA with Regard to Degradation and Transformation Capability in Bacillus Subtilis. Systematic and Applied Microbiology 2003, 26, 495–501. [Google Scholar] [CrossRef]
- Koncan, R.; García-Albiach, R.; Bravo, D.; Cantón, R.; Baquero, F.; Cornaglia, G.; del Campo, R. P571 The Fate of Antibiotic Resistance Genes in Cooked Meat. International Journal of Antimicrobial Agents 2007, 29, S130. [Google Scholar] [CrossRef]
- Le Devendec, L.; Jouy, E.; Kempf, I. Evaluation of Resistance Gene Transfer from Heat-Treated Escherichia Coli. International Journal of Food Microbiology 2018, 270, 39–43. [Google Scholar] [CrossRef]
- Taher, E.M.; Hemmatzadeh, F.; Aly, S.A.; Elesswy, H.A.; Petrovski, K.R. Survival of Staphylococci and Transmissibility of Their Antimicrobial Resistance Genes in Milk after Heat Treatments. LWT 2020, 129, 109584. [Google Scholar] [CrossRef]
- Masters, C.I.; Miles, C.A.; Mackey, B.M. Survival and Biological Activity of Heat Damaged DNA. Letters in Applied Microbiology 1998, 27, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Snyder, H.; Kellogg, S.L.; Skarda, L.M.; Little, J.L.; Kristich, C.J. Nutritional Control of Antibiotic Resistance via an Interface between the Phosphotransferase System and a Two-Component Signaling System. Antimicrobial Agents and Chemotherapy 2014, 58, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Xue, Z.; Villanueva, Y.T.; Durbin-Johnson, B.; Alkan, Z.; Taft, D.H.; Liu, J.; Korf, I.; Laugero, K.D.; Stephensen, C.B.; et al. Association of Diet and Antimicrobial Resistance in Healthy U.S. Adults. mBio 2022, 13. [Google Scholar] [CrossRef]
- Kim, Y.; Leung, M.H.Y.; Kwok, W.; Fournié, G.; Li, J.; Lee, P.K.H.; Pfeiffer, D.U. Antibiotic Resistance Gene Sharing Networks and the Effect of Dietary Nutritional Content on the Canine and Feline Gut Resistome. Animal Microbiome 2020, 2. [Google Scholar] [CrossRef] [PubMed]
- Muurinen, J.; Richert, J.; Wickware, C.L.; Richert, B.; Johnson, T.A. Swine Growth Promotion with Antibiotics or Alternatives Can Increase Antibiotic Resistance Gene Mobility Potential. Scientific Reports 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Su, J.-Q.; An, X.-L.; Huang, F.-Y.; Rensing, C.; Brandt, K.K.; Zhu, Y.-G. Feed Additives Shift Gut Microbiota and Enrich Antibiotic Resistance in Swine Gut. Science of The Total Environment 2018, 621, 1224–1232. [Google Scholar] [CrossRef]
- Poole, K. At the Nexus of Antibiotics and Metals: The Impact of Cu and Zn on Antibiotic Activity and Resistance. Trends in Microbiology 2017, 25, 820–832. [Google Scholar] [CrossRef]
- Yazdankhah, S.; Rudi, K.; Bernhoft, A. Zinc and Copper in Animal Feed – Development of Resistance and Co-Resistance to Antimicrobial Agents in Bacteria of Animal Origin. Microbial Ecology in Health & Disease 2014, 25. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





