Submitted:
02 May 2023
Posted:
02 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
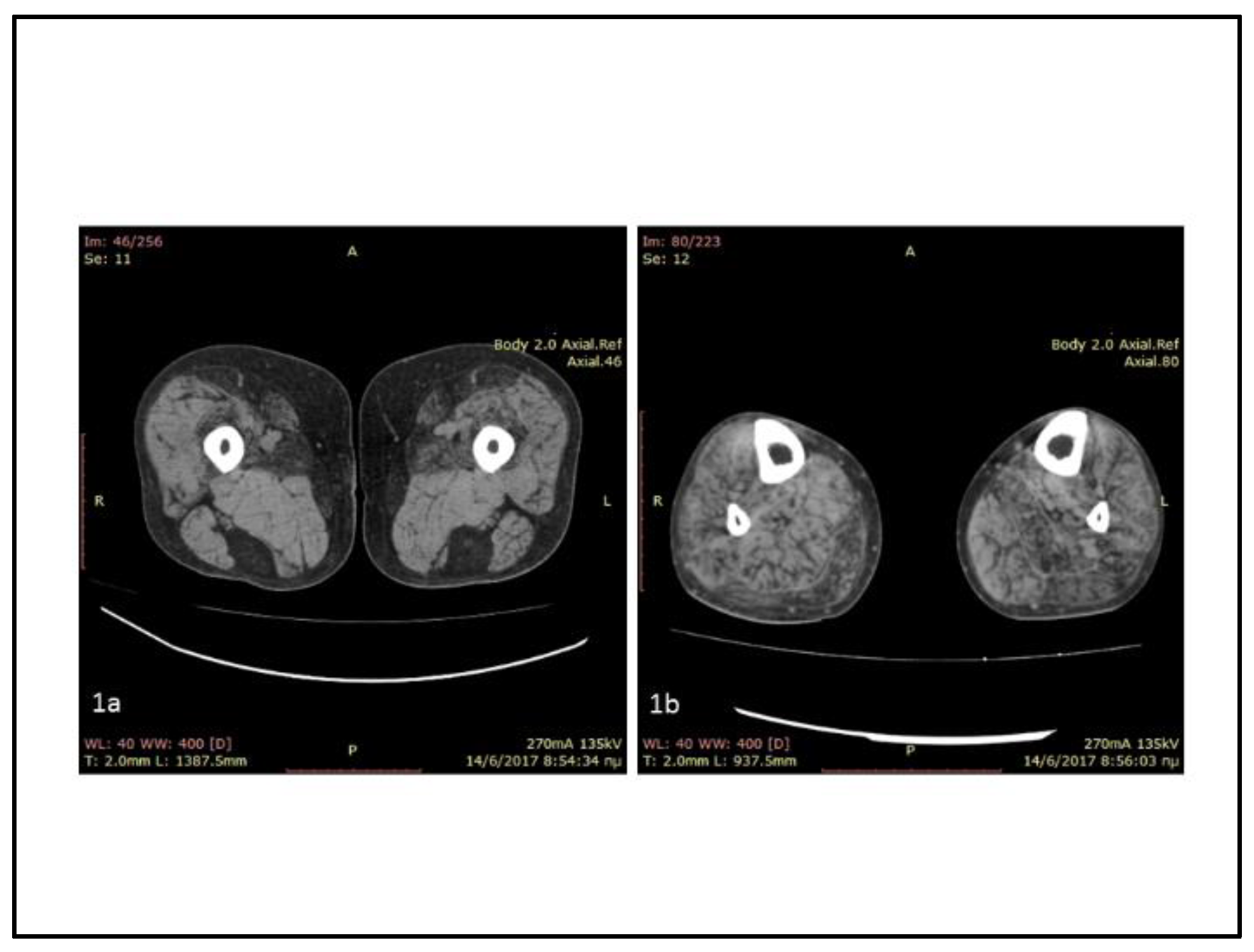
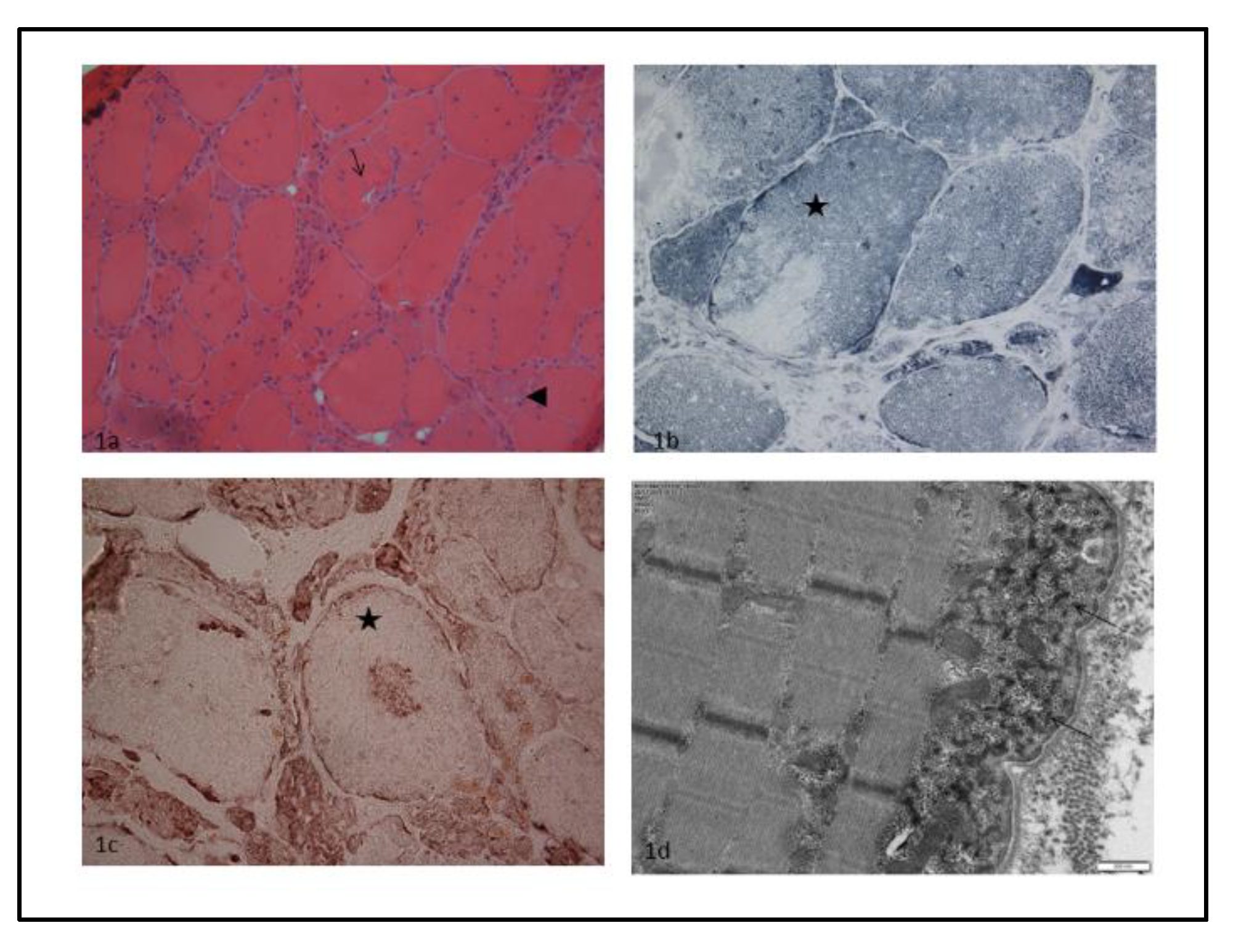
2. Case Report
3. Discussion
4. Materials and Methods
4.1. Muscle Biopsy
4.2. Targeted Gene Enrichment, Next Generation Sequencing NGS (High-Throughput Sequencing)
4.3. Bioinformatics Analysis
4.4. Variants Interpretations
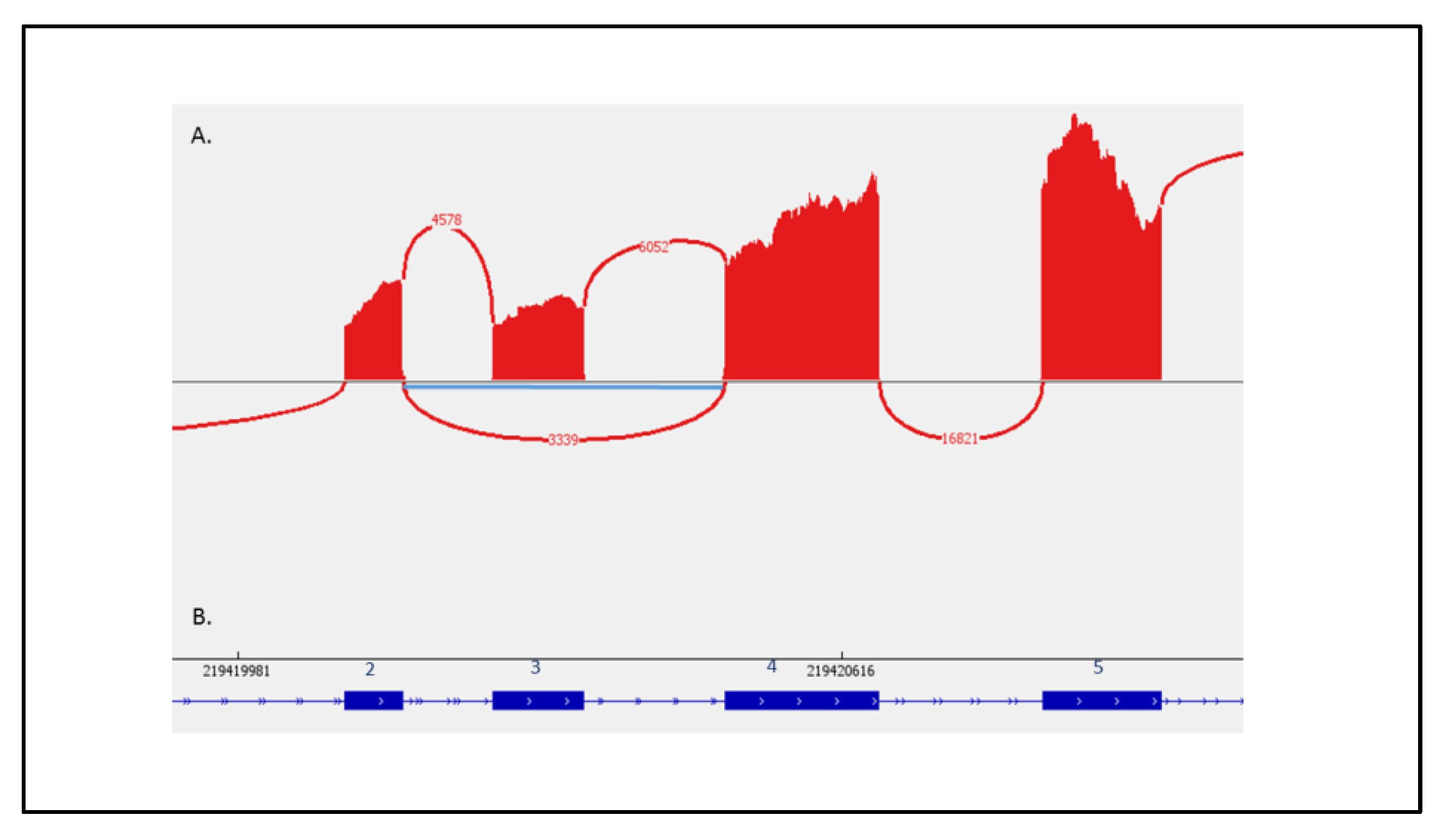
4.5. RNA Sequencing
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
References
- Clemen CS, Herrmann H, Strelkov SV, Schröder R. Desminopathies: pathology and mechanisms. Acta Neuropathol. 2013 Jan;125(1):47-75. Epub 2012 Nov 11. PMID: 23143191; PMCID: PMC3535371m . [CrossRef]
- Capetanaki Y, Papathanasiou S, Diokmetzidou A, Vatsellas G, Tsikitis M. Desmin related disease: a matter of cell survival failure. Curr Opin Cell Biol. 2015 Feb;32:113-20. Epub 2015 Feb 11. PMID: 25680090; PMCID: PMC4365784 . [CrossRef]
- Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem 1994;63: 345–82. [CrossRef]
- Claeys KG, Fardeau M. Myofibrillar myopathies. Handb Clin Neurol. 2013;113:1337-42. [CrossRef]
- Olivé M, Kley RA, Goldfarb LG. Myofibrillar myopathies: new developments. Curr Opin Neurol. 2013;26:527-35. [CrossRef]
- Olivé M, Odgerel Z, Martinez A, et al. Clinical and myopathological evaluation of early- and late-onset subtypes of myofibrillar myopathy. Neuromuscul Disord 2011;21:533-42. [CrossRef]
- van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JD, de Walle HE, Capetanaki Y, van der Kooi AJ, van Langen IM, van den Berg MP, van Tintelen JP. Desmin-related myopathy. Clin Genet. 2011 Oct;80(4):354-66. Epub 2010 Jul 21. PMID: 20718792 . [CrossRef]
- Walter MC, Reilich P, Huebner A, Fischer D, Schröder R, Vorgerd M, Kress W, Born C, Schoser BG, Krause KH, Klutzny U, Bulst S, Frey JR, Lochmüller H. Scapuloperoneal syndrome type Kaeser and a wide phenotypic spectrum of adult-onset, dominant myopathies are associated with the desmin mutation R350P. Brain. 2007 Jun;130(Pt 6):1485-96. Epub 2007 Apr 17. PMID: 17439987 . [CrossRef]
- Schröder R, Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol 2009;19:483-92. [CrossRef]
- Taylor MR, Slavov D, Ku L, Di Lenarda A, Sinagra G, Carniel E et al. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation 2007;115: 1244-51. [CrossRef]
- Wahbi K, Béhin A, Charron P, Dunand M, Richard P, Meune C et al. High cardiovascular morbidity and mortality in myofibrillar myopathies due to DES gene mutations: a 10-year longitudinal study. Neuromuscul Disord. 2012;22:211-8. [CrossRef]
- Béhin A, Salort-Campana E, Wahbi K, Richard P, Carlier RY, Carlier P, Laforêt P, Stojkovic T, Maisonobe T, Verschueren A, Franques J, Attarian S, Maues de Paula A, Figarella-Branger D, Bécane HM, Nelson I, Duboc D, Bonne G, Vicart P, Udd B, Romero N, Pouget J, Eymard B. Myofibrillar myopathies: State of the art, present and future challenges. Rev Neurol (Paris). 2015 Oct;171(10):715-29. Epub 2015 Sep 3. PMID: 26342832 . [CrossRef]
- Dagvadorj A, Goudeau B, Hilton-Jones D, Blancato JK, Shatunov A, Simon-Casteras M, Squier W, Nagle JW, Goldfarb LG, Vicart P. Respiratory insufficiency in desminopathy patients caused by introduction of proline residues in desmin c-terminal alpha-helical segment. Muscle Nerve. 2003 Jun;27(6):669-75. PMID: 12766977 . [CrossRef]
- Schramm N, Born C, Weckbach S, Reilich P, Walter MC, Reiser MF. Involvement patterns in myotilinopathy and desminopathy detected by a novel neuromuscular whole-body MRI protocol. Eur Radiol. 2008 Dec;18(12):2922-36. Epub 2008 Jul 22. PMID: 18648820 . [CrossRef]
- Claeys KG, van der Ven PF, Behin A, Stojkovic T, Eymard B, Dubourg O et al. Differential involvement of sarcomeric proteins in myofibrillar myopathies: a morphological and immunohistochemical study. Acta Neuropathol. 2009;117:293-07. [CrossRef]
- Dalakas MC, Park KY, Semino-Mora C, Lee HS, Sivakumar K, Goldfarb LG. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000 Mar 16;342(11):770-80. PMID: 10717012 . [CrossRef]
- Ojrzyńska N, Bilińska ZT, Franaszczyk M, Płoski R, Grzybowski J. Restrictive cardiomyopathy due to novel desmin gene mutation. Kardiol Pol. 2017;75(7):723. PMID: 28703267 . [CrossRef]
- Kostareva A, Gudkova A, Sjoberg G, Kiselev I, Moiseeva O, Karelkina E, et al. Desmin mutations in a St. Petersburg cohort of cardiomyopathies. Acta Myol. 2006 Dec;25(3):109-15. PMID: 17626518.
- Fan P, Lu CX, Dong XQ, Zhu D, Yang KQ, Liu KQ et al. A novel phenotype with splicing mutation identified in a Chinese family with desminopathy. Chin Med J (Engl). 2019 Jan 20;132(2):127-134. PMID: 30614851; PMCID: PMC6365268 . [CrossRef]
- Nalini A, Gayathri N, Richard P, Cobo AM, Urtizberea JA. New mutation of the desmin gene identified in an extended Indian pedigree presenting with distal myopathy and cardiac disease. Neurol India. 2013 Nov-Dec;61(6):622-6. [CrossRef]
- Arbustini E, Di Toro A, Giuliani L, Favalli V, Narula N, Grasso M. Cardiac Phenotypes in Hereditary Muscle Disorders: JACC State-of-the-Art Review. J Am Coll Cardiol. 2018;72:2485-06. [CrossRef]
- Kostera-Pruszczyk A, Pruszczyk P, Kaminska A, Lee Hee-Suk, Goldfarb LG. Diversity of cardiomyopathy phenotypes caused by mutations in desmin. Int J Cardiol 2008;131:146-7. [CrossRef]
- Strach K, Sommer T, Grohé C, Meyer C, Fischer D, Walter MC, Vorgerd M, Reilich P, Bär H, Reimann J, Reuner U, Germing A, Goebel HH, Lochmüller H, Wintersperger B, Schröder R. Clinical, genetic, and cardiac magnetic resonance imaging findings in primary desminopathies. Neuromuscul Disord. 2008 Jun;18(6):475-82. Epub 2008 May 27. PMID: 18504128 . [CrossRef]
- Leung DG. Magnetic resonance imaging patterns of muscle involvement in genetic muscle diseases: a systematic review. J Neurol. 2017;264:1320-33. [CrossRef]
- Fischer D, Kley RA, Strach K, Meyer C, Sommer T, Eger K, Rolfs A, Meyer W, Pou A, Pradas J, Heyer CM, Grossmann A, Huebner A, Kress W, Reimann J, Schröder R, Eymard B, Fardeau M, Udd B, Goldfarb L, Vorgerd M, Olivé M. Distinct muscle imaging patterns in myofibrillar myopathies. Neurology. 2008 Sep 2;71(10):758-65. PMID: 18765652; PMCID: PMC2583436 . [CrossRef]
- Claeys KG, Fardeau M, Schröder R, Suominen T, Tolksdorf K, Behin A et al. Electron microscopy in myofibrillar myopathies reveals clues to the mutated gene. NeuromusculDisord. 200818:656-66. [CrossRef]
- Vrabie A, Goldfarb LG, Shatunov A, Nägele A, Fritz P, Kaczmarek I et al. The enlarging spectrum of desminopathies: new morphological findings, eastward geographic spread, novel exon 3 desmin mutation. Acta Neuropathol. 2005 Apr;109(4):411-7. Epub 2005 Mar 10. PMID: 15759133 . [CrossRef]
- Park KY, Dalakas MC, Goebel HH, Ferrans VJ, Semino-Mora C, Litvak S, Takeda K, Goldfarb LG. Desmin splice variants causing cardiac and skeletal myopathy. J Med Genet. 2000 Nov;37(11):851-7. PMID: 11073539; PMCID: PMC1734475 . [CrossRef]
- Bang ML, Gregorio C, Labeit S. Molecular dissection of the interaction of desmin with the C-terminal region of nebulin. J Struct Biol. 2002 Jan-Feb;137(1-2):119-27. PMID: 12064939 . [CrossRef]
- Krahn M, Biancalana V, Cerino M, Perrin A, Michel-Calemard L, Nectoux J et al. A National French consensus on gene lists for the diagnosis of myopathies using next-generation sequencing. Eur J Hum Genet. 2019 Mar;27(3):349-352. Epub 2018 Dec 14. PMID: 30552423; PMCID: PMC6460575 . [CrossRef]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J et al; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24. Epub 2015 Mar 5. PMID: 25741868; PMCID: PMC4544753 . [CrossRef]
| ACTA1 (NM_001100.3) |
| BAG3 (NM_004281.3) |
| CRYAB (NM_001885.2) |
| DES (NM_001927.3) |
| DNAJB6 (NM_058246.3) |
| FHL1 (NM_001159702.2) |
| FLNC (NM_001458.4) |
| GNE (NM_001128227.3) |
| HSPB1 (NM_001540.3) |
| HSPB8 (NM_014365.2) |
| MYH2 (NM_017534.5) |
| MYOT (NM_006790.2) |
| RYR1 (NM_000540.2, with partially covered exon 91) |
| SQSTM1 (NM_003900.4) |
| TTN (NM_001267550.1) |
| VCP (NM_007126.3) |
| ZASP/LDB3 (NM_001080114.1, NM_007078.2,NM_001171610.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





