Submitted:
27 April 2023
Posted:
28 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sampath, S.; Khedr, A.; Qamar, S.; Tekin, A.; Singh, R.; Green, R.; Kashyap, R. Pandemics Throughout the History. Cureus 2021, 13, e18136. [Google Scholar] [CrossRef] [PubMed]
- McVernon, J.; Liberman, J. WHO keeps covid-19 a public health emergency of international concern. BMJ 2023, 380, 504. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Liu, S.L. Role of host factors in SARS-CoV-2 entry. J Biol Chem. 2021, 297, 100847. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Zhang, M.; Chang, T.L. ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses 2022, 14, 2535. [Google Scholar] [CrossRef]
- Nocini, R.; Henry, B.M.; Mattiuzzi, C.; Lippi, G. Improving Nasal Protection for Preventing SARS-CoV-2 Infection. Biomedicines 2022, 10, 2966. [Google Scholar] [CrossRef]
- Ligi, D.; Lo Sasso, B.; Henry, B.M.; Ciaccio, M.; Lippi, G.; Plebani, M.; Mannello, F. Deciphering the role of monocyte and monocyte distribution width (MDW) in COVID-19: An updated systematic review and meta-analysis. Clin Chem Lab Med. 2023, 61, 960–973. [Google Scholar] [CrossRef]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021, 590, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M. Cytokine "storm", cytokine "breeze", or both in COVID-19? Clin Chem Lab Med. 2020, 59, 637–639. [Google Scholar] [CrossRef]
- Park, J.; Dean, L.S.; Jiyarom, B.; Gangcuangco, L.M.; Shah, P.; Awamura, T.; Ching, L.L.; Nerurkar, V.R.; Chow, D.C.; Igno, F.; et al. Elevated circulating monocytes and monocyte activation in COVID-19 convalescent individuals. Front Immunol. 2023, 14, 1151780. [Google Scholar] [CrossRef]
- Kilercik, M.; Demirelce, Ö.; Serdar, M.A.; Mikailova, P.; Serteser, M. A new haematocytometric index: Predicting severity and mortality risk value in COVID-19 patients. PLoS ONE 2021, 16, e0254073. [Google Scholar] [CrossRef]
- Ait-Belkacem, I.; Cartagena García, C.; Millet-Wallisky, E.; Izquierdo, N.; Loosveld, M.; Arnoux, I.; Morange, P.E.; Galland, F.; Lambert, N.; Malergue, F.; et al. SARS-CoV-2 spike protein induces a differential monocyte activation that may contribute to age bias in COVID-19 severity. Sci Rep. 2022, 12, 20824. [Google Scholar] [CrossRef] [PubMed]
- Craddock, V.; Mahajan, A.; Spikes, L.; Krishnamachary, B.; Ram, A.K.; Kumar, A.; Chen, L.; Chalise, P.; Dhillon, N.K. Persistent circulation of soluble and extracellular vesicle-linked Spike protein in individuals with postacute sequelae of COVID-19. J Med Virol. 2023, 95, e28568. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Salvagno, G.L.; Montagnana, M.; Lima-Oliveira, G.; Guidi, G.C.; Favaloro, E.J. Quality standards for sample collection in coagulation testing. Semin Thromb Hemost. 2012, 38, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Buoro, S.; Seghezzi, M.; Vavassori, M.; Dominoni, P.; Apassiti Esposito, S.; Manenti, B.; Mecca, T.; Marchesi, G.; Castellucci, E.; Azzarà, G.; et al. Clinical significance of cell population data (CPD) on Sysmex XN-9000 in septic patients with our without liver impairment. Ann Transl Med. 2016, 4, 418. [Google Scholar] [CrossRef]
- Yao, Y.; Subedi, K.; Liu, T.; Khalasawi, N.; Pretto-Kernahan, C.D.; Wotring, J.W.; Wang, J.; Yin, C.; Jiang, A.; Fu, C.; et al. Surface translocation of ACE2 and TMPRSS2 upon TLR4/7/8 activation is required for SARS-CoV-2 infection in circulating monocytes. Cell Discov. 2022, 8, 89. [Google Scholar] [CrossRef]
- Jalloh, S.; Olejnik, J.; Berrigan, J.; Nisa, A.; Suder, E.L.; Akiyama, H.; Lei, M.; Ramaswamy, S.; Tyagi, S.; Bushkin, Y.; et al. CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses. PLoS Pathog. 2022, 18, e1010479. [Google Scholar] [CrossRef]
- Cao, X.; Tian, Y.; Nguyen, V.; Zhang, Y.; Gao, C.; Yin, R.; Carver, W.; Fan, D.; Albrecht, H.; Cui, T.; et al. Spike protein of SARS-CoV-2 activates macrophages and contributes to induction of acute lung inflammation in male mice. FASEB J. 2021, 35, e21801. [Google Scholar] [CrossRef]
- Palestra, F.; Poto, R.; Ciardi, R.; Opromolla, G.; Secondo, A.; Tedeschi, V.; Ferrara, A.L.; Di Crescenzo, R.M.; Galdiero, M.R.; Cristinziano, L.; et al. SARS-CoV-2 Spike Protein Activates Human Lung Macrophages. Int J Mol Sci. 2023, 24, 3036. [Google Scholar] [CrossRef]
- Sukumaran, P.; Nascimento Da Conceicao, V.; Sun, Y.; Ahamad, N.; Saraiva, L.R.; Selvaraj, S.; Singh, B.B. Calcium Signaling Regulates Autophagy and Apoptosis. Cells 2021, 10, 2125. [Google Scholar] [CrossRef]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Chandrasekar, A.P.; Maynes, M.; Natesampillai, S.; Shweta, F.N.U.; Badley, A.D.; Cummins, N.W. SARS-CoV-2 spike protein induces monocyte apoptosis and interleukin-8 production. Top Antivir Med 2021, 29, 61. [Google Scholar]
- Schroeder, J.T.; Bieneman, A.P. The S1 Subunit of the SARS-CoV-2 Spike Protein Activates Human Monocytes to Produce Cytokines Linked to COVID-19: Relevance to Galectin-3. Front Immunol. 2022, 13, 831763. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, T.; Alghanem, B.; Shaibah, H.; Mansour, F.A.; Alamri, H.S.; Akiel, M.A.; Alroqi, F.; Boudjelal, M. SARS-CoV-2 Coronavirus Spike Protein-Induced Apoptosis, Inflammatory, and Oxidative Stress Responses in THP-1-Like-Macrophages: Potential Role of Angiotensin-Converting Enzyme Inhibitor (Perindopril). Front Immunol. 2021, 12, 728896. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Jing, X.; Liu, Y.; Wen, R.; Wang, C. Comparison of laboratory parameters in mild vs. severe cases and died vs. survived patients with COVID-19: Systematic review and meta-analysis. J Thorac Dis. 2022, 14, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Utrero-Rico, A.; González-Cuadrado, C.; Chivite-Lacaba, M.; Cabrera-Marante, O.; Laguna-Goya, R.; Almendro-Vazquez, P.; Díaz-Pedroche, C.; Ruiz-Ruigómez, M.; Lalueza, A.; Folgueira, M.D.; et al. Alterations in Circulating Monocytes Predict COVID-19 Severity and Include Chromatin Modifications Still Detectable Six Months after Recovery. Biomedicines 2021, 9, 1253. [Google Scholar] [CrossRef] [PubMed]
- Kubánková, M.; Hohberger, B.; Hoffmanns, J.; Fürst, J.; Herrmann, M.; Guck, J.; Kräter, M. Physical phenotype of blood cells is altered in COVID-19. Biophys J. 2021, 120, 2838–2847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, R.; Lei, L.; Liu, H.; Wang, Y.; Wang, Y.; Qian, H.; Dai, T.; Zhang, T.; Lai, Y.; et al. Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J Leukoc Biol. 2021, 109, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Malinovska, A.; Hernried, B.; Lin, A.; Badaki-Makun, O.; Fenstermacher, K.; Ervin, A.M.; Ehrhardt, S.; Levin, S.; Hinson, J.S. Monocyte Distribution Width as a Diagnostic Marker for Infection: A Systematic Review and Meta-analysis. Chest, 2023; Epub ahead of print. [Google Scholar] [CrossRef]
- Bajić, D.; Matijašević, J.; Andrijević, L.; Zarić, B.; Lalić-Popović, M.; Andrijević, I.; Todorović, N.; Mihajlović, A.; Tapavički, B.; Ostojić, J. Prognostic Role of Monocyte Distribution Width, CRP, Procalcitonin and Lactate as Sepsis Biomarkers in Critically Ill COVID-19 Patients. J Clin Med. 2023, 12, 1197. [Google Scholar] [CrossRef]
- Lorubbio, M.; Tacconi, D.; Iannelli, G.; Feri, M.; Scala, R.; Montemerani, S.; Mandò, M.; Ognibene, A. The role of Monocyte Distribution Width (MDW) in the prognosis and monitoring of COVID-19 patients. Clin Biochem. 2022, 103, 29–31. [Google Scholar] [CrossRef]
- Riva, G.; Castellano, S.; Nasillo, V.; Ottomano, A.M.; Bergonzini, G.; Paolini, A.; Lusenti, B.; Milić, J.; De Biasi, S.; Gibellini, L.; et al. Monocyte Distribution Width (MDW) as novel inflammatory marker with prognostic significance in COVID-19 patients. Sci Rep. 2021, 11, 12716. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Henry, B.M. Pooled analysis of monocyte distribution width in subjects with SARS-CoV-2 infection. Int J Lab Hematol. 2021, 43, O161–O163. [Google Scholar] [CrossRef] [PubMed]
- Khedmati, M.E.; Sharifi, M.J. Platelet phagocytosis by monocytes. Clin Chem Lab Med. 2022, 60, e204–e206. [Google Scholar] [CrossRef] [PubMed]
- Fadlallah, M.M.; Salman, S.M.; Fadlallah, M.M.; Rahal, H. Hemophagocytic Syndrome and COVID-19: A Comprehensive Review. Cureus 2023, 15, e36140. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, W.; Liu, Z.; Kang, Y.; Zhang, X.; Xu, Z.; Gao, Y.; Qi, Y. A comparative study of spike protein of SARS-CoV-2 and its variant Omicron (B.1.1.529) on some immune characteristics. Sci Rep. 2022, 12, 17058. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Geng, B.; Marcon, E.; Pu, S.; Tang, H.; Merluza, J.; Bello, A.; Snider, J.; Lu, P.; Wood, H.; et al. Omicron Spike Protein Is Vulnerable to Reduction. J Mol Biol. 2023; Epub ahead of print. [Google Scholar] [CrossRef]
- Hu, F.H.; Jia, Y.J.; Zhao, D.Y.; Fu, X.L.; Zhang, W.Q.; Tang, W.; Hu, S.Q.; Wu, H.; Ge, M.W.; Du, W.; et al. Clinical outcomes of the severe acute respiratory syndrome coronavirus 2 Omicron and Delta variant: Systematic review and meta-analysis of 33 studies covering 6 037 144 coronavirus disease 2019-positive patients. Clin Microbiol Infect. 2023; Epub ahead of print. [Google Scholar] [CrossRef]
- Esper, F.P.; Adhikari, T.M.; Tu, Z.J.; Cheng, Y.W.; El-Haddad, K.; Farkas, D.H.; Bosler, D.; Rhoads, D.; Procop, G.W.; Ko, J.S.; et al. Alpha to Omicron: Disease Severity and Clinical Outcomes of Major SARS-CoV-2 Variants. J Infect Dis. 2023, 227, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Varea-Jiménez, E.; Aznar Cano, E.; Vega-Piris, L.; Martínez Sánchez, E.V.; Mazagatos, C.; García San Miguel Rodríguez-Alarcón, L.; Casas, I.; Sierra Moros, M.J.; Iglesias-Caballero, M.; Vazquez-Morón, S.; et al. Comparative severity of COVID-19 cases caused by Alpha, Delta or Omicron SARS-CoV-2 variants and its association with vaccination. Enferm Infecc Microbiol Clin (Engl Ed). 2023; Epub ahead of print. [Google Scholar] [CrossRef]
- Korobova, Z.R.; Arsentieva, N.A.; Liubimova, N.E.; Batsunov, O.K.; Dedkov, V.G.; Gladkikh, A.S.; Sharova, A.A.; Adish, Z.; Chernykh, E.I.; Kaschenko, V.A.; et al. Cytokine Profiling in Different SARS-CoV-2 Genetic Variants. Int J Mol Sci. 2022, 23, 14146. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Rodrigues Gomes, L.G.; Ramalho Pinto, C.H.; Andrade, B.S.; Ahmad, S.; Aljabali, A.A.A.; Alzahrani, K.J.; Banjer, H.J.; Hassan, S.S.; et al. SARS-CoV-2 Variants Show a Gradual Declining Pathogenicity and Pro-Inflammatory Cytokine Stimulation, an Increasing Antigenic and Anti-Inflammatory Cytokine Induction, and Rising Structural Protein Instability: A Minimal Number Genome-Based Approach. Inflammation 2023, 46, 297–312. [Google Scholar] [CrossRef]
- Park, C.; Tavakoli-Tabasi, S.; Sharafkhaneh, A.; Seligman, B.J.; Hicken, B.; Amos, C.I.; Chou, A.; Razjouyan, J. Inflammatory Biomarkers Differ among Hospitalized Veterans Infected with Alpha, Delta, and Omicron SARS-CoV-2 Variants. Int J Environ Res Public Health 2023, 20, 2987. [Google Scholar] [CrossRef]
- McCrindle, B.W.; Harahsheh, A.S.; Handoko, R.; Raghuveer, G.; Portman, M.A.; Khoury, M.; Newburger, J.W.; Lee, S.; Jain, S.S.; Khare, M.; et al. SARS-CoV-2 Variants and Multisystem Inflammatory Syndrome in Children. N Engl J Med. 2023, 388, 1624–1626. [Google Scholar] [CrossRef]
| Spike protein variants | Additional mutations |
|---|---|
| Ancestral | D614G |
| Alpha (B.1.1.7) | AAVal16, Pro1213, HV69-70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| Delta (B.1.617.2) | AAVal16, Pro 1213, T19R, G142D, EF156-157del, R158G, L452R, T478K, D614G, P681R, D950N |
| Omicron (B.1.1.529) | AAVal16, Pro 1213, A67V, HV69-70del, T95I, G142D, VYY143-145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| Parameter | Abbreviation | Clinical significance |
|---|---|---|
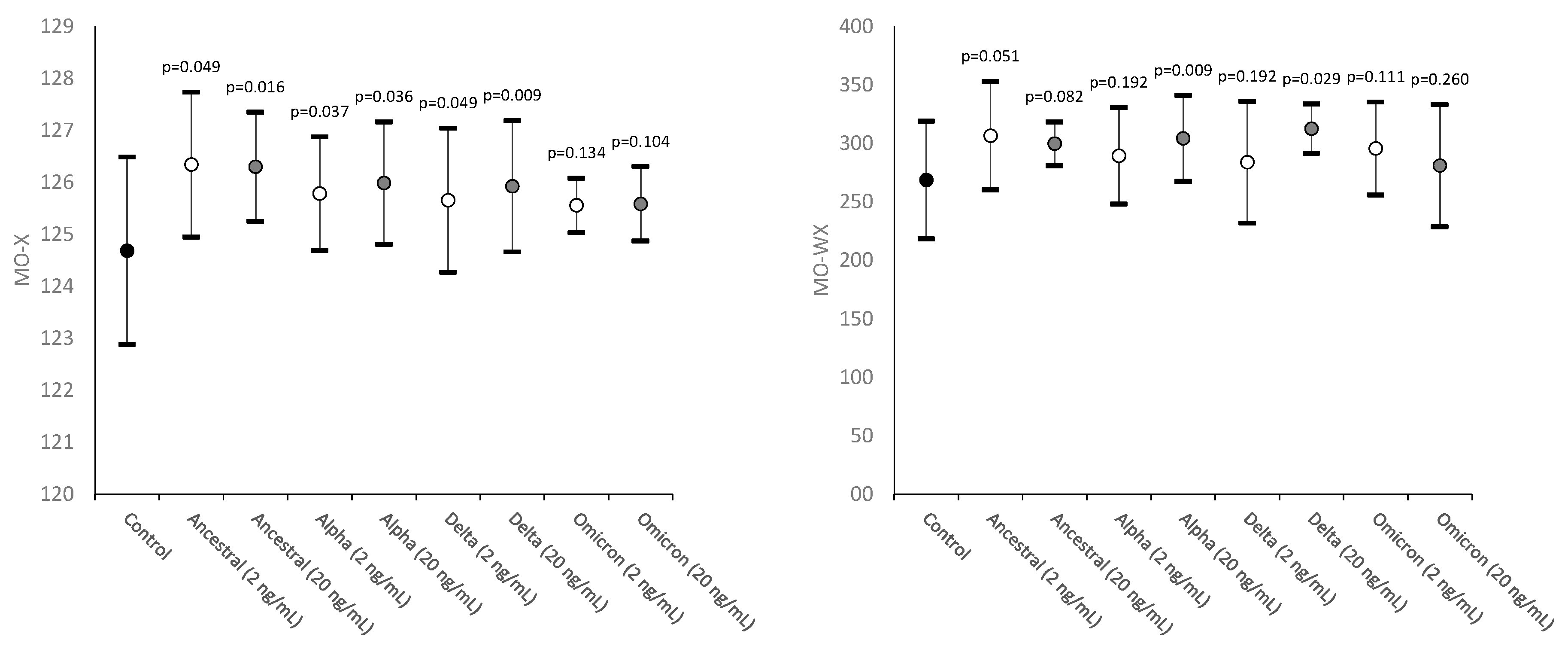
| Monocytes complexity | MO-X | Presence of granules, vacuoles and other cytoplasmic inclusions |
| Width of dispersion of monocytes complexity | MO-WX | Heterogeneity of monocytes complexity |
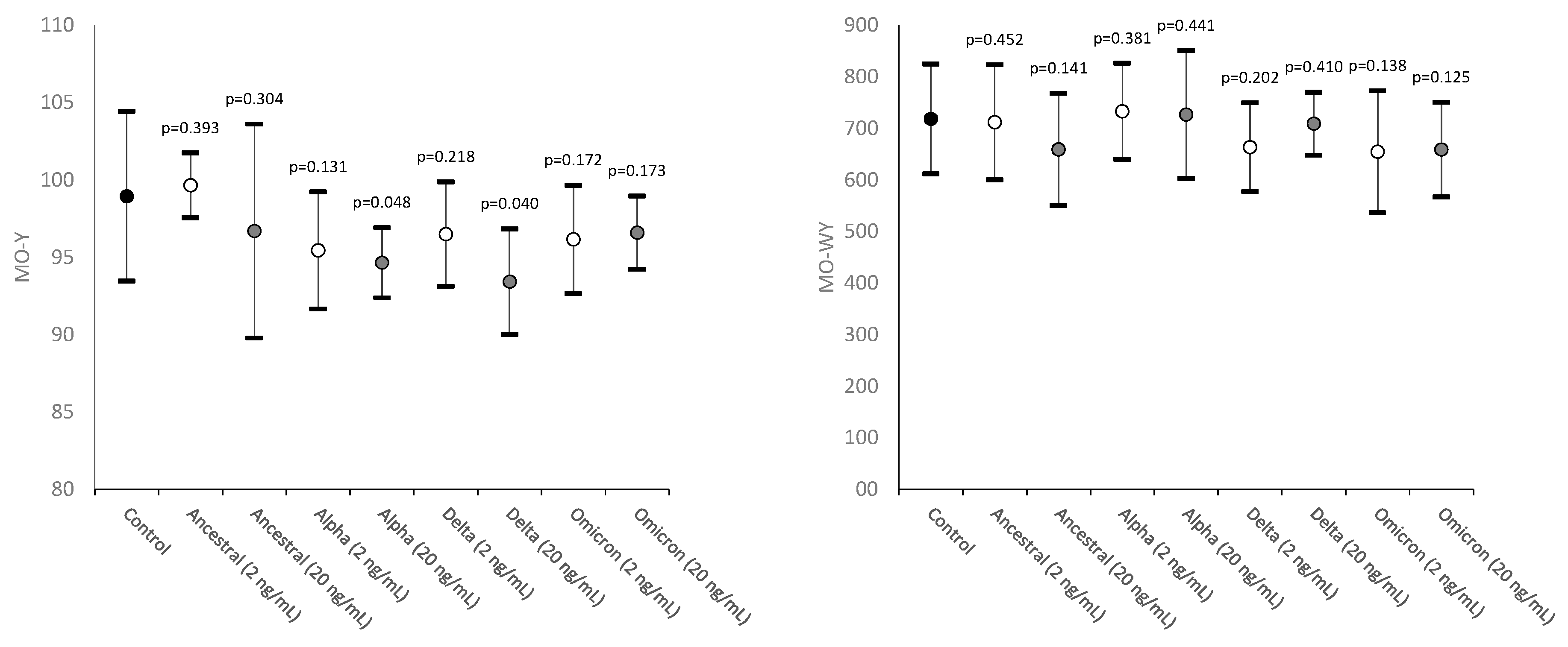
| Monocytes fluorescence intensity | MO-Y | Quantity of cellular DNA and RNA |
| Width of dispersion of monocytes fluorescence intensity | MO-WY | Heterogeneity of monocytes fluorescence intensity |
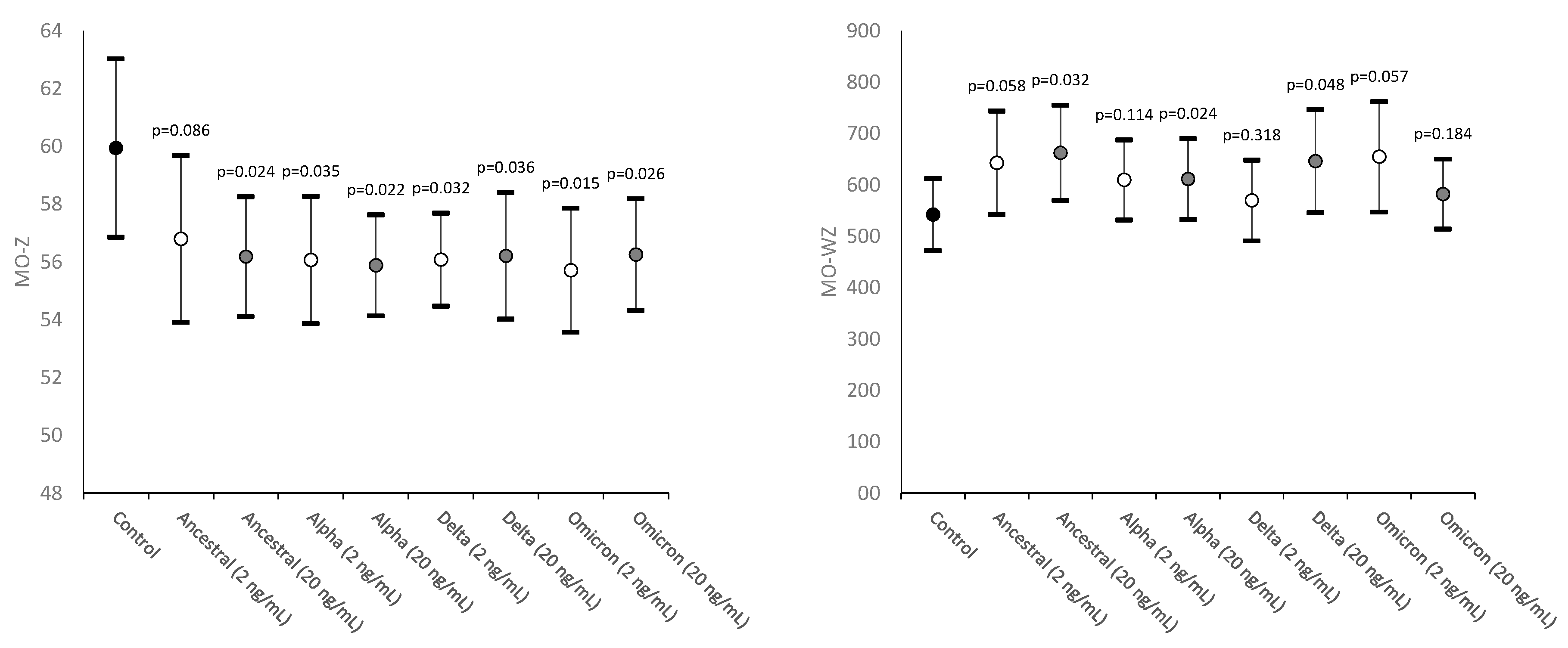
| Monocytes cell size | MO-Z | Abnormal sized cells |
| Width of dispersion of monocytes cell size | MO-WZ | Heterogeneity of monocyte cell size |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





