1. Introduction
Sheep were one of the first species to be domesticated about 8000 to 9000 years ago. The adaptability of this species to different types of weather has allowed their wide geographic distribution. The global population of sheep reached 1.2 billion by the year 2012. Sheep are among the top five most economically important domestic species in the world, and approximately 59 breeds have been registered [
1]. They are easy to manage and maintain, and their pregnancy period is relatively short, which are advantageous for evaluating genetic improvement programs [
2,
3].
Different reproductive biotechnologies focusing sheep include artificial insemination (AI),
in vitro fertilization (IVF), gamete and embryo cryopreservation, and cloning, among others [
4].
In vitro production of cloned embryos involves vertical and horizontal approaches [
5]. The vertical approach includes the generation of monozygotic twins from blastomere separation and embryo bipartition
in vivo or by IVF [
6]. In contrast, the horizontal approach involves somatic cell nuclear transfer (SCNT). In SCNT, an oocyte devoid of its nucleus serves as a cytoplasmic receptor for donated genetic information from a somatic cell [
7]. The first attempts to generate cloned embryos by blastomere separation and embryo bipartition were carried out in sheep, as was the generation of embryos by SCNT [
8]. SCNT is a biotechnology with great potential in the reproduction of sheep with high genetic value [
9], to conserve endangered wild sheep [
10] and to generate transgenic sheep with biomedical purposes [
11].
The scientific and technological advances in the past several decades have been reflected in an increase in scientific information in bibliographic databases for the dissemination of knowledge. This has enhanced the use of bibliometrics [
12]. Bibliometrics is defined as the evaluation of the set of methodological knowledge in published documents through indicators, number of documents published and citations of these documents, according to the region or country of origin, authors, working groups, and research centers, to help evaluate scientific information [
13]. Bibliometric studies have been used to quantify scientific output and to identify groups and areas of excellence, thematic and interdisciplinary emerging disciplines, and thematic collaboration networks [
12]. Governments can use this information to implement policies that benefit the scientific and technological development of their nations [
14].
The evaluation of the different elements of scientific publications can reveal different bibliometric indicators that measure the results of scientific and technological work. The choice of the database to be used in the analysis of scientific information will condition the bibliometric indicators that can be developed [
12].
The aim of this study was to identify regularities of scientific information to provide an overview of scientific research published in mainstream journals on SCNT applied to sheep reproduction. The study used several one-dimensional and multidimensional bibliometric indicators. The data were analyzed using VOSviewer software.
3. Results
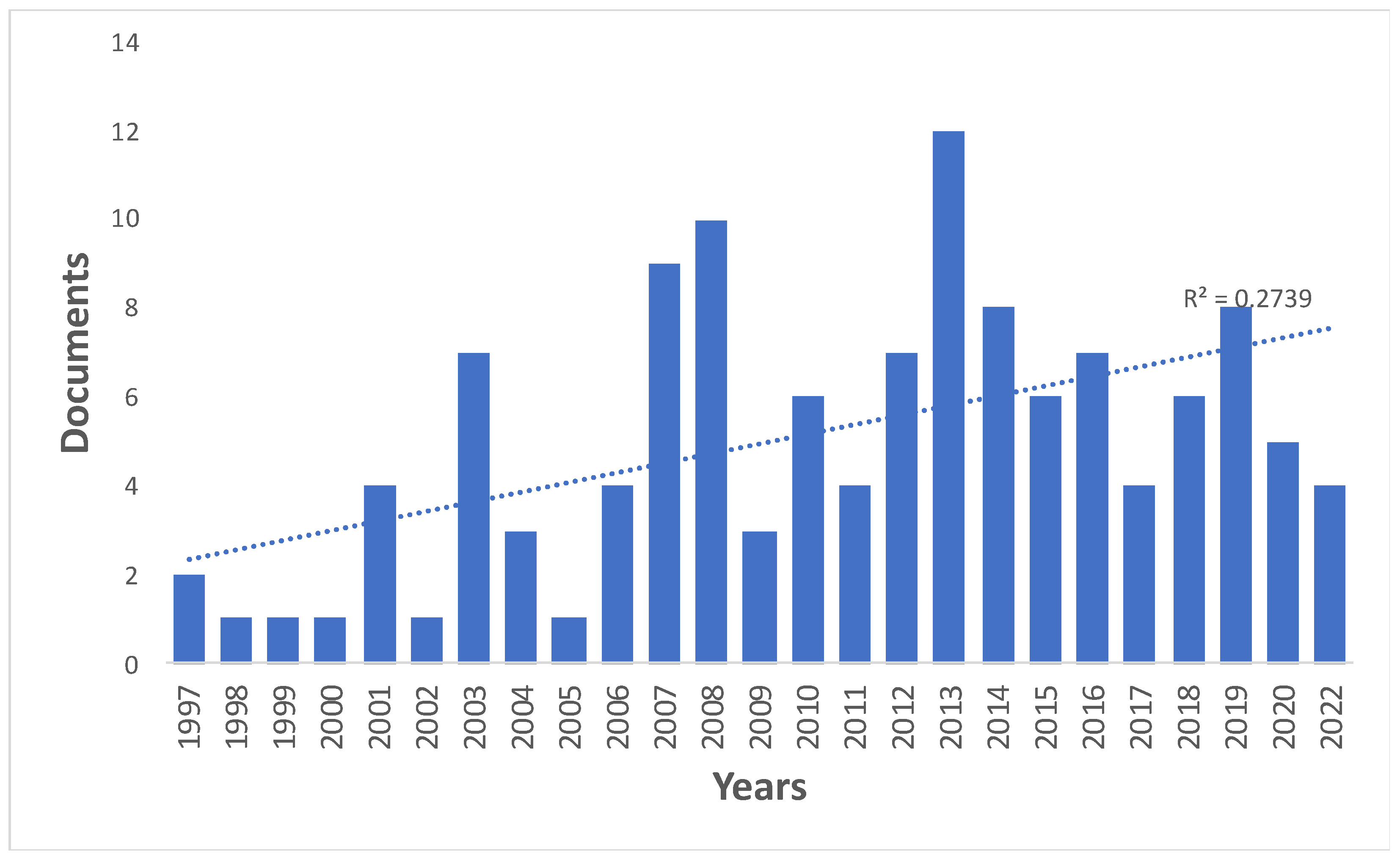
3.1. Growth of literature
The Science Web reference and citation database revealed 107 documents. In addition, 17 documents from the Journal Citation Reports (JCR) that had not been identified in the initial search were included. The total of 124 papers comprised 118 research papers (95.2%), and six review papers (4.8%). The first article on SCNT in sheep titled
Viable offspring derived from fetal and adult mammalian cells was published in 1997 in the journal
Nature [
21]. In subsequent years from 2001, the publication of documents fluctuated from one to 12 every year, with an annual average of five documents. Most documents (n=12) were published in 2013 (
Figure 1).
Figure 1.
Distribution of articles about SCNT in sheep indexed in Web of Science from 1997 to 2022.
Figure 1.
Distribution of articles about SCNT in sheep indexed in Web of Science from 1997 to 2022.
3.2. Most productive author
Keith Henry Stockman Campbell published the most documents about SCNT in sheep. The 15 articles published have been cited 4868 times (H-index of 34) [
22]. Dr. Campbell was affiliated with the University of Nottingham until his death in 2012. One aspect of his research was SCNT in mammals, using sheep as a research model for the generation of cloned or transgenic lambs. The author with the highest scientific productivity is Professor Sir Ian Wilmut, who has published nine articles that have been cited 4992 times (H-index of 69). His research at the University of Edinburgh focuses on cell reprogramming mechanisms and regenerative medicine (
Table 2).
Table 2.
List of leading authors in scientific research of sheep production by SCNT.
Table 2.
List of leading authors in scientific research of sheep production by SCNT.
| Author |
Papers |
Citations |
H-index |
Institution |
Research interests |
| Campbell |
15 |
4868 |
34 |
University of Nottingham |
Cloning mammals, transgenic animals, and stem cells |
| Loi |
13 |
610 |
19 |
University of Teramo |
Developmental biology, reproductive biotechnologies, nuclear reprogramming, and epigenetic modifications |
| Ptak |
11 |
595 |
25 |
Jagiellonian University |
Mechanisms involved in the implantation and placentation of mammals, influence of the environment on the development of organisms |
| Wilmut |
9 |
4992 |
69 |
The University of Edinburgh |
Regenerative medicine and cellular reprogramming mechanisms |
| Hajian |
8 |
139 |
18 |
Royan Institute for Biotechnology |
Reproductive biotechnologies, mammalian cloning, genetics, and molecular biology |
| Hosseini |
8 |
139 |
21 |
Royan Institute for Biotechnology |
Reproductive biotechnologies in wild animals and mammalian cloning |
| Nasr-Esfahani |
8 |
139 |
47 |
Royan Institute for Biotechnology |
Reproductive biotechnologies in ruminants |
| Forouzanfar |
7 |
123 |
16 |
Islamic Azad University |
Reproductive biotechnologies, cloning of mammals and transgenic animals |
| Hou |
7 |
57 |
16 |
Institute of Crops Sciences |
Reproductive biotechnologies and cloning |
| Lee |
7 |
233 |
18 |
Pusan National University |
Cloning of mammals |
| Peura |
7 |
139 |
20 |
Genea Biomedx |
Reproductive biotechnologies |
| Walker |
7 |
284 |
31 |
South Australian Research & Development Institute
|
Reproductive biotechnologies and embryonic development in ruminants |
| Czernik |
6 |
48 |
15 |
University of Teramo |
Assisted reproduction techniques and reproductive biology |
| Guan |
6 |
45 |
13 |
Guangzhou Institute of Energy Conversion |
Reproductive biotechnologies and cloning |
| Moulavi |
6 |
119 |
14 |
Camel Advanced Reproductive Technologies Center |
Reproductive biotechnologies in wild animals and mammalian cloning |
| Choi |
5 |
82 |
13 |
University of Nottingham |
Developmental biology, genome expression, embryonic development, and mammalian cloning |
| Iuso |
5 |
41 |
9 |
National Institute for Biology |
Assisted reproduction techniques and reproductive biology |
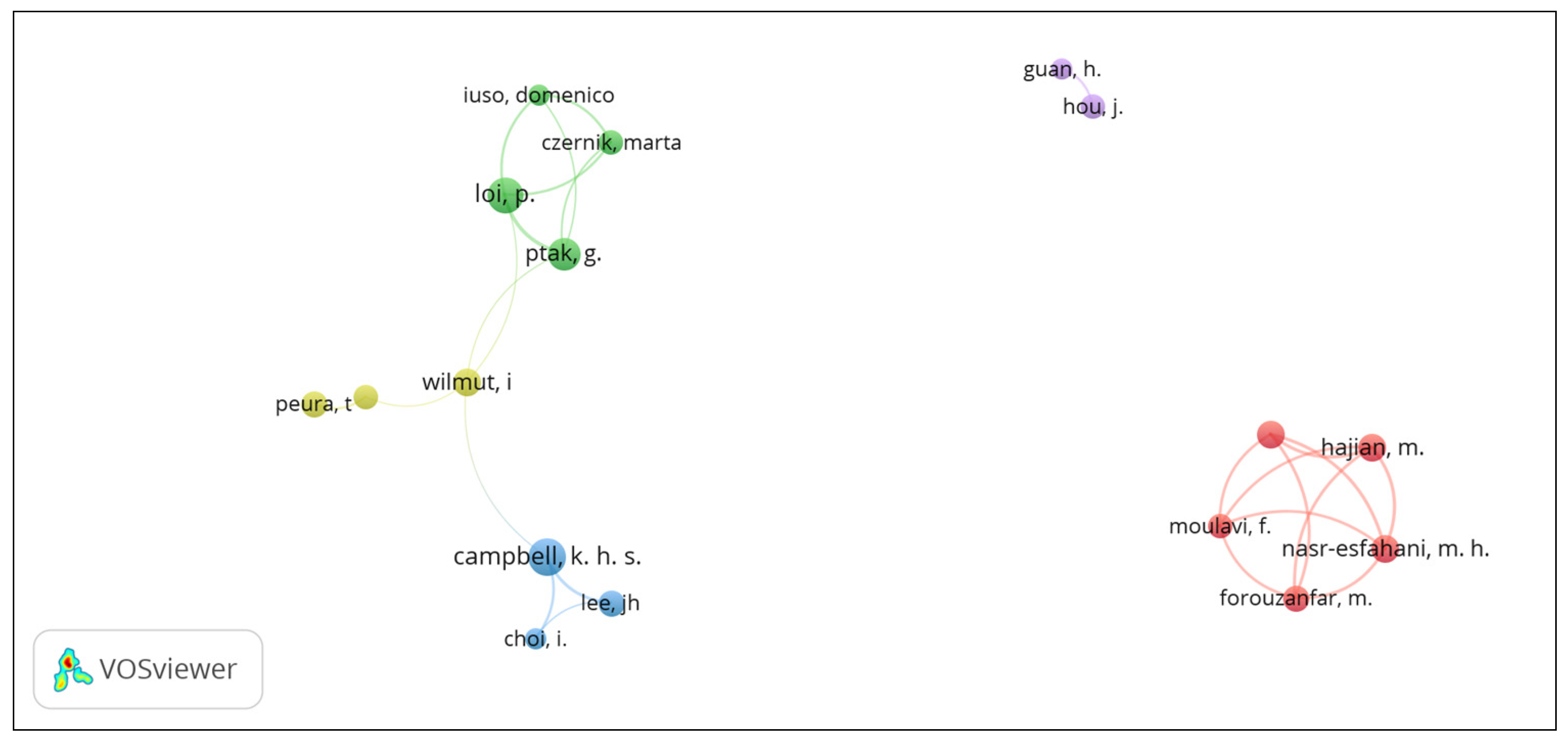
3.3. Cooperation between authors
There are five research groups focused on SCNT in sheep (Figure 2). The group with the largest number of researchers is composed by Samaneh Sadat Hosseini Farahabadi, Mehdi Hajian, Mohammad Hossein Nasr-Esfahani, Forouzafar Mohsen, and Fariba Moulavi. They belong to different institutions in Iran, including the Royan Institute for Biotechnology, the Islamic University of Azad, and the Camels Advanced Reproductive Technologies Center. This research group did not collaborate with others. The second group has four researchers: Pasqualino Loi, Marta Czernik, and Domenico Iuso from the University of Teramo in Italy and Grażyna Ptak from the Jagellonian University in Poland. The third research group includes the aforementioned Dr. Campbell, Joon Hee Lee, and Inchul Choi from the University of Nottingham in England. The fourth group includes Teija Peura of Genea Biomedx (Box Hill, Australia), Simon Walker of the Turretfield Research Centre, (Rosedale, Australia), and the aforementioned Professor Sir Wilmut. Research groups two to four collaborate each other. The final research group was formed by Jian Hou and Hong Guan of the Chinese University of Agriculture, who have no interaction with other research groups.
Figure 2.
Visualization of research groups on SCNT in sheep using VOSviewer software.
Figure 2.
Visualization of research groups on SCNT in sheep using VOSviewer software.
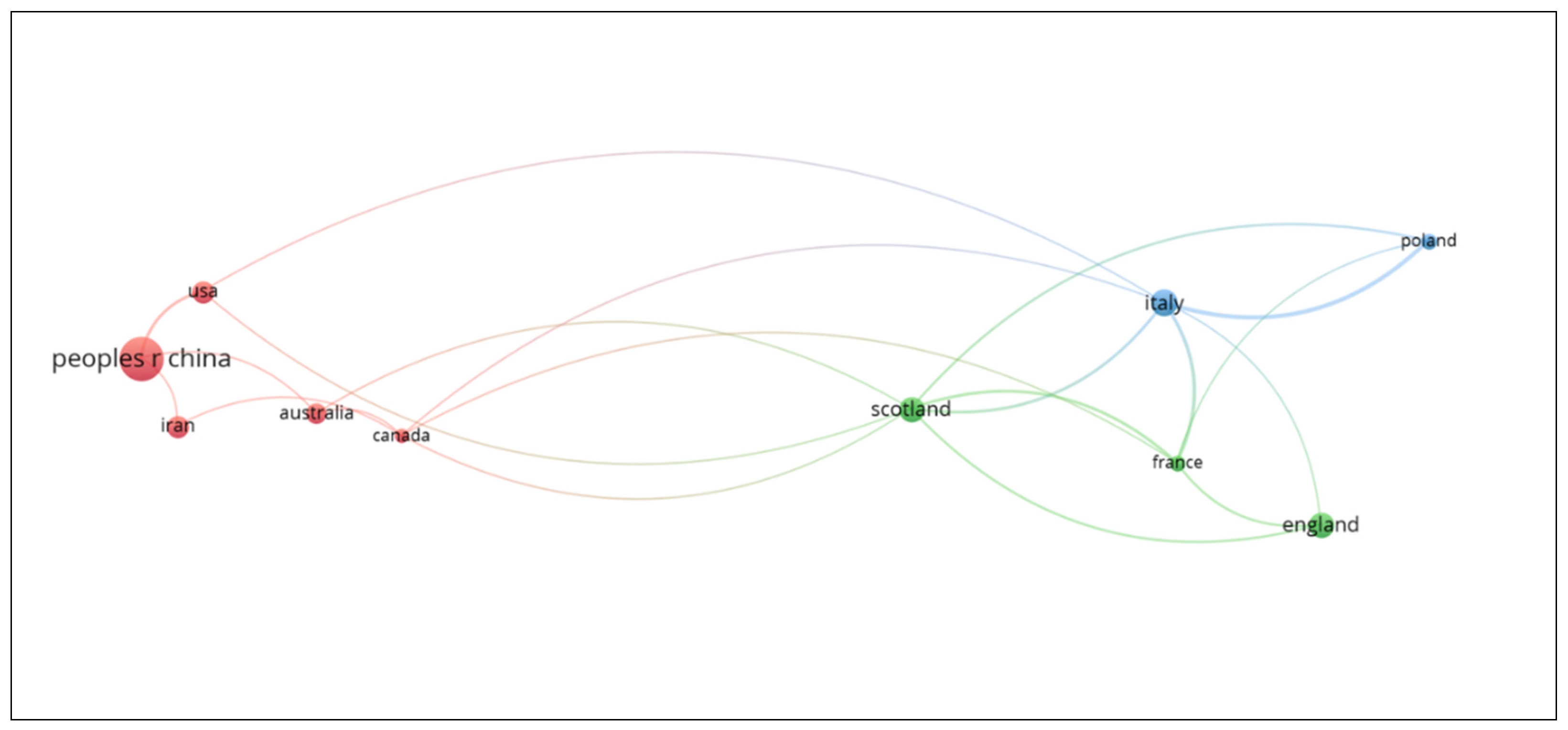
3.4. Publication and cooperation among countries
The countries that have most intensively performed and published research on SCNT involving sheep are shown in Table 3. The collaboration between countries is shown in Figure 3. Scholars from China have collaborated mainly with colleagues from Iran, Australia, Canada and United States. Scholars from England have collaborated with colleagues from Scotland and France. Finally, academics from Italy have collaborated with colleagues from Poland.
Table 3.
Main countries with the most publications concerning SCNT in sheep production.
Table 3.
Main countries with the most publications concerning SCNT in sheep production.
| Country |
Number of publications |
| China |
45 |
| Italy |
16 |
| England |
15 |
| Scotland |
15 |
| United States |
12 |
| Iran |
11 |
| Australia |
10 |
| France |
6 |
| Poland |
6 |
| Canada |
5 |
Figure 3.
Visualization of countries with the greatest contribution of publications about SCNT in sheep and their collaborative relationships using VOSviewer software.
Figure 3.
Visualization of countries with the greatest contribution of publications about SCNT in sheep and their collaborative relationships using VOSviewer software.
3.5. Institutions with the largest contribution of publications
The University of Nottingham has the highest number of published documents concerning SCNT in sheep, with 14 documents. In accordance with the Academic Ranking World of Universities (ARWU) and the World Classification of Universities (QS), it is ranked 101-150th and 114th, respectively. The Roslin Institute of the University of Edinburgh holds the second position, with 13 published documents that have been cited 1844 times, seven of them have been cited more than 100 times each. The University of Edinburgh occupies position 35th of the ARWU and 15 of the QS (Table 4).
Table 4.
Top ten institutions that have contributed to SCNT publications on sheep production.
Table 4.
Top ten institutions that have contributed to SCNT publications on sheep production.
| Institution |
Country |
P |
TC |
AC |
>100 |
>30 |
<30 |
ARWU |
QS |
| 1. University of Nottingham |
United Kingdom |
15 |
510 |
34 |
2 |
3 |
10 |
101-150 |
114 |
| 2. The Roslin Institute |
United Kingdom |
13 |
1844 |
141.8 |
7 |
3 |
3 |
- |
- |
| 3. Università Degli studi di Teramo |
Italy |
12 |
143 |
11.9 |
0 |
3 |
9 |
- |
- |
| 4. China Agricultural University |
China |
11 |
79 |
7.2 |
0 |
0 |
12 |
201-300 |
591-600 |
| 6. Academic Center for Education, Culture & Research (ACECR) |
Iran |
8 |
132 |
16.5 |
0 |
2 |
6 |
- |
- |
| 5. Inner Mongolia Agricultural University |
China |
7 |
37 |
5.2 |
0 |
0 |
7 |
- |
- |
| 7. Shihezi University |
China |
6 |
88 |
14.6 |
0 |
1 |
5 |
- |
- |
| 8. Turretfield Research Centre |
Australia |
5 |
244 |
48.8 |
1 |
0 |
4 |
- |
- |
| 9. The University of Edinburgh |
United Kingdom |
5 |
253 |
50.6 |
1 |
0 |
4 |
35 |
15 |
| 10. Northwest A&F University |
China |
5 |
50 |
10 |
0 |
2 |
3 |
401-500 |
- |
3.6. Journals
The eight main journals whose articles deal with SCNT-related topics in sheep are shown in
Table 5. The information in the table includes supplementary data and Impact Factor (IF) [
23]. It also includes the positions of the journals in accordance with the thematic category, which indicates the quartile according to the JCR. IF is used as an indicator of the relative importance of a journal within a particular area of study and evaluates the frequency with which journal articles are cited during a given period [
24]. The journal
Cellular Reprogramming has published the most publications related to SCNT in sheep (n=11, IF 2.257, JCR position in the fourth quartile of thematic categories). The next journal is
Theriogenology (n=10, IF 2.923, first quartile for the "Veterinary Sciences" category). The journal with the highest number of citations is
Biology of Reproduction (second quartile of the "Reproductive Biology" category), followed by
Theriogenology (first quartile in the "Veterinary Sciences" category).
Table 5.
Main journals with greater contribution of SCNT-related articles in sheep.
Table 5.
Main journals with greater contribution of SCNT-related articles in sheep.
| Journal |
Papers |
Citations |
Impact Factor |
JCR Category |
Rank, Quartile |
| Cellular Reprogramming |
11 |
155 |
2.257 |
Biotechnology and Applied Microbiology |
130/156, Q4 |
| Cell and Tissue Engineering |
27/29, Q4 |
| Genetics and Inheritance |
132/175, Q4 |
| Theriogenology |
10 |
267 |
2.923 |
Reproductive Biology |
20/31, Q3 |
| Veterinary Sciences |
21/145, Q1 |
| Reproduction Fertility and Development |
8 |
88 |
1.973 |
Developmental Biology |
33/39, Q4 |
| Reproductive Biology |
27/31, Q4 |
| Zoology |
59/176, Q2 |
| Molecular Reproduction and Development |
6 |
110 |
2.812 |
Biochemistry and Molecular Biology |
230/297, Q4 |
| Cellular Biology |
159/195, Q4 |
| Developmental Biology |
20/39, Q3 |
| Reproductive Biology |
22/31, Q3 |
| Plos One |
6 |
98 |
3.752 |
Multidisciplinary Sciences |
29/74, Q2 |
| Reproduction in Domestic Animals |
6 |
42 |
1.858 |
Agriculture, Dairy and Animal Science |
33/62, Q3 |
| Reproductive Biology |
29/31, Q4 |
| Veterinary Sciences |
55/145, Q2 |
| Animal Reproduction Sciences |
5 |
171 |
2.22 |
Agriculture, Dairy and Animal Science |
22/62, Q2 |
| Reproductive Biology |
23/31, Q3 |
| Veterinary Sciences |
44/145, Q2 |
| Biology of Reproduction |
5 |
485 |
4.161 |
Reproductive Biology |
10/31, Q2 |
3.7. Most cited documents
The citation structure of documents between 1997 and 2022 included five documents with ≥180 citations (
Table 6). The most cited article refers to the first lamb clone from adult cells obtained from mammary gland, this success demonstrated two facts; first, terminally differentiated cells preserve all their genetic information and it can be restored; second, the SCNT can be applied to superior vertebrates like mammals [
21].
The second most cited article focused on the generation of transgenic lambs from the transfection of fetal fibroblasts with the neomycin resistance marker gene and human coagulation factor IX, for protein-coding in sheep’s milk [
25]. This was followed by an article related to the generation of transgenic lambs. It focused on "gene targeting" in fetal fibroblasts to integrate a therapeutic transgene into the gene locus (COL1A1; collagen type I, alpha 1 chain) in sheep [
26]. The fourth most cited article was related to SCNT as a tool for the recovery of wild species at risk, in which the feasibility of using
Ovis aries oocytes as receptors of
O. orientalis musimon fibroblasts for interspecific SCNT. One of the embryos reached the term of gestation with the birth of an European mouflon [
10].
Table 6.
Most cited SCNT articles concerning the reproduction of sheep.
Table 6.
Most cited SCNT articles concerning the reproduction of sheep.
| Year |
Authors |
Title |
PT |
Source |
Category |
TC |
| 1997 |
Wilmut I. et al. |
Viable offspring derived from fetal and adult mammalian cells [21] |
Article |
Nature |
SCNT |
3476 |
| 1997 |
Schnieke A. E. et al. |
Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts [25] |
Article |
Science |
Transgenic |
650 |
| 2000 |
Mccreath K. J. et al. |
Production of gene-targeted sheep by nuclear transfer from cultured somatic cells [26] |
Article |
Nature |
Transgenic |
450 |
| 2001 |
Loi P. et al. |
Genetic rescue of an endangered mammal by cross-species nuclear transfer using post-mortem somatic cells [10] |
Article |
Nature Biotechnology |
SCNT interspecific |
296 |
| 2004 |
Beaujean N. et al. |
Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer [27] |
Article |
Biology of Reproduction |
DNA methylation in embryos by SCNT |
187 |
| 2001 |
Denning C. et al. |
Deletion of the alpha (1,3) galactosyl transferase (ggta1) gene and the prion protein (prp) gene in sheep [28] |
Article |
Nature Biotechnology |
Transgenic y Xenotransplants |
185 |
| 1999 |
Evans M. J. et al. |
Mitochondrial DNA genotypes in nuclear transfer derived cloned sheep [29] |
Article |
Nature Genetics |
Heteroplasmy in ovine |
169 |
| 2001 |
De Sousa P. A. et al. |
Evaluation of gestational deficiencies in cloned sheep fetuses and placentae [30] |
Article |
Biology of Reproduction |
Placental insufficiency in clone fetuses |
168 |
| 2004 |
Young L. E., Beaujean N. |
DNA methylation in the preimplantation embryo: the differing stories of the mouse and sheep [31] |
Review Article |
Animal Reproduction Sciences |
Methylation process in clone embryos |
121 |
| 2003 |
Young Le. et al. |
Conservation of igf2-h19 and igf2r imprinting in sheep: effects of somatic cell nuclear transfer [32] |
Article |
Mechanisms of development |
Genomic imprinting in sheep |
94 |
| 2006 |
Lee J. H., Campbell K. H. S. |
Effects of enucleation and caffeine on maturation promoting factor (mpf) and mitogen activated protein kinase (mapk) activities in ovine oocytes used as recipient cytoplasts for nuclear transfer [33] |
Article |
Biology of Reproduction |
Nuclear Reprogramming |
69 |
| 2018 |
Fan Z. et al. |
A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene [34] |
Article |
JCI Insight |
Genome Editing |
67 |
| 2007 |
Lagutina I. et al. |
Comparative aspects of somatic cell nuclear transfer with conventional and zona-free method in cattle, horse, pig and sheep [35] |
Article |
Theriogenology |
Modification to conventional SCNT technique |
66 |
| 2006 |
Loi P. et al. |
Placental abnormalities associated with post-natal mortality in sheep somatic cell clones [2] |
Article |
Theriogenology |
Placental abnormalities |
57 |
| 2008 |
Palmieri C. et al. |
A review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep and mice [36] |
Review Article |
Veterinary Pathology |
Placental abnormalities |
56 |
| 2007 |
Bowles E. J. et al. |
Contrasting effects of in vitro fertilization and nuclear transfer on the expression of mtDNA replication factors [37] |
Article |
Genetics |
Mitochondrial DNA replication factors |
47 |
| 2006 |
Alexander B. et al. |
The effects of 6-dimethylaminopurine (6-DMAP) and cycloheximide (CHX) on the development and chromosomal complement of sheep parthenogenetic and nuclear transfer embryos [38] |
Article |
Molecular Reproduction and Development |
Activation of cario-cytoplasmic complexes |
46 |
Five documents were cited 179 to 100 times. For example, Beaujean et al. [
27] evaluated the dynamics of somatic nucleus methylation after SCNT at different stages of early embryonic development, showing that the demethylation process failed in the trophectoderm cells of blastocysts.
Six other documents were cited 99 to 50 times. One study [
32] evaluated the imprinting status of IGF2-H19 (insulin-like growth factor 2 and H19 gene) and IGF2R (insulin-like growth factor 2 receptor) in lambs generated by SCNT. The authors observed that one lamb displayed deregulation in the imprint of the second loci intron of the IGF2R gene.
Three documents were cited 49 to 40 times. One study [
37] evaluated the expression of mitochondrial DNA replication factors encoded by the nucleus and expressed for the first time in the later stages of early embryonic development.
The nine documents with 39 to 30 citations include Peura and Vajta [
39] who described a new SCNT method for cattle and sheep (handmade cloning), characterized by using pelucide-zone-free oocytes and the absence of micromanipulators to enucleate them.
Seven documents were cited 29 to 20 times. They included Zhang et al. [
11] who described generating transgenic lambs with high levels of omega-3 fatty acids using handmade cloning. The authors concluded that handmade cloning efficiency was like that of the conventional technique for the generation of transgenic animals.
Twenty-six documents were cited 19 and 10 times. Among them, Wen et al. [
40] tested two inhibitors of histone deacetylases, trichostatin A, and scriptaid. The latter improved the epigenetic status of ovine embryos via SCNT.
Finally, 52 documents were cited ≤9 times. Choi et al. [
41] combined calcium ionophore, strontium chloride and cytochalasin B to activate cario-cytoplast complexes, improving embryo quality. Among these documents, 10 had no record of citations. The most recent document was by McLean et al. [
42] who studied the effect of embryo aggregation during the vitrification process of cloned sheep embryos. They observed that the aggregated embryos had an
in vivo survival rate similar to that of the group that was not vitrified. The oldest most cited publication is that of Wilmut et al. [
21].
Table 6 includes the articles with the highest number of citations between 1997-2022.
3.8. Identification of research topics
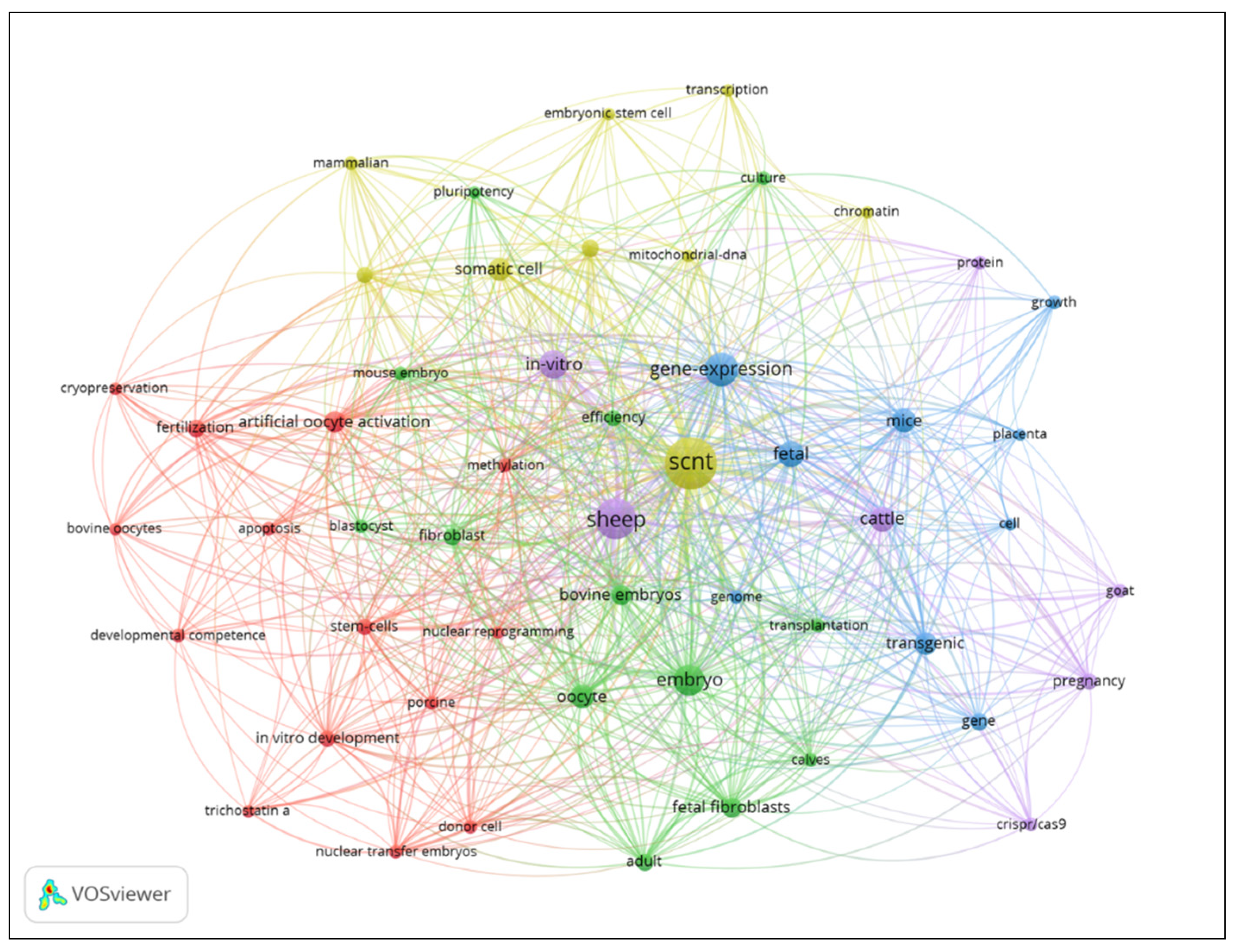
The results of the co-occurrence analysis revealed 553 words or terms that were used most frequently in scientific papers. Only those with more than five co-occurrences were considered. The resulting 52 terms were organized into five clusters. The 14 words or terms most frequently used in the publications are listed in Table 7. This analysis clarified the main topics of interest in the study area.
Table 7.
Main words used most frequently in publications concerning SCNT in sheep.
Table 7.
Main words used most frequently in publications concerning SCNT in sheep.
| Keywords |
Frequency |
Keywords |
Frequency |
| SCNT |
90 |
Mice |
19 |
| Sheep |
56 |
Oocyte |
18 |
| Gene Expression |
36 |
Somatic Cell |
16 |
| Embryo |
32 |
Transgenic |
16 |
| In Vitro |
28 |
Oocyte Activation |
15 |
| Fetal |
22 |
Fetal Fibroblasts |
14 |
| Cattle |
20 |
Bovine Embryo |
13 |
Figure 4 displays the five word-clusters (a cluster is a group of words related to each other) and the relationship of the clusters. Each cluster is represented by one color, cluster one has 14 words (red), cluster two includes 13 words (bright green), cluster three involves 9 words (blue), cluster four contains 9 words (light green) and cluster five has five words (purple).
Figure 4.
Network map of themes of SCNT in sheep, grouped in clusters.
Figure 4.
Network map of themes of SCNT in sheep, grouped in clusters.
4. Discussion
In the present study, 124 documents on the reproduction of sheep by SCNT were counted in the WOS database, this quantity of documents is similar to reported in a review carried out for the 25th anniversary of cloning by SCNT. This review published a survey of all published documents (1997-2020) of SCNT classified by species, among which mice, cattle, pigs, goats, and sheep are the species for which more than 100 publications are registered [
43].
Since the publication of the study on the birth of the Dolly sheep (1997), the number of documents published per year has fluctuated, with an average of 5 documents per year. However, a more detailed analysis, shows that during the first (1997-2006) and second (2007-2016) decade, the number of publications was 25 and 72 documents, respectively; while in the last period (2017-2022) 27 documents have been published, indicating a general growth in research on the reproduction of sheep by cloning.
China, Italy, England, and Scotland are the countries that have generated more than 50% of the scientific production on the reproduction of sheep by SCNT. China is the world’s leading producer of sheep, while Australia, Iran, and the United Kingdom (which includes England and Scotland) are among the ten countries with the highest number of sheep worldwide according to FAO data for 2020 [
44]. This could partly explain the interest of these countries in research on the reproduction of sheep by cloning.
The universities that have shown interest in the reproduction of sheep by cloning, are the University of Nottingham (England), the Roslin Institute of the University of Edinburgh (Scotland), the University of Teramo (Italy), the China Agricultural University (China), which are the affiliation from the most cited authors in this area of research.
The principal journals that publish topics related to sheep reproduction by SCNT are Cellular Reprogramming; Theriogenology, Reproduction, Fertility and Development, Molecular Reproduction and Development, Plos One, and Reproduction in Domestic Animals; these sources published 37.9% of all publications. The most frequent JCR categories in these journals are "reproductive biology", "veterinary sciences", "developmental biology" and "agricultural dairy animal sciences", so these journals are the most suitable for consulting or publishing on this area of research.
The most cited article is the one that reports the birth of Dolly [
21], obtained from a somatic cell from the mammary gland of an adult sheep, an unusual fact that had not been achieved in an upper mammal and that was a watershed for science, for this reason, this article is still widely cited today. The other two most cited articles address issues about the generation of transgenic lambs for the production of human proteins for therapeutic purposes, which means that cell lines expressing a specific gene in the SCNT can be used, this ensures that a lamb is obtained with the desired modification [25, 26].
The words “oocyte” and “somatic cell” were frequent. Both cell types were used for SCNT. Oocytes have been treated with caffeine, which increases the activity of the promoter factor of maturation and of mitogen activated protein kinases that are important for the nuclear reprogramming process during SCNT embryo development [33, 45, 46]. Moreover, the words “embryo”, “gene expression” and “in vitro” were used in connection with each other. Examples include studies that have evaluated the effect of different chemical agents on the state of DNA methylation [47, 48] and inhibitors of histone acetyltransferases [40, 49] in the development of in vitro cloned embryos, especially at the blastocyst stage.
A detailed analysis of
Figure 4 shows that cluster one in red has 14 words related to the competence of
in vitro cloned embryos. Khan et al. [
50] compared the efficiency of conventional SCNT and handmade cloning for the generation of cloned sheep embryos. Better rates of efficiency of enucleation and fusion were obtained with handmade cloning, as well as a higher percentage of segmented and blastocyst stage embryos.
Cluster two in bright green consists of 13 words referring to the development efficiency of cloned embryos at the blastocyst stage using different types of cells as donor of genetic information, most commonly
cumulus cells and fibroblasts. The cells were treated with egg extracts [
51], zebularine [
47], chaetocin [
52] and histone demethylase enzymes [
53] to promote the nuclear reprogramming of somatic cells and improve the epigenetic status of ovine cloned embryos.
Cluster three in blue contains 9 words focusing on the fetal development of cloned sheep and their regulation of gene expression. Ni et al. [
54] evaluated the pregnancies of transgenic lambs produced by SCNT. They observed that the fetal weight, total placenta weight, and mean placentomes weight were greater in pregnancies with live-born lambs but did not survive compared to pregnancies with live-born lambs that survive. Further deregulation was found in miR-21 and miR-16 in the placenta of non-surviving lambs, causing aberrant expression in their targets.
Cluster four in light green consists of 9 words referring to the epigenetic status of cloned sheep embryos. Morphological evaluation is routinely used as the main parameter of embryo quality. However, due to the limited information provided by its unique evaluation, other parameters have focused on evaluating epigenetics [27, 55] and the genetic status of sheep embryos generated by SCNT [9, 52].
Finally, cluster five in purple includes 5 words related to generation of transgenic sheep mediated cloning. Zhang et al. [
56] investigated the effect of suppressing the myostatin (MSTN) gene expression in sheep skeletal muscle satellite cell using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)) technology, to generate lamb clones with better muscle conformation, it has been reported that MSTN gene is responsible for regulating the growth of muscle cells.
It has been 26 years since the first sheep was cloned by SCNT, and since then it has been applied to different domestic and wild species, in some cases with viable newborns. However, SCNT is an inefficient biotechnology, for example, for newborns lambs, have been reported an efficiency range from 5.7 to 15% per transferred blastocyst and from 7.1% to 19.5% per segmented embryo [
57]. Although sheep are easy to handle and have a relatively short gestation period compared with that of the other species of zootechnical interest [
58], their small value and limited potential, do not make them attractive for agriculture use like other livestock species [
59]. If we also take into account that the infrastructure used for SCNT is basically the same for all species, nowadays the SCNT applied to sheep reproduction is not profitable. Therefore, the present bibliometric study shows the areas of research in which cloning research in sheep should be directed, which will help those regions where sheep are an important economic and food source.
5. Conclusions
Bibliometric studies on SCNT in sheep had not been conducted prior to now. This study collected bibliographic data from 124 documents relating to SCNT in sheep. This amount of information in sheep is smaller than that for other species of zootechnical interest, such as cattle and pigs, although sheep were the first large mammal to be successfully cloned. Since 2001, the number of SCNT-related documents published concerning ovine reproduction increased and has fluctuated in ensuing years. The authors that have generated more knowledge in this area are (in alphabetical order) Campbell, Hajian, Hosseini, Loi, Nasr-Esfahani, Ptak, and Wilmut. Five research groups were identified, three of which mutually collaborate. The countries with the largest number of publications were China, Italy and England. The largest collaboration network among countries comprises China, Iran, Australia, Canada and United States. The institutions with the highest productivity of SCNT in sheep are the University of Nottingham and the Roslin Institute at the University of Edinburgh. These two institutions are among the top 150 universities in the world. The principal journals where topics about SCNT in sheep are published have an IF ranging from 1.9 to 4.1, whose quartile position is most often in third and fourth place in the JCR thematic categories. These journals are the most suitable for publishing scientific advances in this area. Articles that have been cited more often address topics related to the generation of transgenic animals, recovery of wild species and xenotransplants. Five main themes were identified in sheep reproduction by SCNT. These themes focused on the competence of in vitro clone embryos, cells used as karyoplasts and their efficiency on embryo development, epigenetic status of clone embryos and their impact on post-implantation development, and generation of transgenic sheep with biomedical and genetic improvement purposes. Concerning SCNT in sheep, these topics are the most relevant, and future studies will have to focus on solutions to the current challenges in this field of study.