Submitted:
14 April 2023
Posted:
17 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. SNP Analysis and Validation
2.3. Statistical Analysis:
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J Clin 2022, 72, 7–33. [Google Scholar] [CrossRef]
- McHugh, J.; Saunders, E. J.; Dadaev, T.; McGrowder, E.; Bancroft, E.; Kote-Jarai, Z.; Eeles, R. Prostate cancer risk in men of differing genetic ancestry and approaches to disease screening and management in these groups. British journal of cancer 2022, 126, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Petrovics, G.; Srivastava, S. A Rich Array of Prostate Cancer Molecular Biomarkers: Opportunities and Challenges. Int J Mol Sci 2019, 20, 1813. [Google Scholar] [CrossRef] [PubMed]
- Conti, D.V.; Darst, B.F.; Moss, L.C.; Saunders, E.J.; Sheng, X.; Chou, A.; Schumacher, F.R.; Olama, A.A.A.; Benlloch, S.; Dadaev, T. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021, 53, 65–75. [Google Scholar] [CrossRef]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 10. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.; Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009, 9, 13. [Google Scholar] [CrossRef]
- Li, F.P.; Fraumeni, J.F.; Mulvihill, J.J.; Blattner, W.A.; Dreyfus, M.G.; Tucker, M.A.; Miller, R.W. A cancer family syndrome in twenty-four kindreds. Cancer Res 1988, 48, 5358–5362. [Google Scholar]
- Malkin, D.; Li, F.P.; Strong, L.C.; Fraumeni, J.F., Jr.; Nelson, C.E.; Kim, D.H.; Kassel, J.; Gryka, M.A.; Bischoff, F.Z.; Tainsky, M.A.; Friend, S.H. Germ line p53 mutation in a familial syndrome of breast cancer. Science 1990, 250, 1233–1238. [Google Scholar] [CrossRef]
- Srivastava, S.; Zou, Z.; Pirollo, K.; Blattner, W.; Chang, E.H. Germ-line transmission of a mutated p53 gene in a cancer prone family with Li-Fraumeni syndrome. Nature 1990, 348, 747–749. [Google Scholar] [CrossRef]
- Zawacka-Pankau, J.E. The Role of p53 Family in Cancer. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Whibley, C.; Pharoah, P.D.; Hollstein, M. p53 polymorphisms: cancer implications. Nat Rev Cancer 2009, 9, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Barnoud, T.; Parris, J.L.D.; Murphy, M.E. Common genetic variants in the TP53 pathway and their impact on cancer. J Mol Cell Biol 2019, 11, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Eiholzer, R.A.; Mehta, S.; Kazantseva, M.; Drummond, C.J.; McKinney, C.; Young, K.; Slater, D.; Morten, B.C.; Avery-Kiejda, K.A.; Lasham, A. et al. Intronic TP53 Polymorphisms Are Associated with Increased Delta133TP53 Transcript, Immune Infiltration and Cancer Risk. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Freedland, S.J.; Isaacs, W.B. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate 2005, 62, 243–252. [Google Scholar] [CrossRef]
- Riviere, P.; Luterstein, E.; Kumar, A.; Vitzthum, L.K.; Deka, R.; Sarkar, R.R.; Bryant, A.K.; Bruggeman, A.; Einck, J.P.; Murphy, J.D. et al. Survival of African American and non-Hispanic white men with prostate cancer in an equal-access health care system. Cancer 2020, 126, 1683–1690. [Google Scholar] [CrossRef]
- Yamoah, K.; Lee, K.M.; Awasthi, S.; Alba, P.R.; Perez, C.; Anglin-Foote, T.R.; Robison, B.; Gao, A.; DuVall, S.L.; Katsoulakis, E. et al. Racial and Ethnic Disparities in Prostate Cancer Outcomes in the Veterans Affairs Health Care System. JAMA Netw Open 2022, 5. Erratum in: JAMA Netw Open 2022, 5. [Google Scholar] [CrossRef]
- Loh, P.R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.A.; Reshef, Y.K.; Finucane, H.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef]
- Taliun, D.; Harris, D.N.; Kessler, M.D.; Carlson, J.; Szpiech, Z.A.; Torres, R.; Taliun, S.A.G.; Corvelo, A.; Gogarten, S.M.; Kang, H.M.; et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 2021, 590, 290–299. [Google Scholar] [CrossRef]
- Dumont, P.; Leu, J.I.; Della Pietra, A.C.; George, D.L.; Murphy, M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003, 33, 357–365. [Google Scholar] [CrossRef]
- Hrstka, R.; Coates, P.J.; Vojtesek, B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med. 2009, 13, 440–453. [Google Scholar] [CrossRef]
- Hu, W.; Feng, Z.; Atwal, G.S.; Levine, A.J. p53: a new player in reproduction. Cell Cycle 2008, 7, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Levine, A.J. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene 2007, 26, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Khalil, A.; Rashid, H. Evaluation of the p53 Arg72Pro polymorphism and its association with cancer risk: A HuGE review and meta-analysis. Genetics Research 2015, 97, e7. [Google Scholar] [CrossRef]
- Huang, S.P.; Huang, C.Y.; Wang, J.S.; Liu, C.C.; Pu, Y.S.; Yu, H.J.; Yu, C.C.; Wu, T.T.; Huang, C.H.; Wu, W.J.; et al. Prognostic significance of p53 and X-ray repair cross-complementing group 1 polymorphisms on prostate-specific antigen recurrence in prostate cancer post radical prostatectomy. Clin Cancer Res 2007, 13, 6632–6638. [Google Scholar] [CrossRef]
- Quiñones, L.A.; Irarrázabal, C.E.; Rojas, C.R.; Orellana, C.E.; Acevedo, C.; Huidobro, C.; Varela, N.E.; Cáceres, D.D. Joint effect among p53, CYP1A1, GSTM1 polymorphism combinations and smoking on prostate cancer risk: an exploratory genotype-environment interaction study. Asian J Androl 2006, 8, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Matsui, H.; Ohtake, N.; Nakata, S.; Takei, T.; Nakazato, H.; Okugi, H.; Koike, H.; Ono, Y.; Ito, K.; et al. A p53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci 2003, 10, 430–435. [Google Scholar] [CrossRef]
- Huang, S.P.; Wu, W.J.; Chang, W.S.; Wu, M.T.; Chen, Y.Y.; Chen, Y.J.; Yu, C.C.; Wu, T.T.; Lee, Y.H.; Huang, J.K.; et al. p53 Codon 72 and p21 codon 31 polymorphisms in prostate cancer. Cancer Epidemiol Biomarkers Prev 2004, 13, 2217–2224. [Google Scholar] [CrossRef]
- Henner, W.D.; Evans, A.J.; Hough, K.M.; Harris, E.L.; Lowe, B.A.; Beer, T.M. Association of codon 72 polymorphism of p53 with lower prostate cancer risk. Prostate 2001, 49, 263–266. [Google Scholar] [CrossRef]
- Cintra, H.S.; Pinezi, J.C.; Machado, G.D.; de Carvalho, G.M.; Carvalho, A.T.; dos Santos, T.E.; Marciano, R.D.; Soares Rde, B. Investigation of genetic polymorphisms related to the outcome of radiotherapy for prostate cancer patients. Dis Markers 2013, 35, 701–710. [Google Scholar] [CrossRef]
- Han, J.Y.; Lee, G.K.; Jang, D.H.; Lee, S.Y.; Lee, J.S. Association of p53 codon 72 polymorphism and MDM2 SNP309 with clinical outcome of advanced nonsmall cell lung cancer. Cancer 2008, 113, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Sohn, S.K.; Chae, Y.S.; Song, H.S.; Kwon, K.Y.; Do, Y.R.; Kim, M.K.; Lee, K.H.; Hyun, M.S.; Lee, W.S.; et al. TP53 codon 72 polymorphism associated with prognosis in patients with advanced gastric cancer treated with paclitaxel and cisplatin. Cancer Chemother Pharmacol 2009, 64, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Zhang, Z.; Nishio, M.; Hamaguchi, M.; Kondo, N.; Iwase, H.; Iwata, H.; Takahashi, S.; Yamashita, H.; Fujii, Y. Association of TP53codon 72 polymorphism and the outcome of adjuvant therapy in breast cancer patients. Breast Cancer Res 2007, 9, R34. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Li, Q.; Chen, Y.; Ravindranath, L.; Young, D.; Ali, A.; Sesterhenn, I.A.; Rosner, I.L.; Cullen, J.; Srivastava, S. Association of germline genetic variants with TMPRSS2-ERG fusion status in prostate cancer. Oncotarget 2020, 11, 1321–1333. [Google Scholar] [CrossRef]
- Teroerde, M.; Nientiedt, C.; Duensing, A.; Hohenfellner, M.; Stenzinger, A.; Duensing, S. Revisiting the Role of p53 in Prostate Cancer. In Prostate Cancer; Bott, S.R.J., Ng, K.L., Eds.; Exon Publications: Brisbane, Australia, 2021; Volume 1, pp. 116–117. ISBN 978-0-6450017-5-4. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Gesztes, W.; Schafer, C.; Young, D.; Fox, J.; Jiang, J.; Chen, Y.; Kuo, H.C.; Mwamukonda, K.B.; Dobi, A.; Burke, A.P.; et al. Focal p53 protein expression and lymphovascular invasion in primary prostate tumors predict metastatic progression. Sci Rep 2022, 12, 5404. [Google Scholar] [CrossRef]
- Levine, A.J. Spontaneous and inherited TP53 genetic alterations. Oncogene 2021, 40, 5975–5983. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Cheng, H.H.; Powers, J.; Gulati, R.; Ledet, E.M.; Morrison, C.; Le, A.; Hausler, R.; Stopfer, J.; Hyman, S. et al. Inherited TP53 Variants and Risk of Prostate Cancer. Eur Urol 2022, 81, 243–250. [Google Scholar] [CrossRef]
- Nicolosi, P.; Ledet, E.; Yang, S.; Michalski, S.; Freschi, B.; O'Leary, E.; Esplin, E.D.; Nussbaum, R.L.; Sartor, O. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol 2019, 5, 523–528. [Google Scholar] [CrossRef]
- Doffe, F.; Carbonnier, V.; Tissier, M.; Leroy, B.; Martins, I.; Mattsson, J.S.M.; Micke, P.; Pavlova, S.; Pospisilova, S.; Smardova, J. et al. Identification and functional characterization of new missense SNPs in the coding region of the TP53 gene. Cell Death Differ 2021, 28, 1477–1492. [Google Scholar] [CrossRef]
- Schaeffer, E.M.; Srinivas, S.; Adra, N.; An, Y.; Barocas, D.; Bitting, R.; Bryce, A.; Chapin, B.; Cheng, H.H.; D'Amico, A.V.; et al. NCCN Guidelines® Insights: Prostate Cancer, Version 1.2023. Journal of the National Comprehensive Cancer Network : JNCCN 2022, 20, 1288–1298. [Google Scholar] [PubMed]
- US Preventive Services Task Force; Grossman, D.C.; Curry, S.J.; Owens, D.K.; Bibbins-Domingo, K.; Caughey, A.B.; Davidson, K.W.; Doubeni, C.A.; Ebell, M.; Epling, J.W.; et al. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018, 319, 1901–1913. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.B.; Albertsen, P.C.; Barry, M.J.; Etzioni, R.; Freedland, S.J.; Greene, K.L.; Holmberg, L.; Kantoff, P.; Konety, B.R.; Murad, M.H.; et al. Early detection of prostate cancer: AUA Guideline. The Journal of urology 2013, 190, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Gnanapradeepan, K.; Leu, J.I.; Basu, S.; Barnoud, T.; Good, M.; Lee, J.V.; Quinn, W.J.; Kung, C.P.; Ahima, R.; Baur, J.A.; et al. Increased mTOR activity and metabolic efficiency in mouse and human cells containing the African-centric tumor-predisposing p53 variant Pro47Ser. Elife 2020, 9, 55994. [Google Scholar] [CrossRef]
- Tutton, S.; Deng, Z.; Gulve, N.; Vladimirova, O.; Beishline, K.; Wiedmer, A.; Murphy, M.; Lieberman, P.M. Elevated telomere dysfunction in cells containing the African-centric Pro47Ser cancer-risk variant of TP53. Oncotarget 2019, 10, 3581–3591. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.R.; Yoshioka, F.K.; Silva, R.L.; Clara, C.A.; Santos, M.J.; Almeida, J.R.; Burbano, R.R.; Rey, J.A.; Casartelli, C. Prognostic value of TP53 Pro47Ser and Arg72Pro single nucleotide polymorphisms and the susceptibility to gliomas in individuals from Southeast Brazil. Genet Mol Res 2008, 7, 207–216. [Google Scholar] [CrossRef]
- Murphy, M.E.; Liu, S.; Yao, S.; Huo, D.; Liu, Q.; Dolfi, SC.; Hirshfield, K.M.; Hong, C.C.; Hu, Q.; Olshan, A.F.; et al. A functionally significant SNP in TP53 and breast cancer risk in African-American women. NPJ Breast Cancer 2017, 3. [Google Scholar] [CrossRef]
- Almeida, L.O.; Custódio, A.C.; Pinto, G.R.; Santos, M.J.; Almeida, J.R.; Clara, C.A.; Rey, J.A.; Casartelli, C. Polymorphisms and DNA methylation of gene TP53 associated with extra-axial brain tumors. Genet Mol Res 2009, 8, 8–18. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Association of p53 codon 248 (exon7) with urinary bladder cancer risk in the North Indian population. Biosci Trend 2011, 5, 205–210. [Google Scholar] [CrossRef]
- Siraj, A.K.; Al-Rasheed, M.; Ibrahim, M.; Siddiqui, K.; Al-Dayel, F.; Al-Sanea, O.; Uddin, S.; Al-Kuraya, K. RAD52 polymorphisms contribute to the development of papillary thyroid cancer susceptibility in Middle Eastern population. J Endocrinol Invest 2008, 31, 893–899. [Google Scholar] [CrossRef]
- Daugherty, C.L.; Curtis, H.; Realini, T.; Charlton, J.F.; Zareparsi, S. Primary open angle glaucoma in a Caucasian population is associated with the p53 codon 72 polymorphism. Mol Vis 2009, 15, 1939–1944. [Google Scholar] [PubMed]
- Ruggeri, R.M.; Vicchio, T.M.; Giovinazzo, S.; Certo, R.; Alibrandi, A.; Trimarchi, F.; Benvenga, S.; Trovato, M. TP53 polymorphism may contribute to genetic susceptibility to develop Hashimoto's thyroiditis. J Endocrinol Invest 2015, 38, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Elsaid, A.; Zahran, R.; Elshazli, R.; El-Sayed, A.; Abou Samra, M.; El-Tarapely, F.; Abdel-Malak, C. Genetic polymorphisms of TP53 Arg72Pro and Pro47Ser among Egyptian patients with colorectal carcinoma. Arch Physiol Biochem 2019, 125, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Sameer, A.S.; Shah, Z.A.; Syeed, N.; Banday, M.Z.; Bashir, S.M.; Bhat, B.A.; Siddiqi, M.A. TP53 Pro47Ser and Arg72Pro polymorphisms and colorectal cancer predisposition in an ethnic Kashmiri population. Genet Mol Res 2010, 9, 651–660. [Google Scholar] [CrossRef]
- Santos, L.E.; Guilhen, A.C.; de Andrade, R.A.; Sumi, L.G.; Ward, L.S. The role of TP53 PRO47SER and ARG72PRO single nucleotide polymorphisms in the susceptibility to bladder cancer. Urol Oncol 2011, 29, 291–294. [Google Scholar] [CrossRef]
- Alawadi, S.; Ghabreau, L.; Alsaleh, M.; Abdulaziz, Z.; Rafeek, M.; Akil, N.; Alkhalaf, M. P53 gene polymorphisms and breast cancer risk in Arab women. Med Oncol 2011, 28, 709–715. [Google Scholar] [CrossRef]
- Mostaid, M.S.; Ahmed, M.U.; Islam, M.S.; Bin Sayeed, M.S.; Hasnat, A. Lung cancer risk in relation to TP53 codon 47 and codon 72 polymorphism in Bangladeshi population. Tumour Biol 2014, 35, 10309–10317. [Google Scholar] [CrossRef]
- Nairuz, T.; Rahman, M.; Bushra, M.U.; Kabir, Y. TP53 Arg72Pro and XPD Lys751Gln Gene Polymorphisms and Risk of Lung Cancer in Bangladeshi Patients. Asian Pac J Cancer Prev 2020, 21, 2091–2098. [Google Scholar] [CrossRef]
- Diakite, B.; Kassogue, Y.; Dolo, G.; Wang, J.; Neuschler, E.; Kassogue, O.; Keita, M.L.; Traore, C.B.; Kamate, B.; Dembele, E.; et al. p.Arg72Pro polymorphism of P53 and breast cancer risk: a meta-analysis of case-control studies. BMC Med Genet 2020, 21, 206. [Google Scholar] [CrossRef]
- Fang, Y.; Wu, X.; Li, L.; Zhu, J.; Wu, H.; Zhou, H.; He, J.; Wang, Y. TP53 Arg72Pro polymorphism and neuroblastoma susceptibility in eastern Chinese children: a three-center case-control study. Biosci Rep 2020, 40. [Google Scholar] [CrossRef]
- Drokow, E.K.; Chen, Y.; Waqas Ahmed, H.A.; Oppong, T.B.; Akpabla, G.S.; Pei, Y.; Kumah, M.A.; Neku, E.A.; Sun, K. The relationship between leukemia and TP53 gene codon Arg72Pro polymorphism: analysis in a multi-ethnic population. Future Oncol 2020, 16, 923–937. [Google Scholar] [CrossRef] [PubMed]
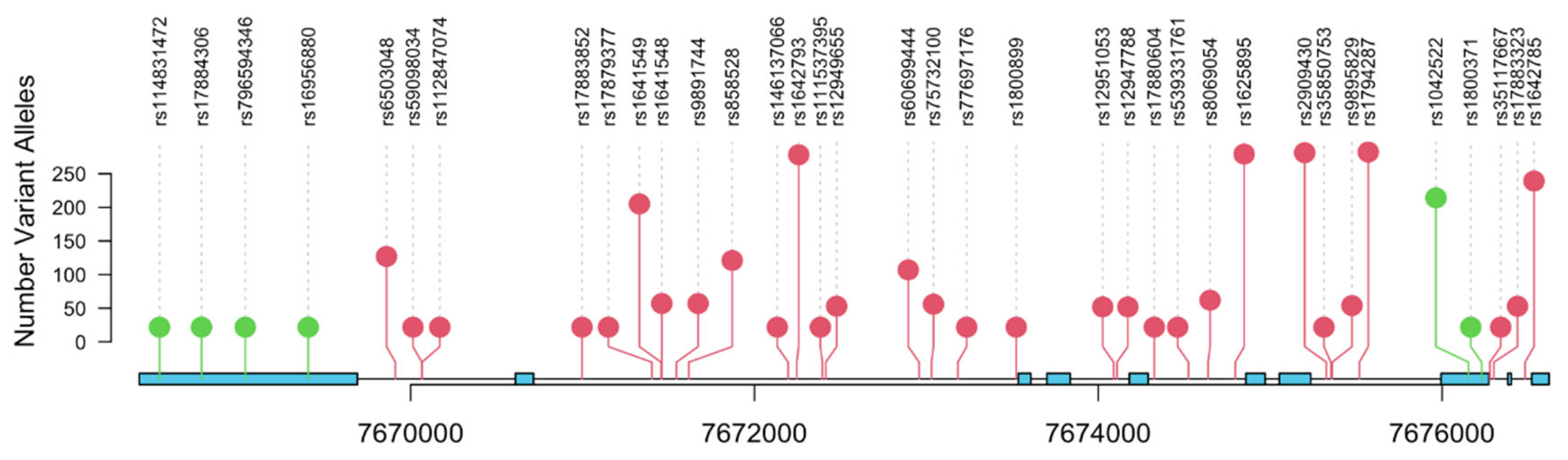
| Characteristic | N = 308 (%) |
|---|---|
| Diagnosis Age (in years) | 57 (9) |
| Unknown | 1 |
| Race | |
| African American | 212 (69%) |
| Caucasian | 95 (31%) |
| Unknown | 1 |
| Diagnosis PSA (ng/ml) | |
| 1:<4 | 72 (24%) |
| 2:4-9 | 193 (63%) |
| 3:10-20 | 31 (10%) |
| 4:>20 | 10 (3.3%) |
| Unknown | 2 |
| Pathologic T Stage | |
| T2 | 228 (74%) |
| T3-4 | 80 (26%) |
| Biopsy Gleason | |
| ≤6 | 204 (70%) |
| 7 | 69 (24%) |
| 8-10 | 17 (5.9%) |
| Unknown | 18 |
| Pathologic Gleason | |
| 3+3 | 169 (57%) |
| 3+4 | 76 (26%) |
| 4+3 | 24 (8.2%) |
| 8-10 | 25 (8.5%) |
| Unknown | 14 |
| BCR | 47 (15%) |
| Unknown | 3 |
| Metastasis | 9 (2.9%) |
| SNP | TP53 rs1800371 SNP | TP53 rs1042522 SNP | |||||
|---|---|---|---|---|---|---|---|
| Patient Characteristics | GG, N = 302 |
GA, N = 6 |
p-value | GG, N=87 | GC, N=137 | CC, N=84 | p-value |
| Diagnosis Age | 57 (9) | 56 (12) | 0.6 | 58 (9) | 57 (8) | 57 (9) | 0.8 |
| Unknown | 1 | 0 | 0 | 1 | 0 | ||
| Race | 0.2 | <0.001 | |||||
| African American | 206 (68%) | 6 (100%) | 81 (93%) | 87 (64%) | 44 (52%) | ||
| Caucasian | 95 (32%) | 0 (0%) |
6 (6.9%) | 49 (36%) | 40 (48%) | ||
| Unknown | 1 | 0 | 0 | 1 | 0 | ||
| Diagnosis PSA | 0.085 | 0.8 | |||||
| 1: <4 | 70 (23%) | 2 (33%) |
22 (25%) | 28 (21%) | 22 (26%) | ||
| 2: 4-9 | 191 (64%) | 2 (33%) |
55 (63%) | 87 (64%) | 51 (61%) | ||
| 3: 10-20 | 30 (10%) | 1 (17%) |
9 (10%) | 14 (10%) | 8 (9.5%) | ||
| 4: >20 | 9 (3.0%) |
1 (17%) |
1 (1.1%) | 6 (4.4%) | 3 (3.6%) | ||
| Unknown | 2 | 0 | 0 | 2 | 0 | ||
| Pathologic T Stage | >0.9 | 0.6 | |||||
| T2 | 223 (74%) | 5 (83%) |
57 (68%) | 104 (76%) | 59 (70%) | ||
| T3-4 | 79 (26%) | 1 (17%) |
22 (25%) | 33 (24%) | 25 (30%) | ||
| Biopsy Gleason | 0.2 | 0.8 | |||||
| ≤6 | 199 (70%) | 5 (83%) |
57 (68%) | 93 (73%) | 54 (69%) | ||
| 7 | 69 (24%) | 0 (0%) |
21 (25%) | 27 (21%) | 21 (27%) | ||
| 8-10 | 16 (5.6%) | 1 (17%) |
6 (7.1%) | 8 (6.2%) | 3 (3.8%) | ||
| Unknown | 18 | 0 | 3 | 9 | 6 | ||
| Pathologic Gleason | 0.6 | >0.9 | |||||
| 3+3 | 164 (57%) | 5 (100%) | 46 (55%) | 74 (57%) | 49 (60%) | ||
| 3+4 | 76 (26%) | 0 (0%) |
22 (26%) | 35 (27%) | 19 (23%) | ||
| 4+3 | 24 (8.3%) | 0 (0%) |
7 (8.3%) | 10 (7.8%) | 7 (8.6%) | ||
| 8-10 | 25 (8.7%) | 0 (0%) |
9 (11%) | 10 (7.8%) | 6 (7.4%) | ||
| Unknown | 13 | 1 | 3 | 8 | 3 | ||
| BCR | 47 (16%) | 0 (0%) |
0.6 | 13 (15%) | 20 (15%) | 14 (17%) | >0.9 |
| Unknown | 3 | 0 | 1 | 1 | 1 | ||
| Metastasis | 8 (2.6%) |
1 (17%) |
0.2 | 3 (3.4%) | 6 (4.4%) | 0 (0%) |
0.2 |
| Position | SNP | Location | HR | P |
|---|---|---|---|---|
| chr17:7667612:A:G | rs4968186 | downstream | 1.753 | 0.004 |
| chr17:7676963:G:A | rs9894227 | intronic | 1.720 | 0.007 |
| chr17:7676734:C:T | rs8079544 | intronic | 1.632 | 0.020 |
| chr17:7676154:G:C | rs1042522 | exonic | 1.523 | 0.046 |
| chr17:7664197:C:T | rs35119871 | intergenic | 1.461 | 0.046 |
| chr17:7675519:A:G | rs1794287 | intronic | 1.508 | 0.058 |
| chr17:7676230:G:A | rs1800371 | exonic | 1.473 | 0.067 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





