Submitted:
13 April 2023
Posted:
14 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
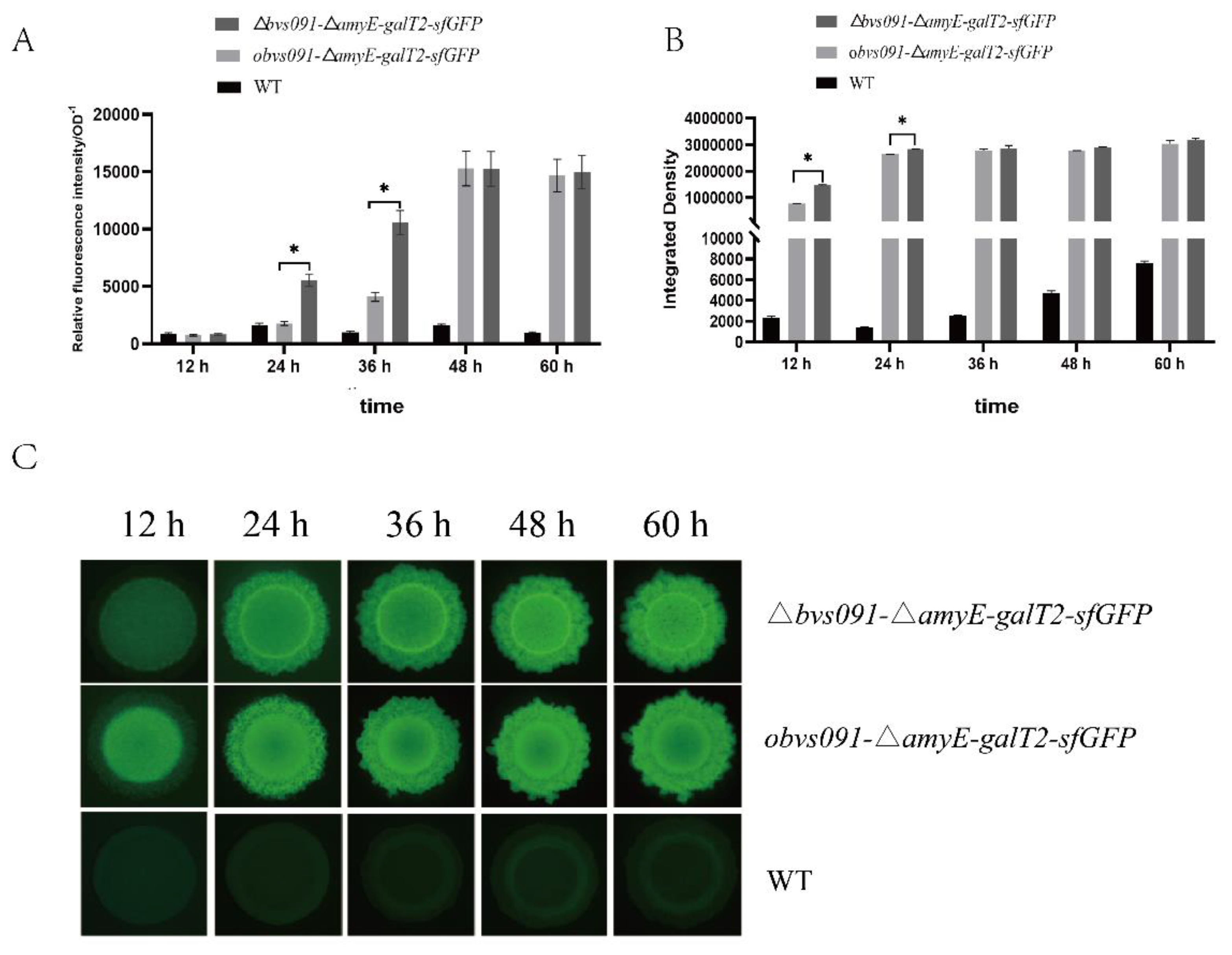
2. Results
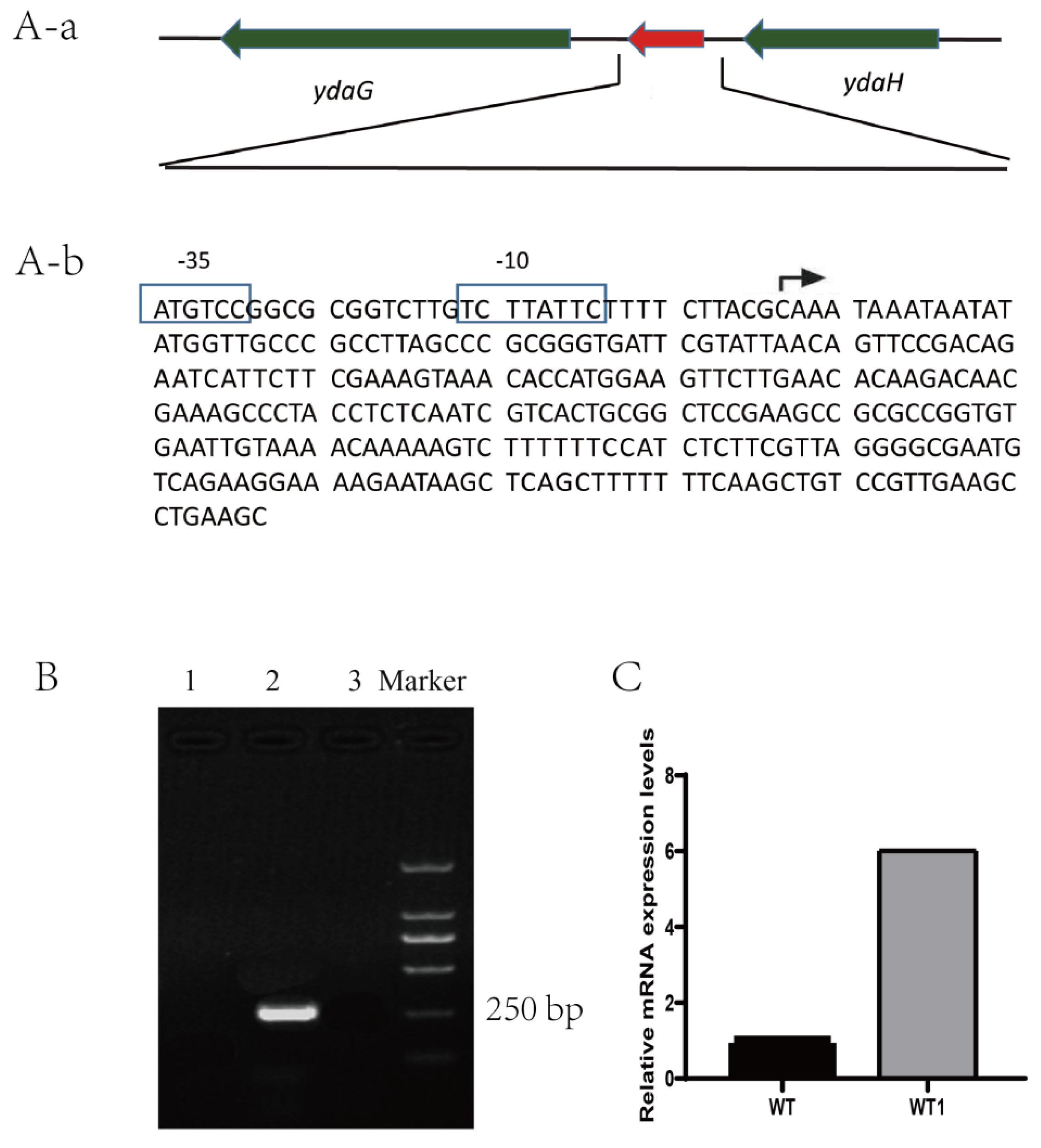
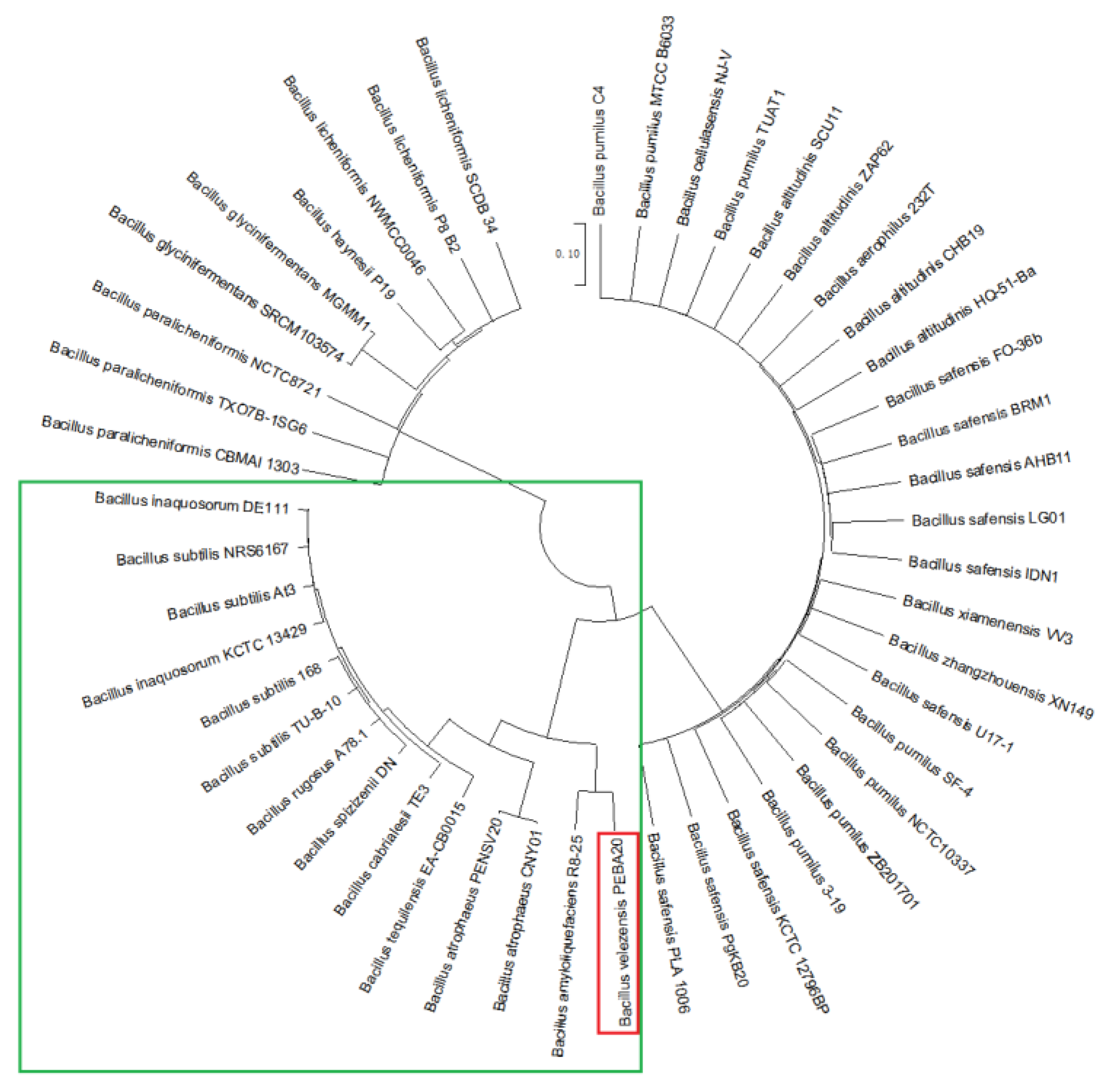
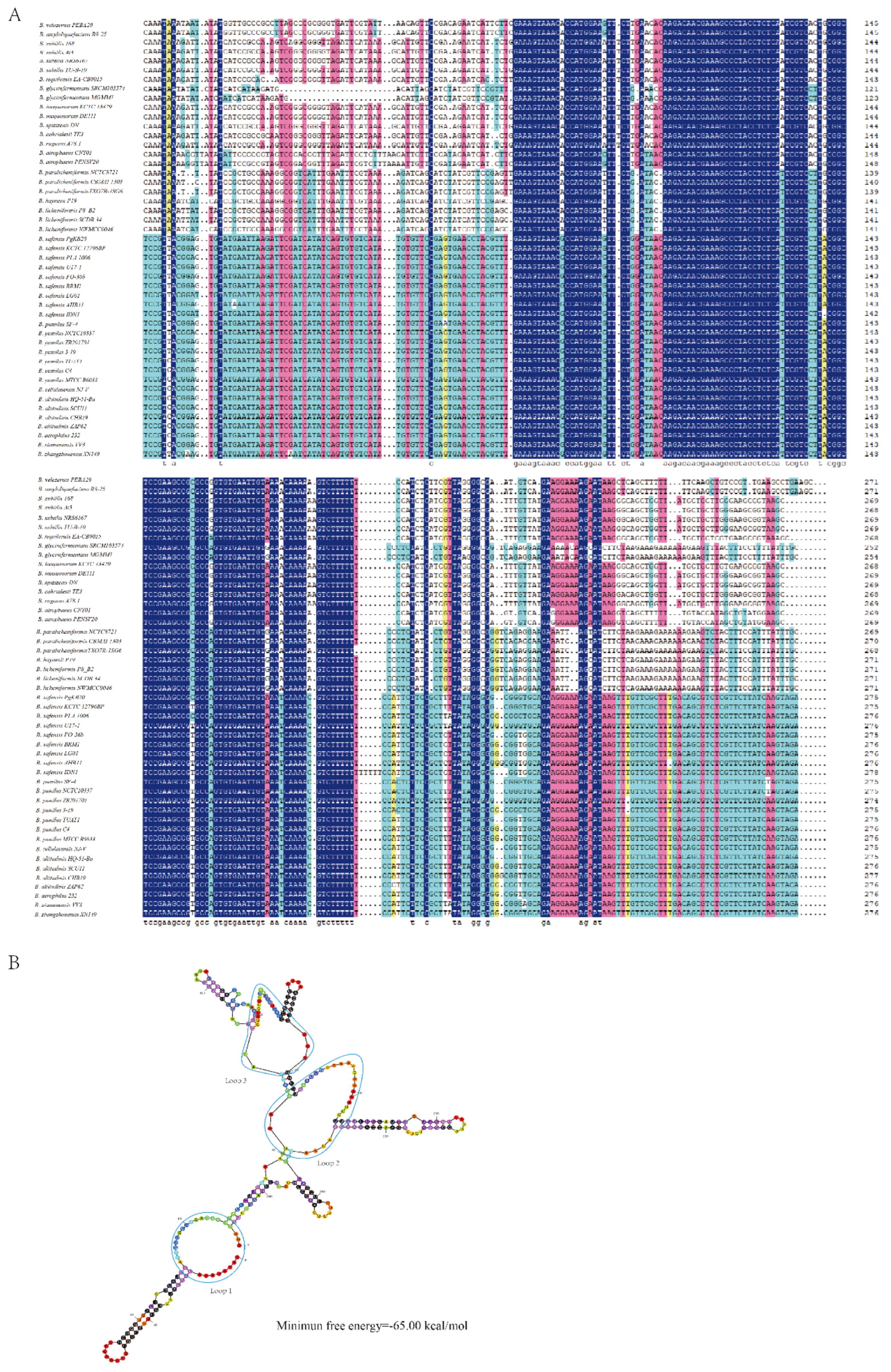
2.1. Characterization of sRNA Bvs091
2.2. Effect of sRNA Bvs091 on the phenotype of B. velezensis
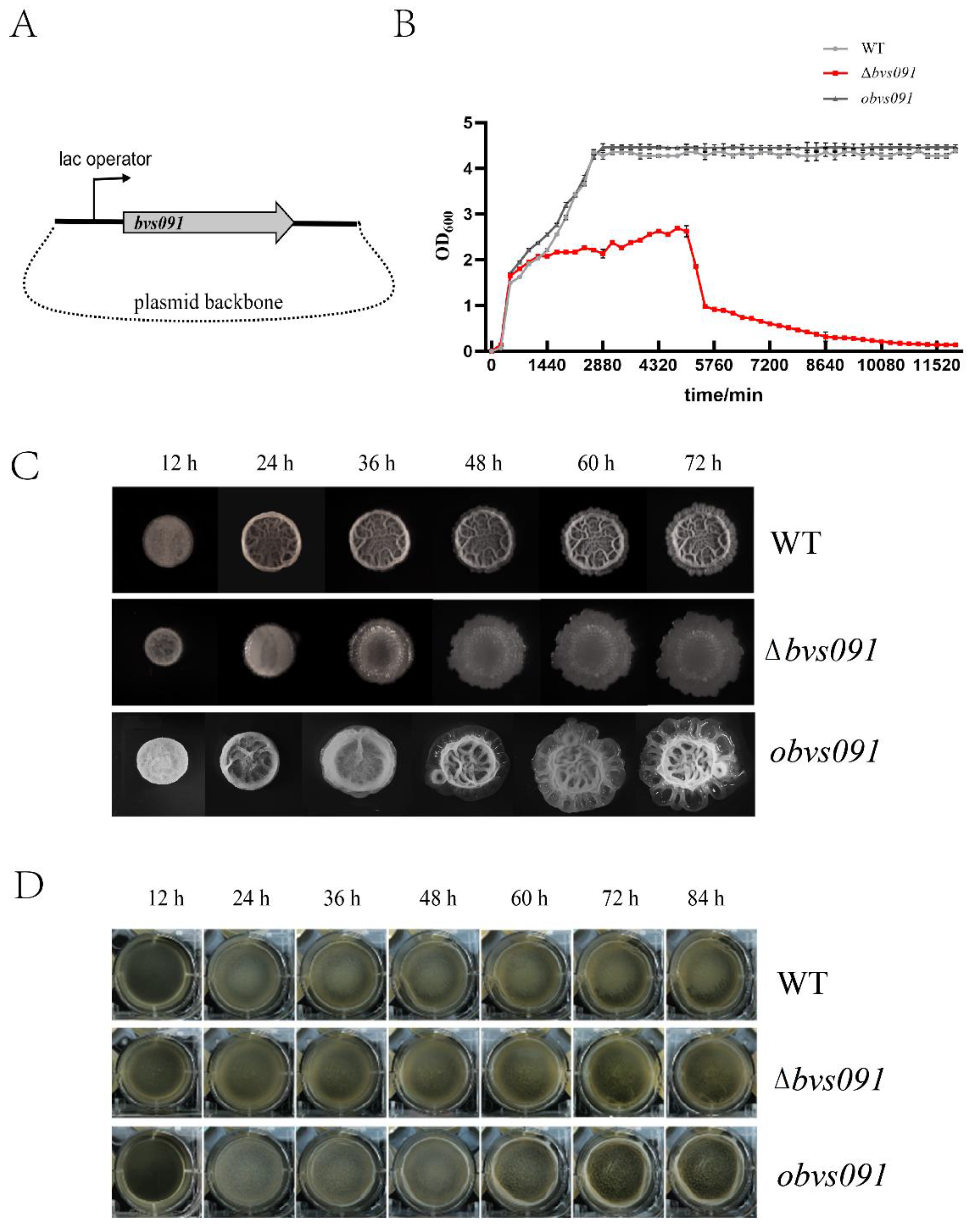
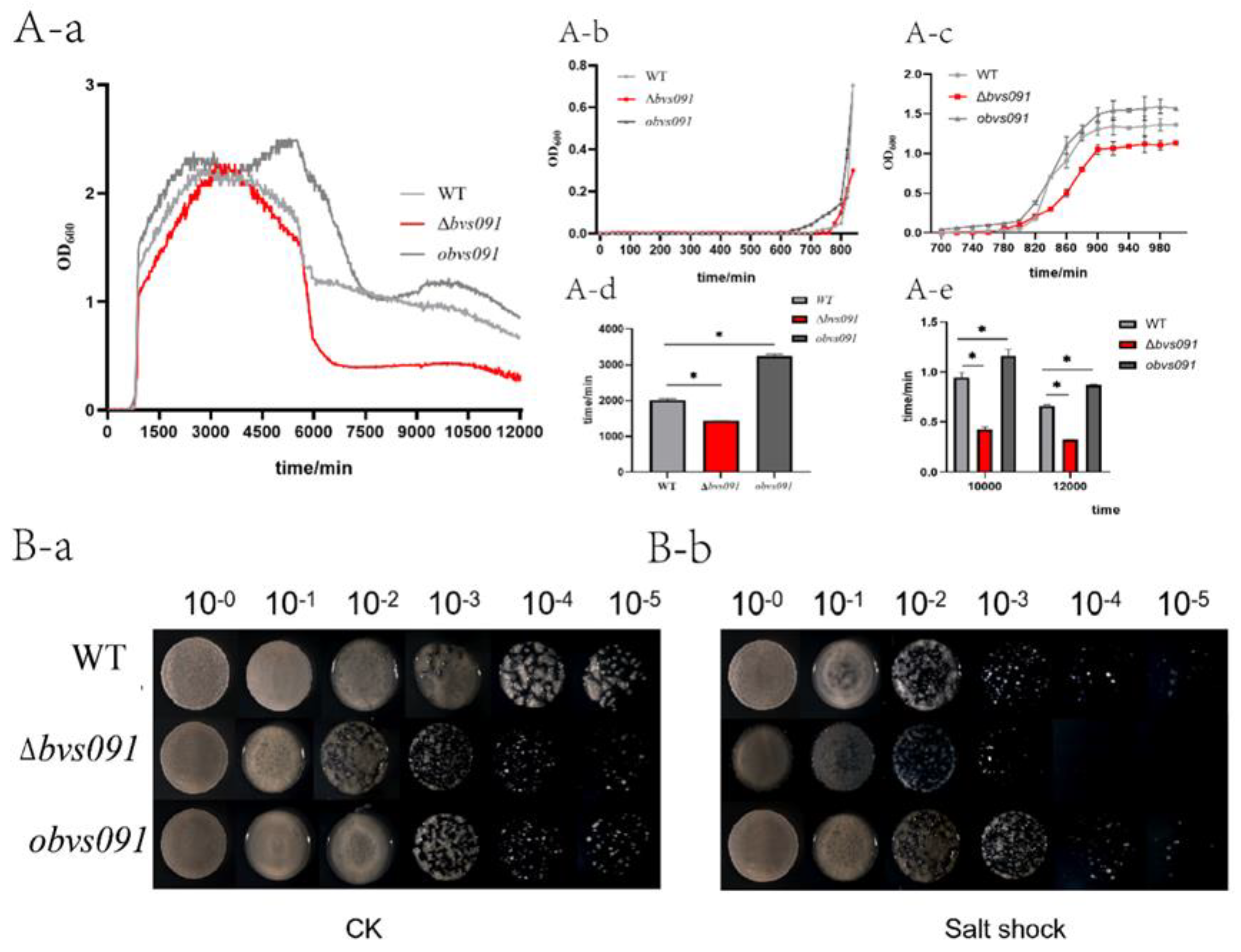
2.2.1. sRNA Bvs091’s effect on B. velezensis growth
2.2.2. sRNA Bvs091 enhances salt tolerance of B. velezensis
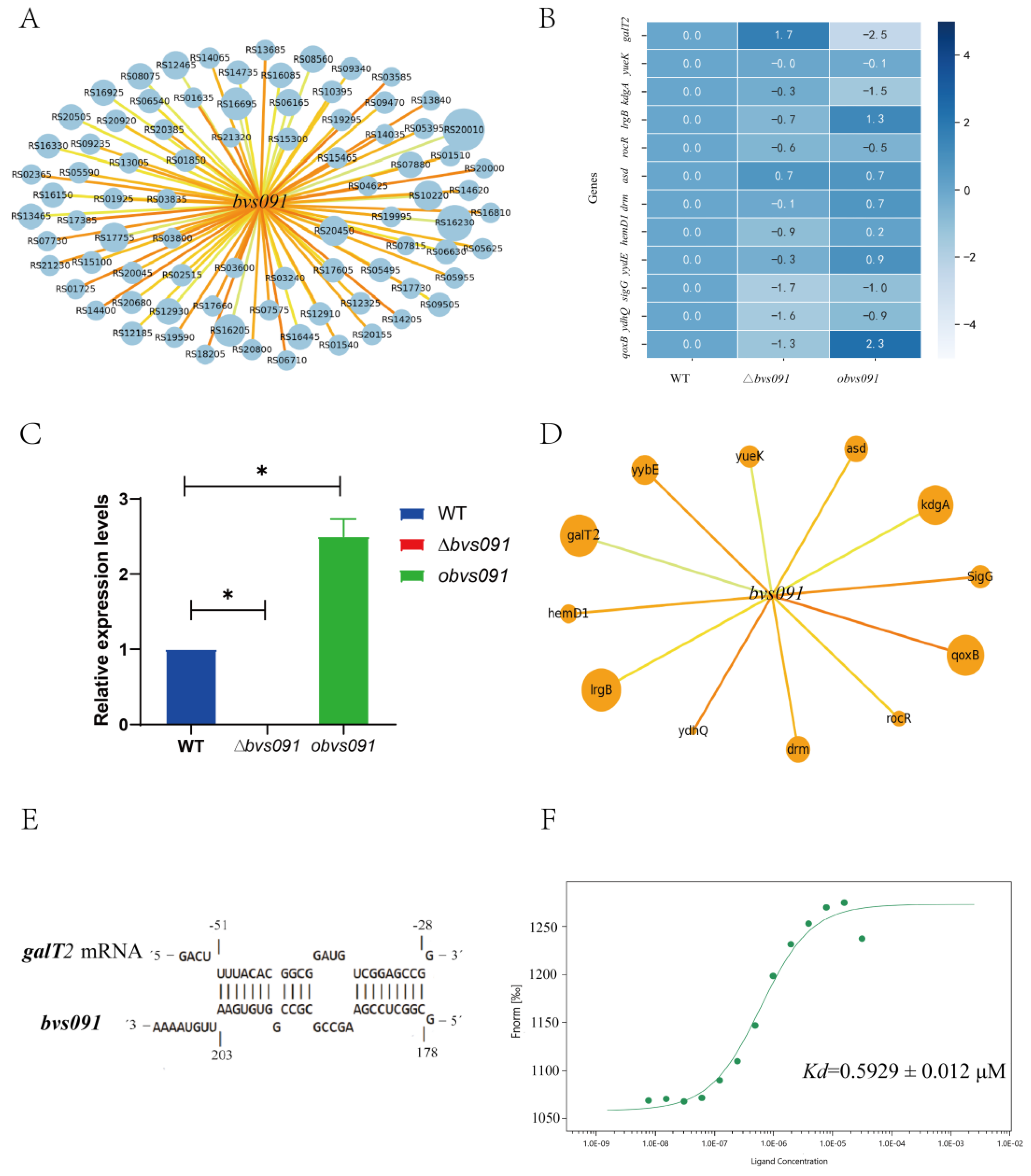
2.3. Target prediction and validation of sRNA Bvs091
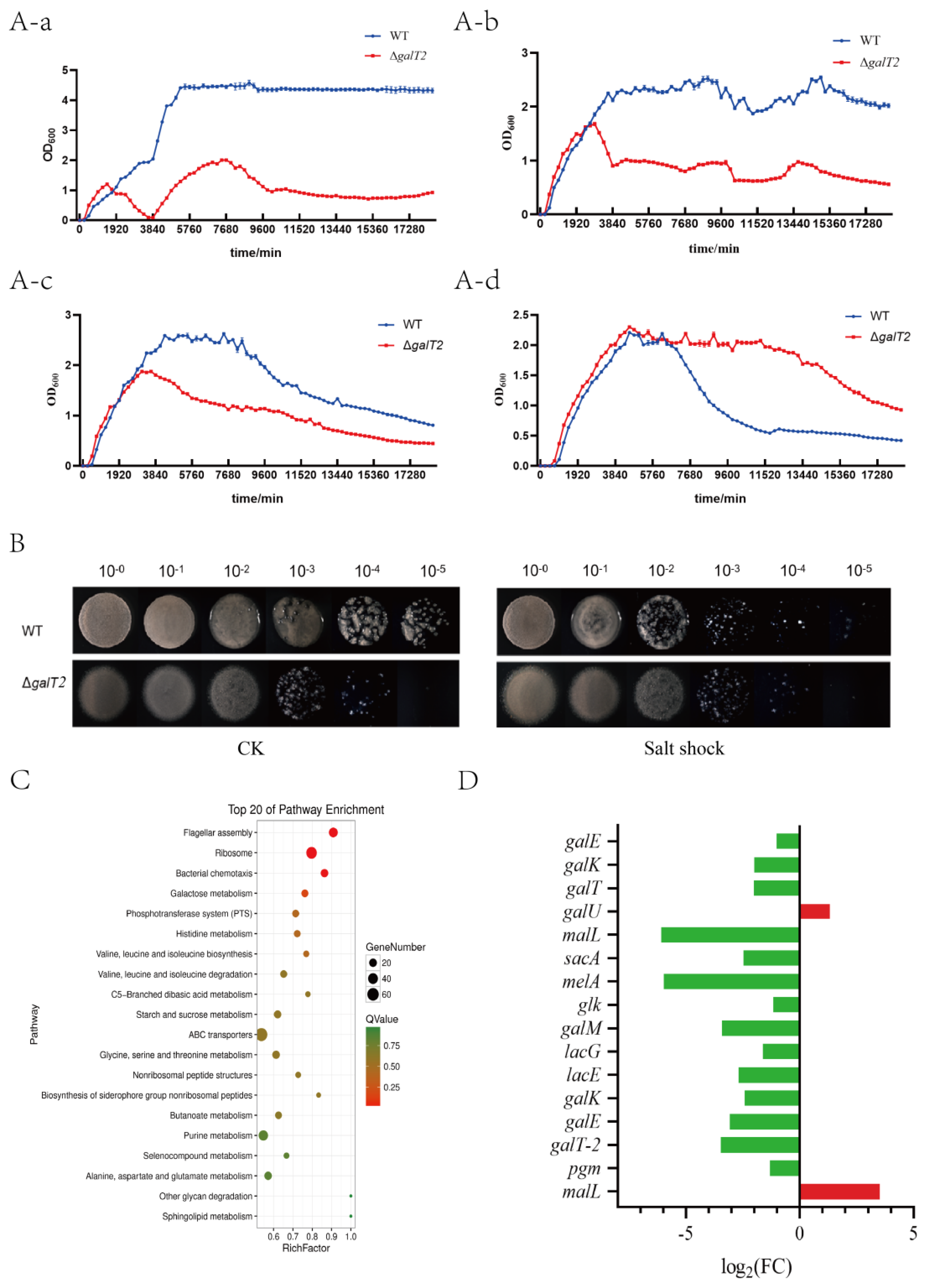
2.3. The function of galT2 contributing to salt tolerance
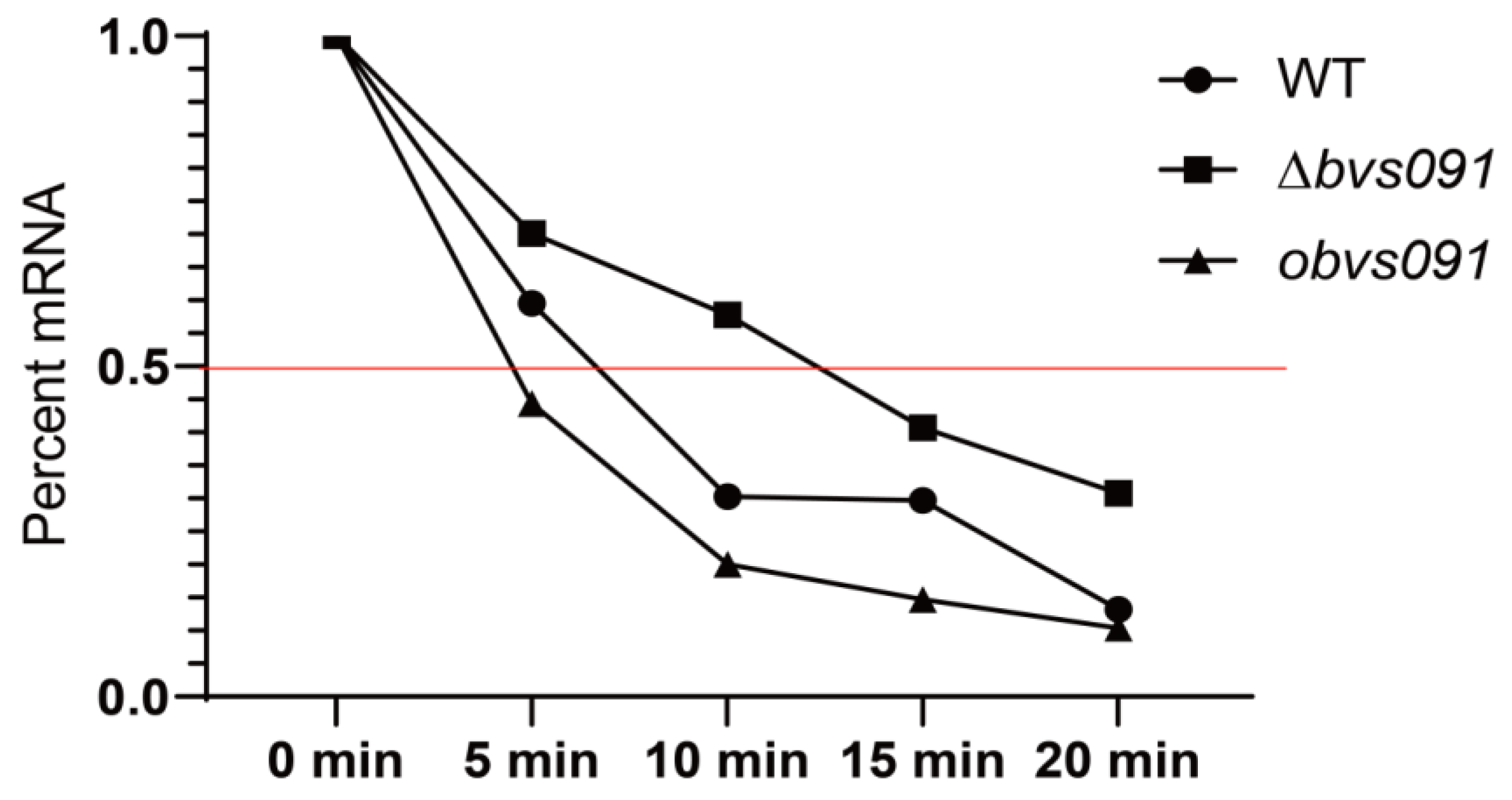
2.4.1. Bvs091 reduces the stability of galT2 mRNA
2.4.2. Bvs091 reduces the protein synthesis of GalT2
2.4.3. Bvs091 and galT2 mRNA matching binding site.
3. Discussion
4. Materials and Methods
4.1. Bacteria strain, growth conditions, and salt stress treatment.
4.2. RNA extraction
4.3. Mutant strain construction
4.4. Half-life experiment
4.5. Bioinformatics analysis
4.5. Microscale thermophoresis (MST) measurements
4.6. Solid‒gas interface and gas‒liquid biofilm phenotype measurement
4.7. RNA extraction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Richmond, C.S.; Glasner, J.D.; Mau, R.; Jin, H.; Blattner, F.R. Genome-Wide Expression Profiling in Escherichia Coli K-12. Nucleic Acids Research. 1999, 27, 3821–3835. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, P.; Eichenberger, P.; Losick, R.; Youngman, P. The Transcriptional Profile of Early to Middle Sporulation in Bacillus Subtilis. Proc Natl Acad Sci U S A. 2000, 97, 8063–8068. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell. 2009, 136, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell. 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Pain, A.; Ott, A.; Amine, H.; Rochat, T.; Bouloc, P.; Gautheret, D. An Assessment of Bacterial Small RNA Target Prediction Programs. RNA Biol. 2015, 12, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Peer, A.; Faigenbaum-Romm, R.; Gatt, Y.E.; Reiss, N.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol Cell. 2016, 63, 884–897. [Google Scholar] [CrossRef] [PubMed]
- Dar, D.; Sorek, R. Bacterial Noncoding RNAs Excised from within Protein-Coding Transcripts. mBio. 2018, 9, e01730–18. [Google Scholar] [CrossRef]
- Rasmussen, S.; Nielsen, H.B.; Jarmer, H. The Transcriptionally Active Regions in the Genome of Bacillus Subtilis. Mol Microbiol. 2009, 73, 1043–1057. [Google Scholar] [CrossRef]
- Irnov, I.; Sharma, C.M.; Vogel, J.; Winkler, W.C. Identification of Regulatory RNAs in Bacillus Subtilis. Nucleic Acids Res. 2010, 38, 6637–6651. [Google Scholar] [CrossRef]
- Saito, S.; Kakeshita, H.; Nakamura, K. Novel Small RNA-Encoding Genes in the Intergenic Regions of Bacillus Subtilis. Gene. 2009, 428, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, N, Chinali, A, Gerth, U, Brantl, S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol. 2006, 62, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Genes for Small, Noncoding RNAs under Sporulation Control in Bacillus Subtilis. J Bacteriol, 2006, 188, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus Amyloliquefaciens, Bacillus Velezensis, and Bacillus Siamensis Form an “Operational Group B. Amyloliquefaciens” within the B. Subtilis Species Complex. Front Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative Analysis of the Complete Genome Sequence of the Plant Growth-Promoting Bacterium Bacillus Amyloliquefaciens FZB42. Nat Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.J.; Yan, Y.C.; Li, X.Y.; Liu, Z.Y. Draft Genome Sequence of Bacillus Velezensis PEBA20, a Strain with a Plant Growthpromoting Effect and Biocontrol Potential. Genome Announc. 2018, 6, e00286–18. [Google Scholar] [CrossRef]
- Xu, T.; Li, X.; Chen, K.; Qin, H.; Yi, Z.; Meng, Y.; Liu, Z. A Sporulation-Specific SRNA Bvs196 Contributing to the Developing Spore in Bacillus Velezensis. Microorganisms. 2022, 10, E3487–3496. [Google Scholar] [CrossRef]
- Ul Haq, I.; Müller, P.; Brantl, S. Intermolecular Communication in Bacillus Subtilis: RNA-RNA, RNA-Protein and Small Protein-Protein Interactions. Front Mol Biosci. 2020, 7, 178. [Google Scholar] [CrossRef]
- Brantl, S.; Müller, P. Cis-and Trans-Encoded Small Regulatory RNAs in Bacillus Subtilis. Microorganisms. 2021, 9, 1865. [Google Scholar] [CrossRef]
- Brantl, S. Regulatory Mechanisms Employed by Cis-Encoded Antisense RNAs. Curr Opin Microbiol. 2007, 10, 102–109. [Google Scholar] [CrossRef]
- Brantl, S. Plasmid Replication Control by Antisense RNAs. Microbiol Spectr. 2014, 2, PLAS-0001-2013. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Gilet, L.; Condon, C. The Essential Function of B. Subtilis RNase III Is to Silence Foreign Toxin Genes. PLoS Genet. 2012, 8, e1003181. [Google Scholar] [CrossRef] [PubMed]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Small Untranslated RNA Antitoxin in Bacillus Subtilis. J Bacteriol. 2005, 187, 6641–6650. [Google Scholar] [CrossRef] [PubMed]
- Jahn N, Preis H, Wiedemann C, Brantl S. BsrG_SR4 from Bacillus Subtilis—The First Temperature-Dependent Type I Toxin-Antitoxin System. Mol Microbiol. 2012, 83, 579–598. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Jahn, N.; Ring, C.; Maiwald, C.; Neubert, R.; Meißner, C.; Brantl, S. A Multistress Responsive Type I Toxin-Antitoxin System: BsrE/SR5 from the B. Subtilis Chromosome. RNA Biol. 2016, 13, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Reif, C.; Löser, C.; Brant, S. Bacillus Subtilis Type I Antitoxin SR6 Promotes Degradation of Toxin Yont MRNA and Is Required to Prevent Toxic YoyJ Overexpression. Toxins (Basel). 2018, 10, 74. [Google Scholar] [CrossRef]
- Brantl, S.; Müller, P. Toxin-Antitoxin Systems in Bacillus Subtilis. Toxins (Basel). 2019, 11, 262. [Google Scholar] [CrossRef] [PubMed]
- Rath, H.; Reder, A.; Hoffmann, T.; Hammer, E.; Seubert, A.; Bremer, E.; Völker, U.; Mäder, U. Management of Osmoprotectant Uptake Hierarchy in Bacillus Subtilis via a SigB-Dependent Antisense RNA. Front Microbiol. 2020, 11, 622. [Google Scholar] [CrossRef]
- Brantl, S. Acting Antisense: Plasmid- and Chromosome-Encoded SRNAs from Gram-Positive Bacteria. Future Microbiol. 2012, 7, 853–871. [Google Scholar] [CrossRef]
- Morita, T, Mochizuki, Y, Aiba, H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci. 2006, 103, 4858–4863. [Google Scholar] [CrossRef]
- Licht, A.; Preis, S.; Brantl, S. Implication of CcpN in the Regulation of a Novel Untranslated RNA (SR1) in Bacillus Subtilis. Mol Microbiol. 2005, 58, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; dé rique Braun, F.; Helfer, A.-C.; Romby, P.; Condon, C. SRNA-Mediated Activation of Gene Expression by Inhibition of 5’-3’ Exonucleolytic MRNA Degradation. Elife. 2017, 6, e23602. [Google Scholar] [CrossRef] [PubMed]
- Grainger, D.C.; Lee, D.J.; Busby, S.J. Direct Methods for Studying Transcription Regulatory Proteins and RNA Polymerase in Bacteria. Curr Opin Microbiol. 2009, 12, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A. Prokaryotic Genome Regulation: Multifactor Promoters, Multitarget Regulators and Hierarchic Networks. FEMS Microbiol Rev. 2010, 34, 628–645. [Google Scholar] [CrossRef] [PubMed]
- Arrieta-Ortiz, M.L.; Hafemeister, C.; Bate, A.R.; Chu, T.; Greenfield, A.; Shuster, B.; Barry, S.N.; Gallitto, M.; Liu, B.; Kacmarczyk, T.; et al. An Experimentally Supported Model of the Bacillus Subtilis Global Transcriptional Regulatory Network. Mol Syst Biol. 2015, 11, 839. [Google Scholar] [CrossRef]
- Nicolas, P.; Mäder, U.; Dervyn, E.; Rochat, T.; Leduc, A.; Pigeonneau, N.; Bidnenko, E.; Marchadier, E.; Hoebeke, M.; Aymerich, S.; et al. Condition-Dependent Transcriptome Reveals High-Level Regulatory Architecture in Bacillus Subtilis. Science (1979) 2012, 335, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Maaß, S.; Wachlin, G.; Bernhardt, J.; Eymann, C.; Fromion, V.; Riedel, K.; Becher, D.; Hecker, M. Highly Precise Quantification of Protein Molecules per Cell during Stress and Starvation Responses in Bacillus Subtilis. Molecular and Cellular Proteomics. 2014, 13, 2260–2276. [Google Scholar] [CrossRef]
- Meeske, A.J.; Sham, L.T.; Kimsey, H.; Koo, B.M.; Gross, C.A.; Bernhardt, T.G.; Rudner, D.Z. MurJ and a Novel Lipid II Flippase Are Required for Cell Wall Biogenesis in Bacillus Subtilis. Proc Natl Acad Sci U S A. 2015, 112, 6437–6442. [Google Scholar] [CrossRef]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Man, S.; Cheng, R.; Miao, C.; Gong, Q.; Gu, Y.; Lu, X.; Han, F.; Yu, W. Artificial Trans-Encoded Small Non-Coding RNAs Specifically Silence the Selected Gene Expression in Bacteria. Nucleic Acids Res. 2011, 39, e50. [Google Scholar] [CrossRef]
- Backofen, R.; Hess, W.R. Computational Prediction of SRNAs and Their Targets in Bacteria. RNA Biol 2010, 7, 33–42. [Google Scholar] [CrossRef]
- Chai, Y.; Beauregard, P.B.; Vlamakis, H.; Losick, R.; Kolter, R. Galactose Metabolism Plays a Crucial Role in Biofilm Formation by Bacillus Subtilis. mBio. 2012, 3, e00184–12. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, N.; Moll, I.; Brantl, S. In Vitro Analysis of the Interaction between the Small RNA SR1 and Its Primary Target AhrC MRNA. Nucleic Acids Res 2007, 35, 4331–4346. [Google Scholar] [CrossRef] [PubMed]
- Wick, L.M.; Egli, T. Molecular Components of Physiological Stress Responses in Escherichia Coli. Adv Biochem Eng Biotechnol. 2004, 89, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Brill, J.; Hoffmann, T.; Bleisteiner, M.; Bremer, E. Osmotically Controlled Synthesis of the Compatible Solute Proline Is Critical for Cellular Defense of Bacillus Subtilis against High Osmolarity. J Bacteriol. 2011, 193, 5335–5346. [Google Scholar] [CrossRef] [PubMed]
- Asally, M.; Kittisopikul, M.; Rué, P.; Du, Y.; Hu, Z.; Çaǧatay, T.; Robinson, A.B.; Lu, H.; Garcia-Ojalvo, J.; Süel, G.M. Localized Cell Death Focuses Mechanical Forces during 3D Patterning in a Biofilm. Proc Natl Acad Sci U S A. 2012, 109, 18891–18896. [Google Scholar] [CrossRef]
- Finking, R.; Marahiel, M.A. Biosynthesis of Nonribosomal Peptides. Annu Rev Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, W.; Wang, Y.; Wu, L.; Liu, X.; Yan, T.; Alm, E.; Arkin, A.; Thompson, D.K.; Fields, M.W.; et al. Transcriptome Analysis of Shewanella Oneidensis MR-1 in Response to Elevated Salt Conditions. J Bacteriol. 2005, 187, 2501–2507. [Google Scholar] [CrossRef]
- Gancz, H.; Merrell, D.S. The Helicobacter Pylori Ferric Uptake Regulator (Fur) Is Essential for Growth under Sodium Chloride Stress. Journal of Microbiology. 2011, 49, 294–298. [Google Scholar] [CrossRef]
- Krispin,O, Allmansberger, R. The Bacillus subtilis galE gene is essential in the presence of glucose and galactose. J Bacteriol. 1998, 180, 2265–2270. [Google Scholar] [CrossRef]
- Irnov, I.; Winkler, W.C. A Regulatory RNA Required for Antitermination of Biofilm and Capsular Polysaccharide Operons in Bacillales. Mol Microbiol. 2010, 76, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.; Chastanet, A.; Débarbouillé, M. New Vector for Efficient Allelic Replacement in Naturally Nontransformable, Low-GC-Content, Gram-Positive Bacteria. Appl Environ Microbiol. 2004, 70, 6887–6891. [Google Scholar] [CrossRef] [PubMed]
- Pedelacq, J.D.; Cabantous, S. Development and Applications of Superfolder and Split Fluorescent Protein Detection Systems in Biology. Int J Mol Sci. 2019, 20, 3479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhan, Y.; Yan, Y.; Liu, Y.; Hu, G.; Wang, S.; Yang, H.; Qiu, X.; Liu, Y.; Li, J.; et al. The Pseudomonas Stutzeri-Specific Regulatory Noncoding RNA Nfis Targets Katb MRNA Encoding a Catalase Essential for Optimal Oxidative Resistance and Nitrogenase Activity. J Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate Boundary Prediction and Improved Detection of Structural RNAs. RNA. 2012, 18, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.R.; Richter, A.S.; Papenfort, K.; Mann, M.; Vogel, J.; Hess, W.R.; Backofen, R.; Georg, J. Comparative Genomics Boosts Target Prediction for Bacterial Small RNAs. Proc Natl Acad Sci U S A. 2013, 110, E3487–3496. [Google Scholar] [CrossRef] [PubMed]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-Binding Assays in Biological Liquids Using Microscale Thermophoresis. Nat Commun. 2010, 1, 100. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction Analysis and Beyond. J Mol Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics. 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics. 2009, 26, 139–140. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




