Submitted:
22 February 2023
Posted:
23 February 2023
You are already at the latest version
Abstract
Keywords:
Introduction
- -
- storage footprint: the raw data, the intermediate analysis files and the results of their analysis occupy increasingly more disk space (i.e. the raw sequence of a Human whole-genome might vary between 100GB to 150GB depending on the coverage);
- -
- number of data points: high-throughput technologies allow to process thousands of samples in a relatively short time, thus multiplying the number of data points collected during each experiment; this further increases when single-cell technologies are employed [4];
- -
- range of data types: genomic, epigenetic, transcriptomic, proteomic data - among others - can be generated from each single sample, with the intent of combining their information for the analysis [5].
- (a)
- Transparency and trust: machine learning and deep learning are considered “black boxes” and this rarely fits with contexts (healthcare, for example) where a justification should be provided for decisions. Explainability offers a solution, i.e. opens the black box and provides the means to connect the outcome of a model to the information it received as an input, or to reveal the inner mechanisms of the model, i.e. how a decision or a prediction has been reached.
- (b)
- Quality control or troubleshooting: mapping the outcomes of an artificial neural network to those features in the input which contributed most to the activation of the model, and therefore to reaching its decisions, helps identifying any bias in the datasets, and to spot input features which should not be expected to influence the model to a certain extent.
- (c)
- Novel insights: assigning an importance score to those input features that best contribute to the outcome of a deep neural network, provides a way to identify relationships in the data that other methods might not be able to reveal.
Explainable DL and Genome Engineering
Explainable DL and Drug Design
Explainable DL and Radiomics
Explainable DL and genomics
- -
- deep layers in a neural network increase the abstraction in the representation of the input data, using non-linear activation functions
- -
- non-linear relationships in the data are therefore captured by a neural network, enhancing the ability to unmask different patterns in the data [92]
- -
- latent representations offer new insights into networks and interactions at different level in genomics, epigenomics and functional dimensions of the data [93]
Conclusions and learning resources
Abbreviations
References
- Visvikis D, Cheze Le Rest C, Jaouen V, Hatt M. Artificial intelligence, machine (deep) learning and radio(geno)mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging 2019;46:2630–7. [CrossRef]
- Wang, P. On Defining Artificial Intelligence. J Artif Gen Intell 2019;10:1–37. [CrossRef]
- Jiang Y, Li X, Luo H, Yin S, Kaynak O. Quo vadis artificial intelligence? Discov Artif Intell 2022;2:4. [CrossRef]
- Chen W, Zhao Y, Chen X, Yang Z, Xu X, Bi Y, et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat Biotechnol 2021;39:1103–14. [CrossRef]
- Hassan M, Awan FM, Naz A, deAndrés-Galiana EJ, Alvarez O, Cernea A, et al. Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review. Int J Mol Sci 2022;23:4645. [CrossRef]
- Goyal I, Singh A, Saini JK. Big Data in Healthcare: A Review. 2022 1st Int. Conf. Inform. ICI, Noida, India: IEEE; 2022, p. 232–4. [CrossRef]
- Mor U, Cohen Y, Valdés-Mas R, Kviatcovsky D, Elinav E, Avron H. Dimensionality reduction of longitudinal ’omics data using modern tensor factorizations. PLOS Comput Biol 2022;18:e1010212. [CrossRef]
- Picard M, Scott-Boyer M-P, Bodein A, Périn O, Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput Struct Biotechnol J 2021;19:3735–46. [CrossRef]
- Samek W, Montavon G, the SLP of, 2021. Explaining deep neural networks and beyond: A review of methods and applications. IeeexploreIeeeOrg n.d. https://doi.org/10.1109/JPROC.2021.3060483&url_ctx_fmt=info:ofi/fmt:kev:mtx:ctx&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.atitle=Explaining.
- Montavon G, Samek W, Processing KMDS, 2018. Methods for interpreting and understanding deep neural networks. Elsevier n.d. [CrossRef]
- Watson, DS. Interpretable machine learning for genomics. Hum Genet 2022;141:1499–513. [CrossRef]
- Roscher R, Bohn B, Duarte MF, Garcke J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2020;8:42200–16. [CrossRef]
- Carrieri AP, Haiminen N, Maudsley-Barton S, Gardiner L-J, Murphy B, Mayes AE, et al. Explainable AI reveals changes in skin microbiome composition linked to phenotypic differences. Sci Rep 2021;11:4565. [CrossRef]
- Holzinger A, Saranti A, Molnar C, Biecek P, Samek W. Explainable AI Methods - A Brief Overview. In: Holzinger A, Goebel R, Fong R, Moon T, Müller K-R, Samek W, editors. XxAI - Explain. AI, vol. 13200, Cham: Springer International Publishing; 2022, p. 13–38. [CrossRef]
- Bach S, Binder A, Montavon G, Klauschen F, Müller K-R, Samek W. On Pixel-Wise Explanations for Non-Linear Classifier Decisions by Layer-Wise Relevance Propagation. PLOS ONE 2015;10:e0130140. [CrossRef]
- Shrikumar A, Greenside P, Kundaje A. Learning Important Features Through Propagating Activation Differences 2017. [CrossRef]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. 2017 IEEE Int. Conf. Comput. Vis. ICCV, Venice: IEEE; 2017, p. 618–26. [CrossRef]
- Lundberg SM, Lee S-I. A Unified Approach to Interpreting Model Predictions. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, et al., editors. Adv. Neural Inf. Process. Syst., vol. 30, Curran Associates, Inc.; 2017.
- Wang J, Wiens J, Lundberg S. Shapley Flow: A Graph-based Approach to Interpreting Model Predictions. Proc Mach Learn Res 2021;130:721–9.
- Loh HW, Ooi CP, Seoni S, Barua PD, Molinari F, Acharya UR. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput Methods Programs Biomed 2022;226:107161. [CrossRef]
- Rajabi E, Etminani K. Towards a Knowledge Graph-Based Explainable Decision Support System in Healthcare. Stud Health Technol Inform 2021;281:502–3. [CrossRef]
- Chaddad A, Peng J, Xu J, Bouridane A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023;23:634. [CrossRef]
- Newman SJ, Furbank RT. Explainable machine learning models of major crop traits from satellite-monitored continent-wide field trial data. Nat Plants 2021;7:1354–63. [CrossRef]
- Ryo, M. Explainable artificial intelligence and interpretable machine learning for agricultural data analysis. Artif Intell Agric 2022;6:257–65. [CrossRef]
- Sapoval N, Aghazadeh A, Nute MG, Antunes DA, Balaji A, Baraniuk R, et al. Current progress and open challenges for applying deep learning across the biosciences. Nat Commun 2022;13:1728. [CrossRef]
- Woźniak S, Pantazi A, Bohnstingl T, Eleftheriou E. Deep learning incorporating biologically inspired neural dynamics and in-memory computing. Nat Mach Intell 2020;2:325–36. [CrossRef]
- Wang J, Zhang X, Cheng L, Luo Y. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol 2019;17:13–22. [CrossRef]
- Zhang G, Zeng T, Dai Z, Dai X. Prediction of CRISPR/Cas9 single guide RNA cleavage efficiency and specificity by attention-based convolutional neural networks. Comput Struct Biotechnol J 2021;19:1445–57. [CrossRef]
- Chuai G, Ma H, Yan J, Chen M, Hong N, Xue D, et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol 2018;19:80. [CrossRef]
- O’Brien AR, Burgio G, Bauer DC. Domain-specific introduction to machine learning terminology, pitfalls and opportunities in CRISPR-based gene editing. Brief Bioinform 2020;22:308–14. [CrossRef]
- Jiménez-Luna J, Grisoni F, Schneider G. Drug discovery with explainable artificial intelligence. Nat Mach Intell 2020;2:573–84. [CrossRef]
- Chen H, Gomez C, Huang C-M, Unberath M. Explainable medical imaging AI needs human-centered design: guidelines and evidence from a systematic review. Npj Digit Med 2022;5:1–15. [CrossRef]
- Singh A, Sengupta S, Lakshminarayanan V. Explainable Deep Learning Models in Medical Image Analysis. J Imaging 2020;6:52. [CrossRef]
- Novakovsky G, Dexter N, Libbrecht MW, Wasserman WW, Mostafavi S. Obtaining genetics insights from deep learning via explainable artificial intelligence. Nat Rev Genet 2022:1–13. [CrossRef]
- Wang JY, Doudna JA. CRISPR technology: A decade of genome editing is only the beginning. Science 2023;379:eadd8643. [CrossRef]
- Xue L, Tang B, Chen W, Luo J. Prediction of CRISPR sgRNA Activity Using a Deep Convolutional Neural Network. J Chem Inf Model 2019;59:615–24. [CrossRef]
- Xiang X, Corsi GI, Anthon C, Qu K, Pan X, Liang X, et al. Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat Commun 2021;12:3238. [CrossRef]
- Yang Q, Wu L, Meng J, Ma L, Zuo E, Sun Y. EpiCas-DL: Predicting sgRNA activity for CRISPR-mediated epigenome editing by deep learning. Comput Struct Biotechnol J 2022;21:202–11. [CrossRef]
- Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids 2015;4:e264. [CrossRef]
- Xiao L-M, Wan Y-Q, Jiang Z-R. AttCRISPR: a spacetime interpretable model for prediction of sgRNA on-target activity. BMC Bioinformatics 2021;22:589. [CrossRef]
- Liu Q, He D, Xie L. Prediction of off-target specificity and cell-specific fitness of CRISPR-Cas System using attention boosted deep learning and network-based gene feature. PLoS Comput Biol 2019;15:e1007480. [CrossRef]
- Mathis N, Allam A, Kissling L, Marquart KF, Schmidheini L, Solari C, et al. Predicting prime editing efficiency and product purity by deep learning. Nat Biotechnol 2023:1–9. [CrossRef]
- Wang D, Zhang C, Wang B, Li B, Wang Q, Liu D, et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat Commun 2019;10:4284. [CrossRef]
- Jayatunga MKP, Xie W, Ruder L, Schulze U, Meier C. AI in small-molecule drug discovery: a coming wave? Nat Rev Drug Discov 2022;21:175–6. [CrossRef]
- The National Genomics Research and Healthcare Knowledgebase 2017. [CrossRef]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. [CrossRef]
- Ozerov IV, Lezhnina KV, Izumchenko E, Artemov AV, Medintsev S, Vanhaelen Q, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat Commun 2016;7:13427. [CrossRef]
- Ivanenkov YA, Polykovskiy D, Bezrukov D, Zagribelnyy B, Aladinskiy V, Kamya P, et al. Chemistry42: An AI-Driven Platform for Molecular Design and Optimization. J Chem Inf Model 2023;63:695–701. [CrossRef]
- Pan X, Lin X, Cao D, Zeng X, Yu PS, He L, et al. Deep learning for drug repurposing: methods, databases, and applications 2022.
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–82. [CrossRef]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2023 update. Nucleic Acids Res 2023;51:D1373–80. [CrossRef]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [CrossRef]
- Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019;47:D930–40. [CrossRef]
- Karimi M, Wu D, Wang Z, Shen Y. DeepAffinity: interpretable deep learning of compound–protein affinity through unified recurrent and convolutional neural networks. Bioinformatics 2019;35:3329–38. [CrossRef]
- Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 2016;44:D1045–53. [CrossRef]
- Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 2008;36:D684–8. [CrossRef]
- Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, the UniProt Consortium. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015;31:926–32. [CrossRef]
- Rodríguez-Pérez R, Bajorath J. Interpretation of machine learning models using shapley values: application to compound potency and multi-target activity predictions. J Comput Aided Mol Des 2020;34:1013–26. [CrossRef]
- Pope PE, Kolouri S, Rostami M, Martin CE, Hoffmann H. Explainability Methods for Graph Convolutional Neural Networks. 2019 IEEECVF Conf. Comput. Vis. Pattern Recognit. CVPR, Long Beach, CA, USA: IEEE; 2019, p. 10764–73. [CrossRef]
- Mastropietro A, Pasculli G, Feldmann C, Rodríguez-Pérez R, Bajorath J. EdgeSHAPer: Bond-centric Shapley value-based explanation method for graph neural networks. IScience 2022;25:105043. [CrossRef]
- Dey S, Luo H, Fokoue A, Hu J, Zhang P. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinformatics 2018;19:476. [CrossRef]
- Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res 2016;44:D1075–9. [CrossRef]
- Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng 2018;2:749–60. [CrossRef]
- van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 2020;11:91. [CrossRef]
- Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer 2022;3:1026–38. [CrossRef]
- Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun 2020;11:3877. [CrossRef]
- Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 2021;594:106–10. [CrossRef]
- Gehrung M, Crispin-Ortuzar M, Berman AG, O’Donovan M, Fitzgerald RC, Markowetz F. Triage-driven diagnosis of Barrett’s esophagus for early detection of esophageal adenocarcinoma using deep learning. Nat Med 2021;27:833–41. [CrossRef]
- Yee E, Popuri K, Beg MF, Initiative the ADN. Quantifying brain metabolism from FDG-PET images into a probability of Alzheimer’s dementia score. Hum Brain Mapp 2020;41:5–16. [CrossRef]
- Etminani K, Soliman A, Davidsson A, Chang JR, Martínez-Sanchis B, Byttner S, et al. A 3D deep learning model to predict the diagnosis of dementia with Lewy bodies, Alzheimer’s disease, and mild cognitive impairment using brain 18F-FDG PET. Eur J Nucl Med Mol Imaging 2022;49:563–84. [CrossRef]
- Oppedal K, Borda MG, Ferreira D, Westman E, Aarsland D. European DLB consortium: diagnostic and prognostic biomarkers in dementia with Lewy bodies, a multicenter international initiative. Neurodegener Dis Manag 2019;9:247–50. [CrossRef]
- Qiu S, Miller MI, Joshi PS, Lee JC, Xue C, Ni Y, et al. Multimodal deep learning for Alzheimer’s disease dementia assessment. Nat Commun 2022;13:3404. [CrossRef]
- Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158–64. [CrossRef]
- Cuadros J, Bresnick G. EyePACS: An Adaptable Telemedicine System for Diabetic Retinopathy Screening. J Diabetes Sci Technol Online 2009;3:509–16.
- Madsen B, Browning S. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet 2009;5:e1000384.
- Zawistowski M, Reppell M, Wegmann D, St Jean PL, Ehm MG, Nelson MR, et al. Analysis of rare variant population structure in Europeans explains differential stratification of gene-based tests. Eur J Hum Genet 2014;22:1137–44. [CrossRef]
- Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Publ Group 2016;17:392–406. [CrossRef]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. [CrossRef]
- Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet 2014;15:765–76. [CrossRef]
- Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, et al. Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am J Hum Genet 2018;102:1204–11. [CrossRef]
- Wainschtein P, Jain D, Zheng Z, TOPMed Anthropometry Working Group, Aslibekyan S, Becker D, et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat Genet 2022;54:263–73. [CrossRef]
- Young, AI. Discovering missing heritability in whole-genome sequencing data. Nat Genet 2022;54:224–6. [CrossRef]
- Weiner DJ, Nadig A, Jagadeesh KA, Dey KK, Neale BM, Robinson EB, et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature 2023. [CrossRef]
- McCaw ZR, Colthurst T, Yun T, Furlotte NA, Carroll A, Alipanahi B, et al. DeepNull models non-linear covariate effects to improve phenotypic prediction and association power. Nat Commun 2022;13:241. [CrossRef]
- Gusareva ES, Van Steen K. Practical aspects of genome-wide association interaction analysis. Hum Genet 2014;133:1343–58. [CrossRef]
- Lescai F, Franceschi C. The Impact of Phenocopy on the Genetic Analysis of Complex Traits. PLOS ONE 2010;5:e11876. [CrossRef]
- Wei W-H, Hemani G, Haley CS. Detecting epistasis in human complex traits. Nat Publ Group 2014;15:722–33. [CrossRef]
- Domingo J, Baeza-Centurion P, Lehner B. The Causes and Consequences of Genetic Interactions (Epistasis). Annu Rev Genomics Hum Genet 2019;20:433–60. [CrossRef]
- Niel C, Sinoquet C, Dina C, Rocheleau G. A survey about methods dedicated to epistasis detection. Front Genet 2015;6:25. [CrossRef]
- Sailer ZR, Harms MJ. Detecting High-Order Epistasis in Nonlinear Genotype-Phenotype Maps. Genetics 2017;205:1079–88. [CrossRef]
- Sailer ZR, Harms MJ. High-order epistasis shapes evolutionary trajectories. PLOS Comput Biol 2017;13:e1005541. [CrossRef]
- Luo P, Li Y, Tian L-P, Wu F-X. Enhancing the prediction of disease–gene associations with multimodal deep learning. Bioinformatics 2019;35:3735–42. [CrossRef]
- Eraslan G, Avsec Ž, Gagneur J, Theis FJ. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet 2019;278:1. [CrossRef]
- Uppu S, Krishna A, Gopalan RP. A Deep Learning Approach to Detect SNP Interactions. J Softw 2016;11:965–75. [CrossRef]
- Romagnoni A, Jégou S, Van Steen K, Wainrib G, Hugot J-P. Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Rep 2019;9:1–18. [CrossRef]
- Mieth B, Rozier A, Rodriguez JA, Höhne MM-C, Görnitz N, Müller K-R. DeepCOMBI: explainable artificial intelligence for the analysis and discovery in genome-wide association studies. NAR Genomics Bioinforma 2021;3:lqab065. [CrossRef]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-Dimensionality Reduction Reveals High-Order Interactions among Estrogen-Metabolism Genes in Sporadic Breast Cancer. Am J Hum Genet 2001;69:138–47. [CrossRef]
- Chen G-B, Lee SH, Montgomery GW, Wray NR, Visscher PM, Gearry RB, et al. Performance of risk prediction for inflammatory bowel disease based on genotyping platform and genomic risk score method. BMC Med Genet 2017;18:94. [CrossRef]
- Mieth B, Kloft M, Rodríguez JA, Sonnenburg S, Vobruba R, Morcillo-Suárez C, et al. Combining Multiple Hypothesis Testing with Machine Learning Increases the Statistical Power of Genome-wide Association Studies. Sci Rep 2016;6:36671. [CrossRef]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78. [CrossRef]
- Greenside P, Shimko T, Fordyce P, Kundaje A. Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics 2018;34:i629–37. [CrossRef]
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 2015;33:831–8. [CrossRef]
- Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods 2015;12:931–4. [CrossRef]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [CrossRef]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [CrossRef]
- Yap M, Johnston RL, Foley H, MacDonald S, Kondrashova O, Tran KA, et al. Verifying explainability of a deep learning tissue classifier trained on RNA-seq data. Sci Rep 2021;11:2641. [CrossRef]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [CrossRef]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci 2018;27:233–44. [CrossRef]
- Zeng W, Wang Y, Jiang R. Integrating distal and proximal information to predict gene expression via a densely connected convolutional neural network. Bioinformatics 2020;36:496–503. [CrossRef]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017;171:1573-1588.e28. [CrossRef]
- Zhou Q, Wong WH. CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc Natl Acad Sci U S A 2004;101:12114–9. [CrossRef]
- Avsec Ž, Agarwal V, Visentin D, Ledsam JR, Grabska-Barwinska A, Taylor KR, et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat Methods 2021;18:1196–203. [CrossRef]
- Kelley, DR. Cross-species regulatory sequence activity prediction. PLOS Comput Biol 2020;16:e1008050. [CrossRef]
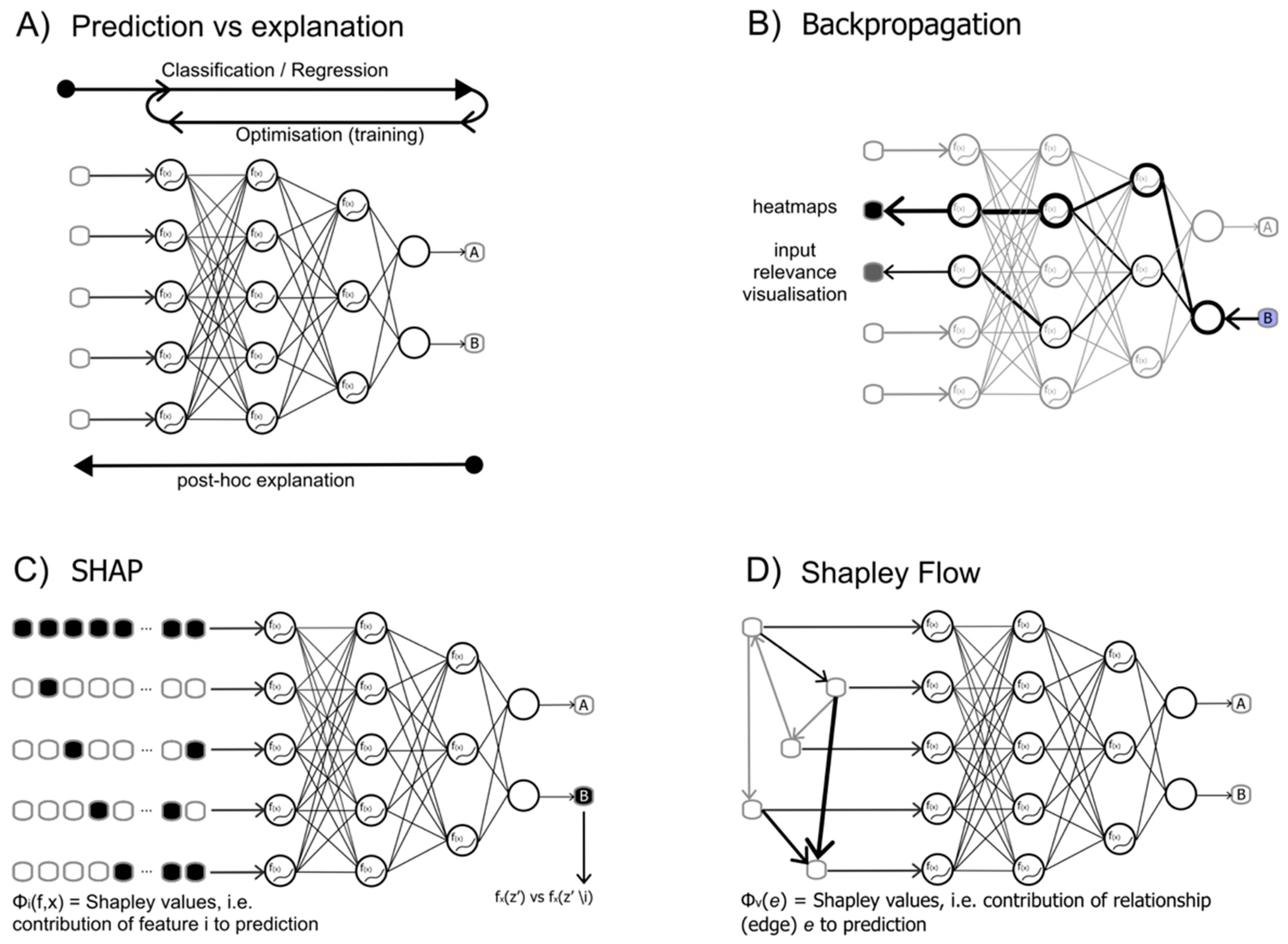
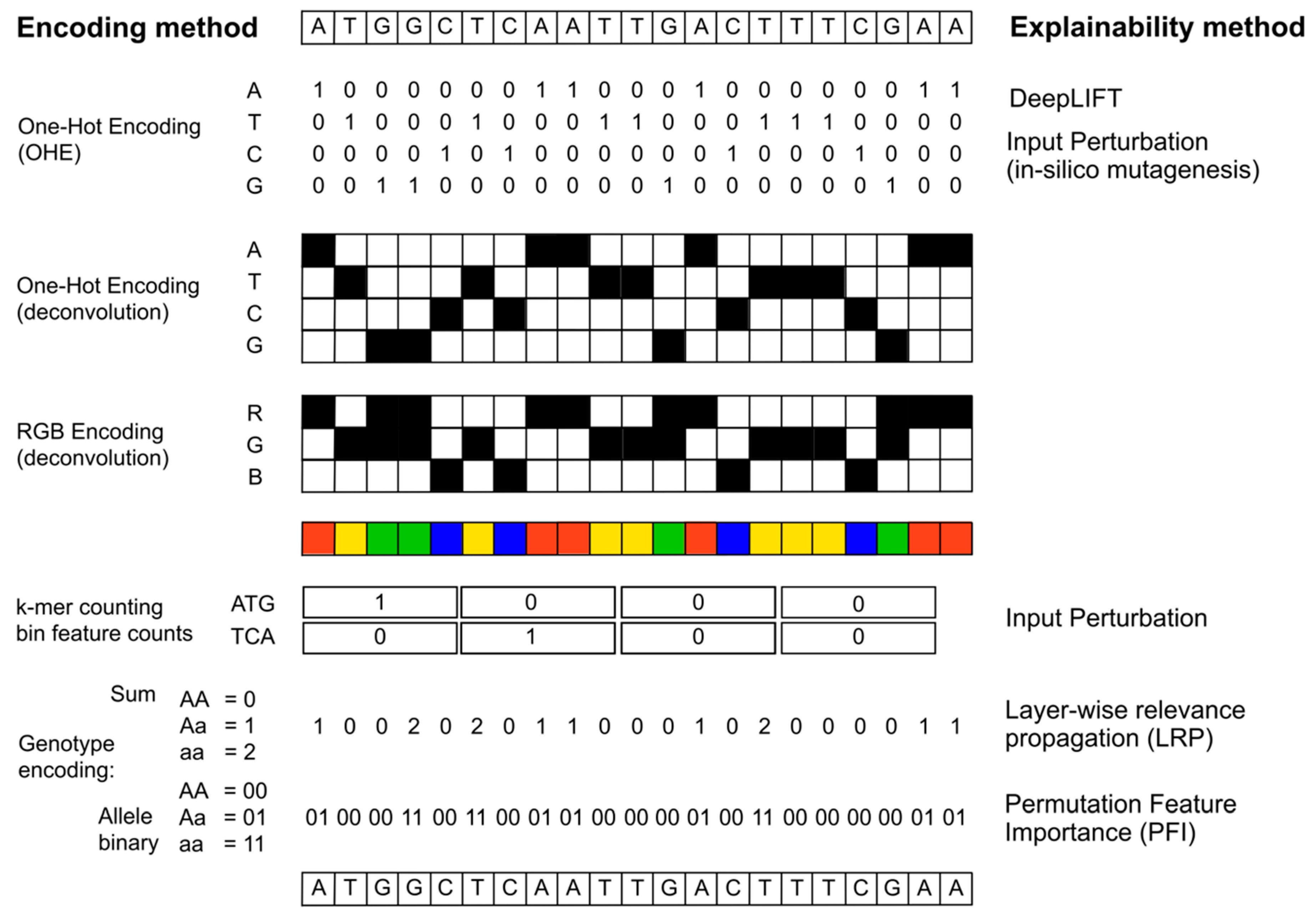
| Method | Description | Code | Tutorial | Reference |
|---|---|---|---|---|
| LRP | The acronym stands for Layer-wise Relevance Propagation, and is a method that explains graphs (originally) and deep neural networks by propagating the outcome decision backward across the neural network. | https://github.com/sebastian-lapuschkin/lrp_toolbox | https://colab.research.google.com/drive/166FYIwxblfrEltkYqY_jiJoAm9VLMweJ?usp=sharing | Bach et al. 2015, Lapuschkin et al. 2016 |
| https://git.tu-berlin.de/gmontavon/lrp-tutorial | ||||
| DeepLift | The acronym stands for "Deep Learning Important FeaTures" and works in a similar way to LRP furher implementing additional rules on how to distribute the relevance during backpropagation. | https://github.com/kundajelab/deeplift | https://www.youtube.com/playlist?list=PLJLjQOkqSRTP3cLB2cOOi_bQFw6KPGKML | Shrikumar A. et al |
| GradCAM | The name stands for Gradient-weighted Class Activation Mapping, and it is a method that exploits the gradients of any output to produce a localisation map highlighting the most important regions in an input image for predicting the output. | https://keras.io/examples/vision/grad_cam/ | https://colab.research.google.com/drive/1bA2Fg8TFbI5YyZyX3zyrPcT3TuxCLHEC?usp=sharing | Selvaraju et al. 2017 |
| https://github.com/ismailuddin/gradcam-tensorflow-2/blob/master/notebooks/GradCam.ipynb | ||||
| SHAP | SHAP stands for "SHapley Additive exPlanations", a method derived from game theory and aimed at measuring the contribution of each input feature into a prediction. The method helps with both local and global interpretability. | https://github.com/slundberg/shap | https://www.kaggle.com/code/dansbecker/shap-values | Lundberg and Lee, 2017 |
| https://towardsdatascience.com/a-complete-shap-tutorial-how-to-explain-any-black-box-ml-model-in-python-7538d11fae94 | ||||
| Shapely Flow | A further development on the SHAP algorithm, where relationships (dependency structure) between input variables are described with a graph: Shapley values are then attributed asimmetrically using this information, but are assigned to the relationships rather than the variables. | https://github.com/nathanwang000/Shapley-Flow | https://github.com/nathanwang000/Shapley-Flow/blob/master/notebooks/tutorial.ipynb | Wang et al. 2021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





