Submitted:
21 February 2023
Posted:
23 February 2023
You are already at the latest version
Abstract
Keywords:
Glossary
sORFs and SEPs: small in size but not in importance
Studying bacterial biology in the genomics era
Sequencing revolution demands annotation evolution
Automatic genome annotation requires manual curation: a frustrating paradox
sORFs: the weak spots of automated bacterial genome annotation

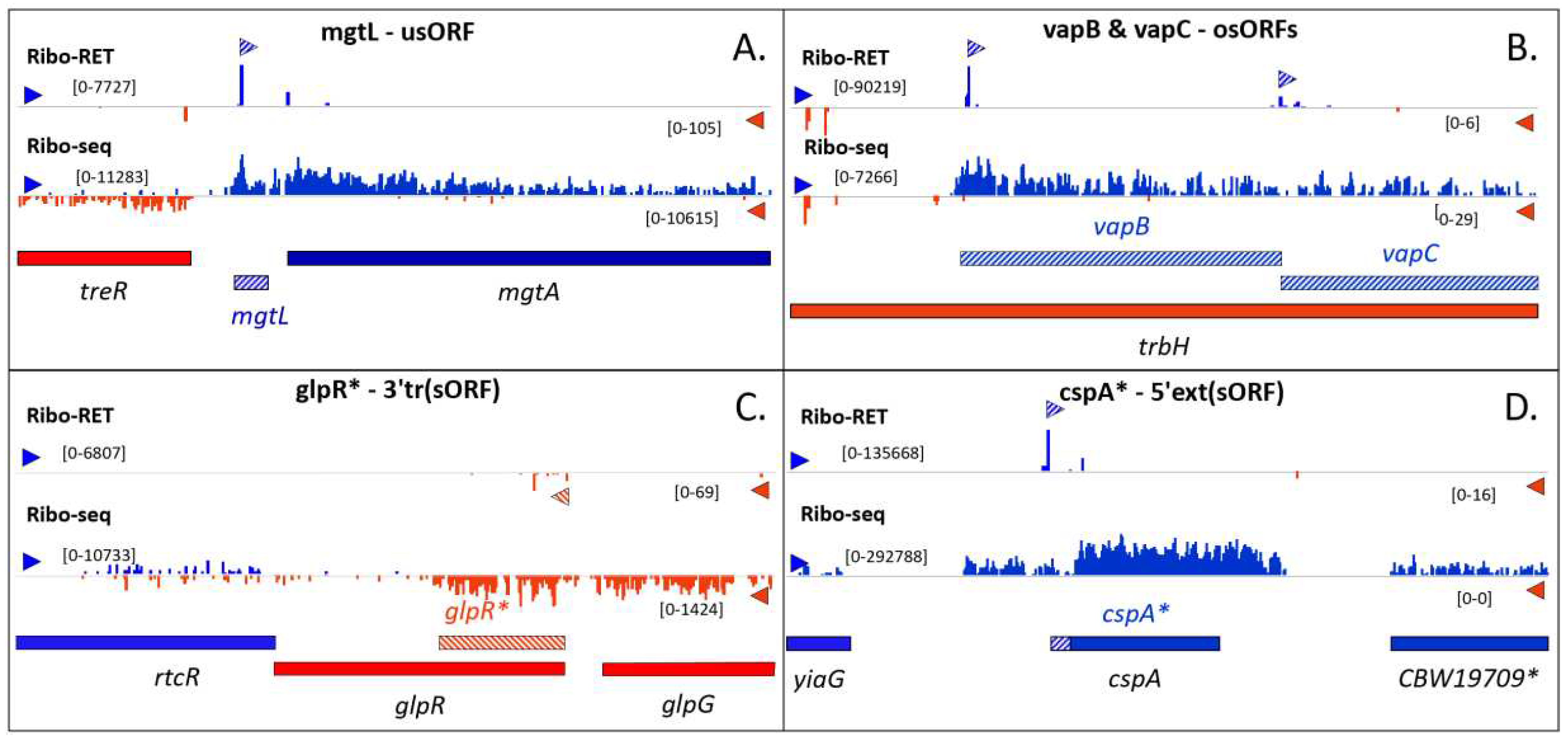
SEPs as a novel research hotspot for the study of bacterial biology
SEPs as accomplices in bacterial (infection) biology
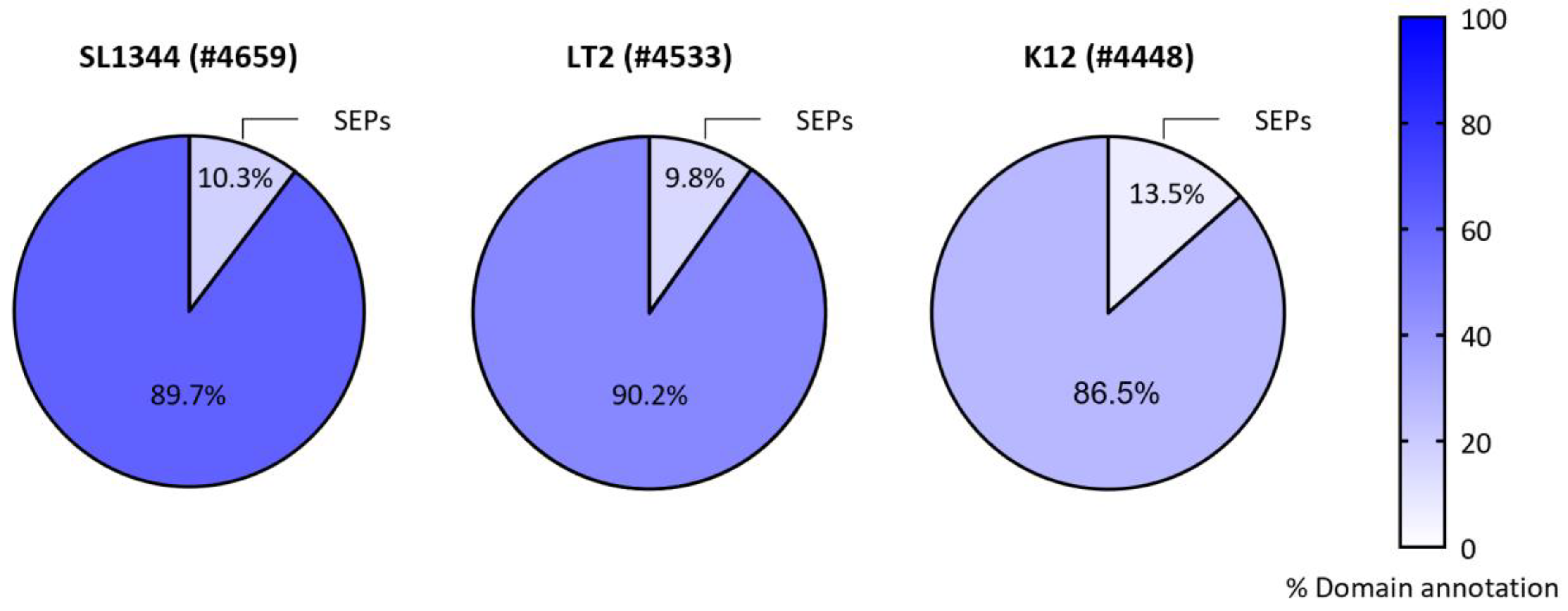
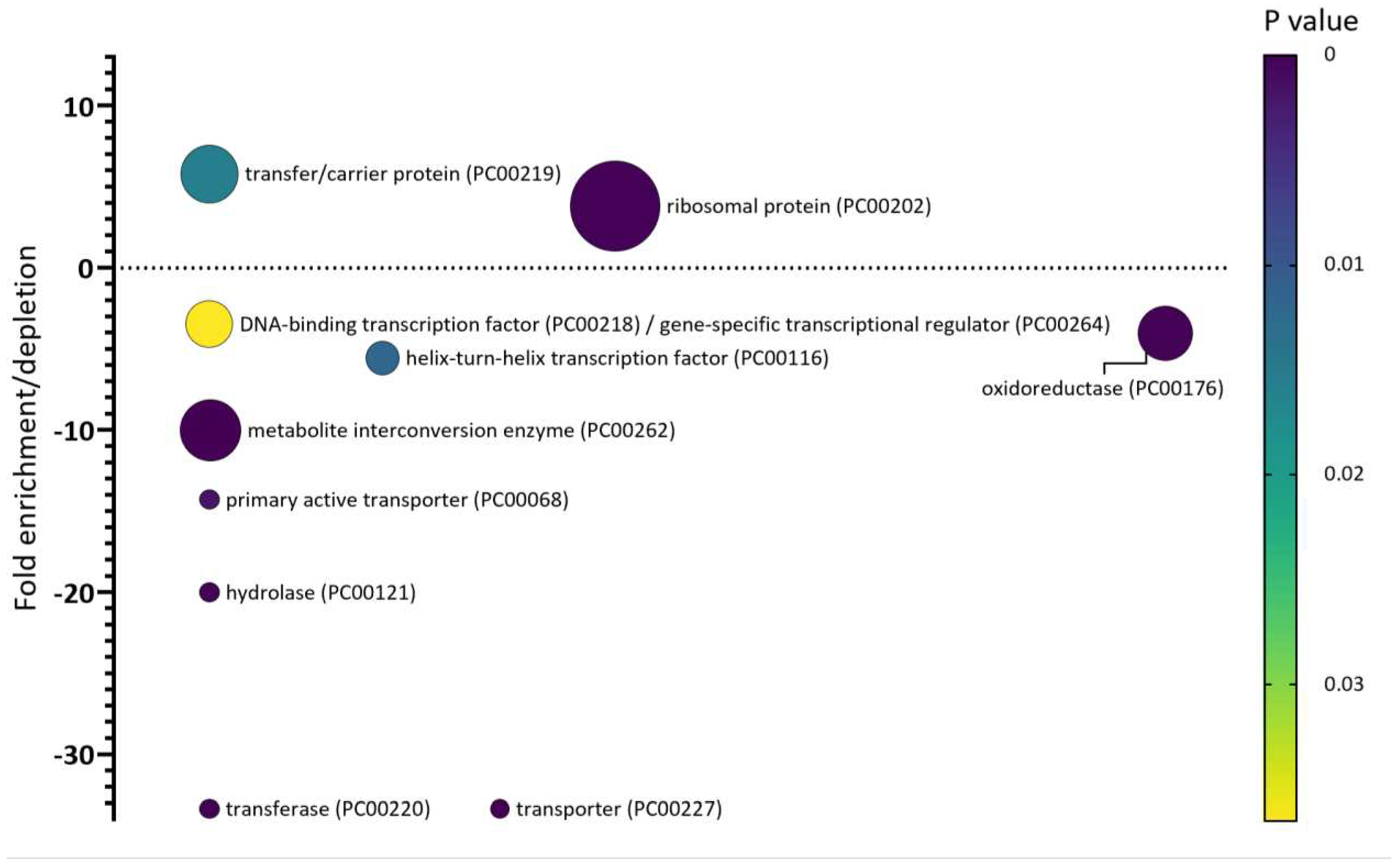
The SEP arsenal of Salmonella
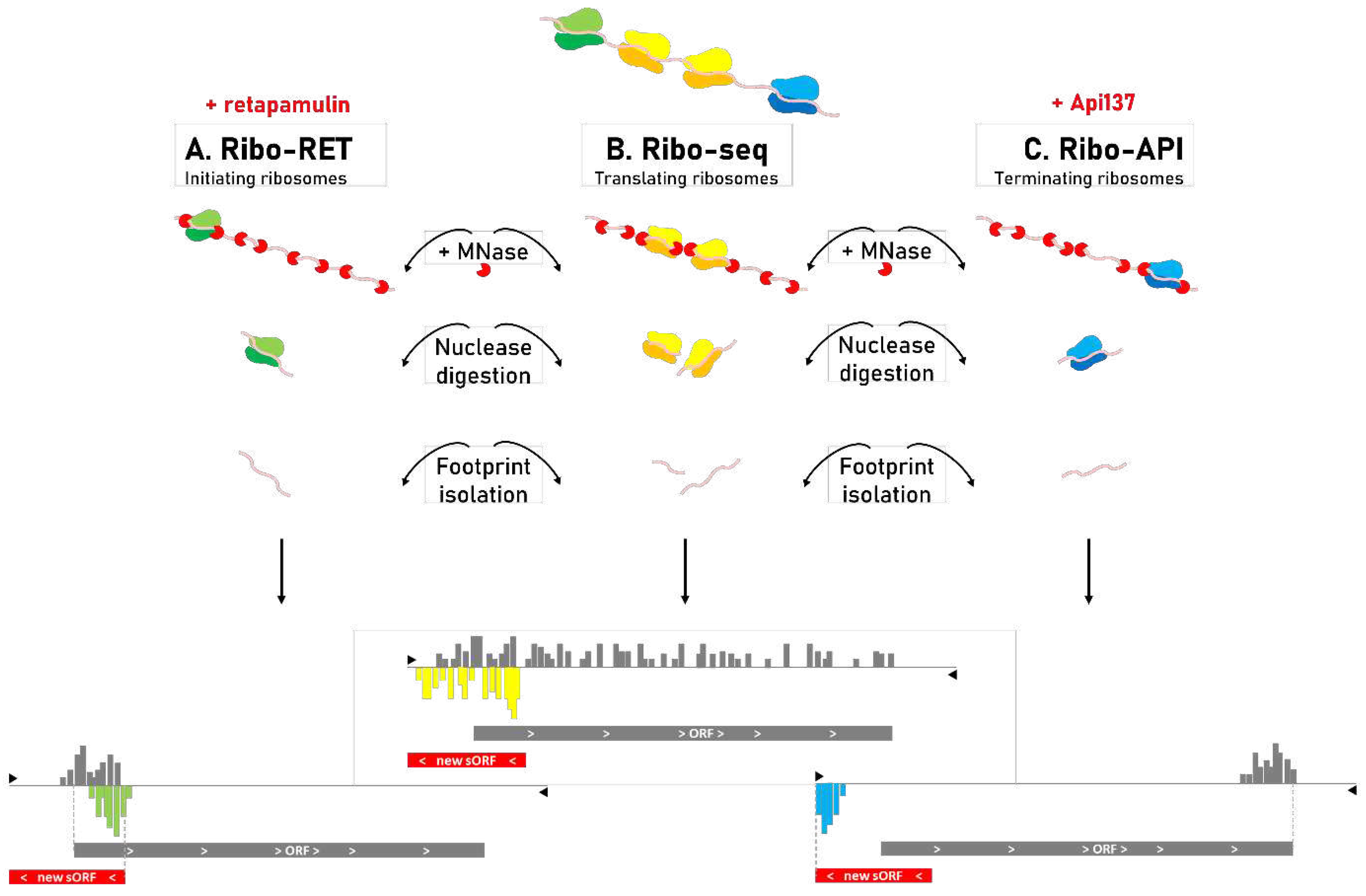
Ribo-seq: a game changer for genome (re)annotation
SEPs: the thorns in the eye of standard protein detection methods
Empirical SEP discovery is hindered by biochemical peculiarities
Experimental SEP validation suffers the same flaws
Current trends in dealing with sORFs and their encoded SEPs
State-of-the-art in the genomic discovery of sORFs
State-of-the-art in experimental SEP validation and functional SEP studies
The ultimate aim: functional characterization of validated bacterial SEPs
Concluding Remarks and Future Perspectives
| State-of-the-art | Pros | Cons | Suggested improvements | |
|---|---|---|---|---|
| sORF prediction and annotation | Ribo-seq | - Genome-wide - Independent from existing annotations - Indicative of ribosomal activity - Broadly applicable - Improved resolution for detection of start (Ribo-RET) and stop codons (Ribo-API) |
- Requirement for experimental SEP validation - Computationally intensive and complex - Poor data resolution inherent to bacterial Ribo-seq |
Refinement of bacterial Ribo-seq protocols and data-analysis |
| SEP expression validation | MS | - SEP abundance data - Proteome-wide technique suited for empirical SEP discovery |
- Limited number of tryptic SEP peptides - Hydrophobic character of peptides - Sensitivity of detection |
- Use of alternative proteases (e.g. chymotrypsin) - High-MW protein depletion or low-MW protein enrichment |
| (Immuno) blotting | - Information on MW and thus SEP integrity - Quantification of SEP expression |
- Tag interference on SEP function/localization - Small SEP size - Sensitivity not adequate for low SEP abundances |
- Use of smaller, charge-neutral tags (e.g. HiBiT) - SEP specific customization (e.g., blotting membrane (type, pore size), blotting buffer and method) - In solution detection of SEPs |
|
| SEP functional characterization | Conservation analysis | - first impression of SEP functioning - High-throughput screening |
Lower conservation of SEPs | - Interrogation of gene co-occurrence - RNA secondary structure analysis |
| Domain prediction | First impression of SEP functioning and localization | Too short primary SEP sequences for domain prediction | Motif prediction (e.g. transmembrane motifs) | |
| Mutation analysis | Targeted and multiplex approach | Laboursome | ||
| Expression analysis | Conditional impact of expression unknown | Conditional expression maps |
Acknowledgments
References
- Gualerzi, C.O. and Pon, C.L. (2015) Initiation of mRNA translation in bacteria: Structural and dynamic aspectsCellular and Molecular Life Sciences, 72Birkhauser Verlag AG, 4341–4367.
- Gawron, D. et al. (2014) The proteome under translational controlProteomics, 14Wiley-VCH Verlag, 2647–2662.
- Weaver, J. et al. (2019) Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10.
- Meydan, S. et al. (2019) Retapamulin-Assisted Ribosome Profiling Reveals the Alternative Bacterial Proteome. Mol Cell 74, 481-493.e6. [CrossRef]
- Vazquez-Laslop, N. et al. (2022) Identifying Small Open Reading Frames in Prokaryotes with Ribosome Profiling. in Journal of Bacteriology, 204. [CrossRef]
- Willems, P. et al. (2022) To New Beginnings: Riboproteogenomics Discovery of N-Terminal Proteoforms in Arabidopsis Thaliana. Front Plant Sci 12, 1–18. [CrossRef]
- Ingolia, N.T. et al. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (1979) 324, 218–223. [CrossRef]
- Buskirk, A.R. and Green, R. (2017) Ribosome pausing, arrest and rescue in bacteria and eukaryotesPhilosophical Transactions of the Royal Society B: Biological Sciences, 372Royal Society.
- Guo, H. et al. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. [CrossRef]
- Ingolia, N.T. et al. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [CrossRef]
- Li, G.W. et al. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. [CrossRef]
- Hucker, S.M. et al. (2017) Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome. PLoS One 12. [CrossRef]
- VanOrsdel, C.E. et al. (2018) Identifying New Small Proteins in Escherichia coli. Proteomics 18, 1–8.
- Li, G.W. et al. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541. [CrossRef]
- Ndah, E. et al. (2017) REPARATION: Ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acids Res 45. [CrossRef]
- Baek, J. et al. (2017) Identification of unannotated small genes in Salmonella. G3: Genes, Genomes, Genetics 7, 983–989.
- Venturini, E. et al. (2020) A global data-driven census of Salmonella small proteins and their potential functions in bacterial virulence . microLife 1, 1–20. [CrossRef]
- Laczkovich, I. et al. (2022) Discovery of Unannotated Small Open Reading Frames in Streptococcus pneumoniae D39 Involved in Quorum Sensing and Virulence Using Ribosome Profiling. mBio 13. [CrossRef]
- Smith, C. et al. (2022) Pervasive translation in Mycobacterium tuberculosis. 11, 73980. [CrossRef]
- Bartholomaus, A. et al. (2021) SmORFer: A modular algorithm to detect small ORFs in prokaryotes. Nucleic Acids Res 49. [CrossRef]
- Froschauer, K. et al. Complementary Ribo-seq approaches map the translatome and provide a small protein census in the foodborne pathogen Campylobacter jejuni. [CrossRef]
- Michel, A.M. et al. (2014) GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res 42. [CrossRef]
- Araújo-Bazán, L. et al. (2019) Synthetic developmental regulator MciZ targets FtsZ across Bacillus species and inhibits bacterial division. Mol Microbiol 111, 965–980. [CrossRef]
- Burby, P.E. and Simmons, L.A. (2020) Regulation of cell division in bacteria by monitoring genome integrity and DNA replication statusJournal of Bacteriology, 202American Society for Microbiology.
- Sweet, M.E. et al. (2021) Structural basis for potassium transport in prokaryotes by KdpFABC. Proc Natl Acad Sci U S A 118, 1–9. [CrossRef]
- Wang, H. et al. (2017) Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc Natl Acad Sci U S A 114, 5689–5694. [CrossRef]
- Yoshitani, K. et al. (2019) Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control. Biosci Biotechnol Biochem 83, 684–694.
- Xu, J. et al. (2019) MgrB affects the acid stress response of Escherichia coli by modulating the expression of iraM. FEMS Microbiol Lett 366.
- Olvera, M.R. et al. (2019) Synthetic hydrophobic peptides derived from MgtR weaken Salmonella pathogenicity and work with a different mode of action than endogenously produced peptides. Sci Rep 9, 1–13. [CrossRef]
- Yadavalli, S.S. et al. (2020) Functional determinants of a small protein controlling a broadly conserved bacterial sensor kinase. J Bacteriol 202. [CrossRef]
- Williams, A.H. et al. (2019) The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat Commun 10. [CrossRef]
- Sur, V.P. et al. (2022) Dynamic study of small toxic hydrophobic proteins PepA1 and PepG1 of Staphylococcus aureus. Int J Biol Macromol. [CrossRef]
- Gray, T. et al. (2021) Small Proteins; Big Questions. J Bacteriol. [CrossRef]
- Fijalkowski, I. et al. (2021) Small Protein Enrichment Improves Proteomics Detection of sORF Encoded Polypeptides. Front Genet 12, 2042. [CrossRef]
- Fijalkowski, I. et al. (2022) Hidden in plain sight : challenges in proteomics detection of small ORF-encoded polypeptides. [CrossRef]
- Stringer, A. et al. (2022) Identification of Novel Translated Small Open Reading Frames in Escherichia coli Using Complementary Ribosome Profiling Approaches. [CrossRef]
- Loman, N.J. and Pallen, M.J. (2015) Twenty years of bacterial genome sequencing. Nat Rev Microbiol 13, 787–794. [CrossRef]
- Land, M. et al. (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15, 141–161. [CrossRef]
- Dorado, G. et al. (2021) Analyzing modern biomolecules: The revolution of nucleic-acid sequencing-review. Biomolecules 11, 1–18. [CrossRef]
- van Dijk, E.L. et al. (2018) The Third Revolution in Sequencing Technology. Trends in Genetics 34, 666–681.
- Ye, C. et al. (2020) Unculturable and culturable periodontal-related bacteria are associated with periodontal inflammation during pregnancy and with preterm low birth weight delivery. Sci Rep 10. [CrossRef]
- Fels, U. et al. (2020) Bacterial Genetic Engineering by Means of Recombineering for Reverse Genetics. Front Microbiol 11, 1–19. [CrossRef]
- Tanizawa, Y. et al. (2018) DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. [CrossRef]
- Schwengers, O. et al. (2021) Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7. [CrossRef]
- Syberg-Olsen, M.J. et al. (2021) Pseudofinder : detection of pseudogenes in prokaryotic genomes. bioRxiv 39, 1–7. [CrossRef]
- Zhao, Y. et al. (2018) PGAP-X: Extension on pan-genome analysis pipeline. BMC Genomics 19. [CrossRef]
- Fijalkowska, D. et al. (2020) Bacterial riboproteogenomics: The era of N-terminal proteoform existence revealed. FEMS Microbiol Rev 44, 418–431. [CrossRef]
- Cao, S. et al. (2021) Proteogenomic discovery of sORF-encoded peptides associated with bacterial virulence in Yersinia pestis. Commun Biol 4.
- Dong, Y. et al. (2021) Genome annotation of disease-causing microorganisms. Brief Bioinform 22, 845–854. [CrossRef]
- Danchin, A. et al. (2018) No wisdom in the crowd: genome annotation in the era of big data – current status and future prospects. Microb Biotechnol 11, 588–605. [CrossRef]
- Dziurzynski, M. et al. (2021) Simple, Reliable, and Time-Efficient Manual Annotation of Bacterial Genomes with MAISEN. Methods in Molecular Biology 2242, 221–229.
- Lobb, B. et al. (2020) An assessment of genome annotation coverage across the bacterial tree of life. Microb Genom 6. [CrossRef]
- Hemm, M.R. et al. (2010) Small stress response proteins in Escherichia coli: Proteins missed by classical proteomic studies. J Bacteriol 192, 46–58. [CrossRef]
- Storz, G. et al. (2014) Small proteins can no longer be ignored. Annu Rev Biochem 83, 753–777. [CrossRef]
- Andrews, S.J. and Rothnagel, J.A. (2014) Emerging evidence for functional peptides encoded by short open reading framesNature Reviews Genetics, 15Nature Publishing Group, 193–204.
- Mudge, J.M. et al. (2022) Standardized annotation of translated open reading framesNature Biotechnology, 40Nature Research, 994–999.
- Willems, P. et al. (2020) Lost and Found: Re-searching and Re-scoring Proteomics Data Aids Genome Annotation and Improves Proteome Coverage. [CrossRef]
- Finkel, Y. et al. (2018) Viral Short ORFs and Their Possible Functions. Proteomics 18, 1–8. [CrossRef]
- Lluch-Senar, M. et al. (2015) Defining a minimal cell: essentiality of small ORF s and nc RNA s in a genome-reduced bacterium . Mol Syst Biol 11, 780. [CrossRef]
- Martin, J.E. et al. (2015) The Escherichia coli Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese. PLoS Genet 11, 1–31. [CrossRef]
- Wassarman, K.M. et al. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15, 1637–1651. [CrossRef]
- Hemm, M.R. et al. (2020) Escherichia coli Small Proteome . EcoSal Plus 9.
- Gaßel, M. et al. (1999) The KdpF Subunit Is Part of the K-translocating Kdp Complex of Escherichia coli and Is Responsible for Stabilization of the Complex in Vitro*. [CrossRef]
- Andresen, L. et al. (2020) The small toxic salmonella protein timp targets the cytoplasmic membrane and is repressed by the small rna timr. mBio 11, 1–16. [CrossRef]
- Wang, N. et al. (2021) sORF-encoded polypeptide SEP1 Is a novel virulence factor of phytophthora pathogens. Molecular Plant-Microbe Interactions 34, 157–167.
- Fozo, E.M. et al. (2008) Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol 70, 1076–1093. [CrossRef]
- Impens, F. et al. (2017) N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat Microbiol 2.
- Miravet-Verde, S. et al. (2019) Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol 15, 1–17. [CrossRef]
- Garai, P. and Blanc-Potard, A. (2020) Uncovering small membrane proteins in pathogenic bacteria: Regulatory functions and therapeutic potential. Mol Microbiol 114, 710–720. [CrossRef]
- Yadavalli, S.S. and Yuan, J. (2022) Bacterial Small Membrane Proteins: The Swiss Army Knife of Regulators at the Lipid Bilayer. in Journal of Bacteriology, 204. [CrossRef]
- Giess, A. et al. (2017) Ribosome signatures aid bacterial translation initiation site identification. BMC Biol 15, 1–14. [CrossRef]
- Ochman, H. et al. (1999) Calibrating bacterial evolution. Proc Natl Acad Sci U S A 96, 12638–12643.
- Chaudhuri, D. et al. (2018) Salmonella Typhimurium infection leads to colonization of the mouse brain and is not completely cured with antibiotics. Front Microbiol 9, 1–12. [CrossRef]
- Michaux, C. et al. (2017) RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc Natl Acad Sci U S A 114, 6824–6829. [CrossRef]
- Wang, Y. et al. (2020) Recent advances in ribosome profiling for deciphering translational regulation. Methods 176, 46–54. [CrossRef]
- Pavesi, A. et al. (2018) Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes. PLoS One 13, 1–24. [CrossRef]
- Wright, B.W. et al. (2022) Overlapping genes in natural and engineered genomes. Nat Rev Genet 23, 154–168. [CrossRef]
- Olexiouk, V. et al. (2018) An update on sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 46, D497–D502. [CrossRef]
- Mohammad, F. et al. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. [CrossRef]
- Fremin, B.J. and Bhatt, A.S. (2020) Structured RNA Contaminants in Bacterial Ribo-Seq. mSphere 5. [CrossRef]
- Meleady, P. (2018) Two-dimensional gel electrophoresis and 2D-DIGE. Methods in Molecular Biology 1664, 3–14.
- Lee, P.Y. et al. (2020) The evolution of two-dimensional gel electrophoresis - from proteomics to emerging alternative applications. J Chromatogr A 1615, 460763. [CrossRef]
- Kielkopf, C.L. et al. (2021) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins. Cold Spring Harb Protoc 2021, 494–504. [CrossRef]
- Gao, Z. et al. (2019) AP3: An Advanced Proteotypic Peptide Predictor for Targeted Proteomics by Incorporating Peptide Digestibility. Anal Chem 91, 8705–8711. [CrossRef]
- Cassidy, L. et al. (2019) Depletion of High-Molecular-Mass Proteins for the Identification of Small Proteins and Short Open Reading Frame Encoded Peptides in Cellular Proteomes. J Proteome Res 18, 1725–1734. [CrossRef]
- Becher, D. et al. (2020) Optimized proteomics workflow for the detection of small proteins. J Proteome Res 19, 4004–4018. [CrossRef]
- Ahrens, C.H. et al. (2022) A Practical Guide to Small Protein Discovery and Characterization Using Mass Spectrometry. [CrossRef]
- Kurien, B.T. and Hal Scofield, R. (2015) Western Blotting: An Introduction. Methods Mol Biol 1312, 17.
- Tomisawa, S. et al. (2013) A new approach to detect small peptides clearly and sensitively by Western blotting using a vacuum-assisted detection method. Biophysics (Japan) 9, 79–83. [CrossRef]
- Olexiouk, V. et al. (2016) SORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 44, D324–D329. [CrossRef]
- Peeters, M.K.R. and Menschaert, G. (2020) The hunt for sORFs: A multidisciplinary strategy. Exp Cell Res 391. [CrossRef]
- Kubatova, N. et al. (2020) Rapid Biophysical Characterization and NMR Spectroscopy Structural Analysis of Small Proteins from Bacteria and Archaea. ChemBioChem 21, 1178–1187. [CrossRef]
- Uversky, V.N. (2017) Paradoxes and wonders of intrinsic disorder: Stability of instability. Intrinsically Disord Proteins 5, e1327757. [CrossRef]
- Yu, J. et al. (2021) Comprehensive evaluation of protein-coding sORFs prediction based on a random sequence strategy. Frontiers in Bioscience - Landmark 26, 272–278. [CrossRef]
- Gelhausen, R. et al. (2022) RiboReport - Benchmarking tools for ribosome profiling-based identification of open reading frames in bacteria. Brief Bioinform 23, 1–22. [CrossRef]
- Hopp, T.P. et al. (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology 6, 1204–1210. [CrossRef]
- Zeghouf, M. et al. (2004) Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res 3, 463–468. [CrossRef]
- Vandemoortele, G. et al. (2019) Pick a Tag and Explore the Functions of Your Pet Protein. Trends Biotechnol 37, 1078–1090. [CrossRef]
- Booth, W.T. et al. (2018) Impact of an N-terminal polyhistidine tag on protein thermal stability. ACS Omega 3, 760–768. [CrossRef]
- Munadziroh, E. et al. (2020) Effect of poly-histidine tag position toward inhibition activity of secretory leukocyte protease inhibitor as candidate for material wound healing. Avicenna J Med Biotechnol 12, 32–36.
- Zhou, C. et al. (2022) Probing the sORF-Encoded Peptides of Deinococcus radiodurans in Response to Extreme Stress. Molecular & Cellular Proteomics 21, 100423.
- Xiong, J. (2006) Protein Motifs and Domain Prediction. Essential Bioinformatics. [CrossRef]
- Srikumar, S. et al. (2015) RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella Typhimurium. PLoS Pathog 11. [CrossRef]
- Kitata, R.B. et al. (2022) Advances in data-independent acquisition mass spectrometry towards comprehensive digital proteome landscapeMass Spectrometry ReviewsJohn Wiley and Sons Inc.
- Keeling, D.M. et al. The meanings of “function” in biology and the problematic case of de novo gene emergence. [CrossRef]
- Liu, X. et al. (2020) Combined proximity labeling and affinity purification−mass spectrometry workflow for mapping and visualizing protein interaction networks, 15. [CrossRef]
- Samavarchi-Tehrani, P. et al. (2020) Proximity dependent biotinylation: Key enzymes and adaptation to proteomics approaches. Molecular and Cellular Proteomics 19, 757–773. [CrossRef]
- Santin, Y.G. et al. (2018) In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat Microbiol 3, 1304–1313. [CrossRef]
- Guard, J. (2022) Through the Looking Glass: Genome, Phenome, and Interactome of Salmonella entericaPathogens, 11MDPI.
- Todor, H. et al. (2021) Bacterial CRISPR screens for gene functionCurrent Opinion in Microbiology, 59Elsevier Ltd, 102–109.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





