Submitted:
12 April 2023
Posted:
13 April 2023
You are already at the latest version
Abstract
Keywords:
Introduction
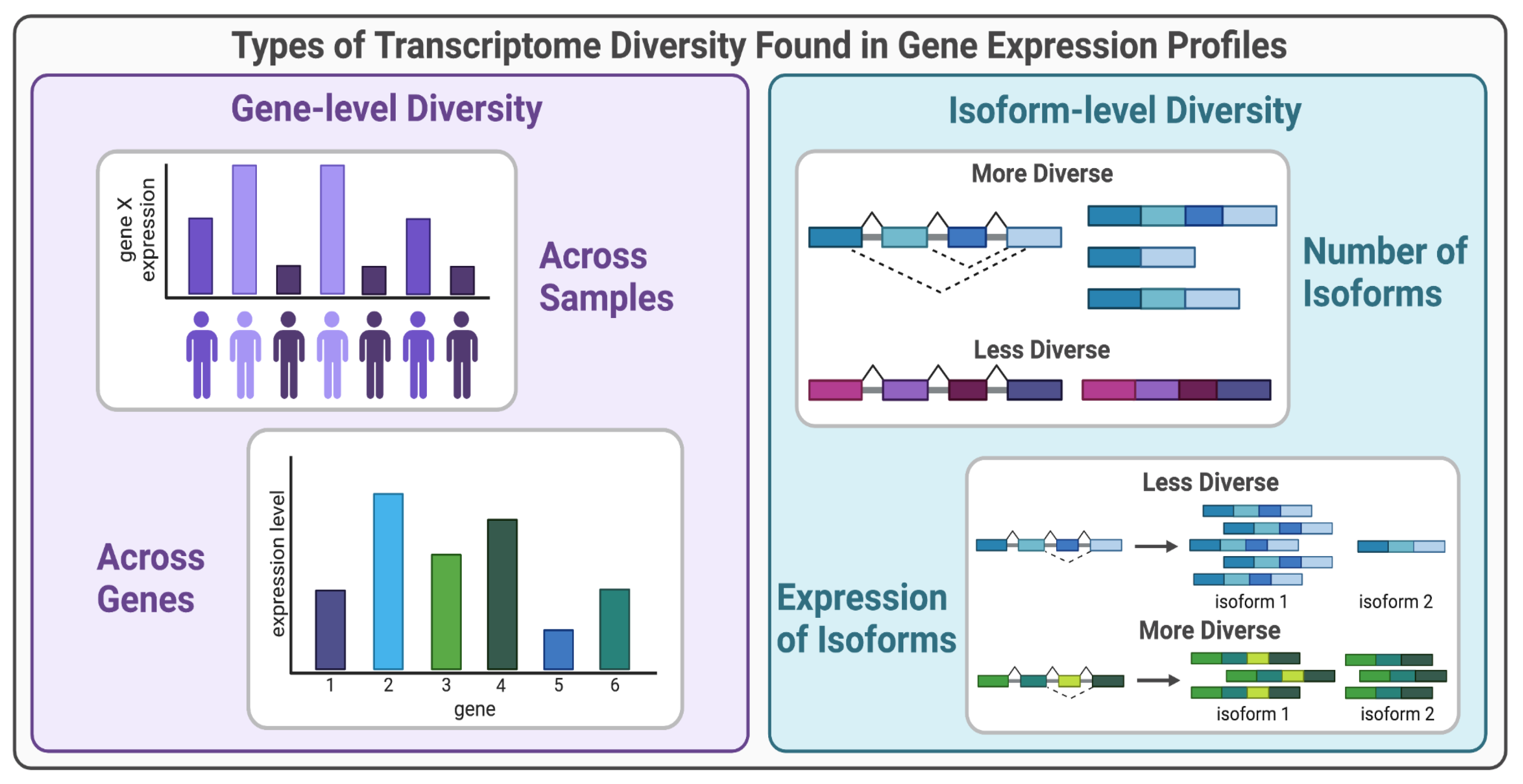
I. Gene-level Diversity in Gene Expression Profiles
A. Biological Processes that Lead to Gene-level Diversity
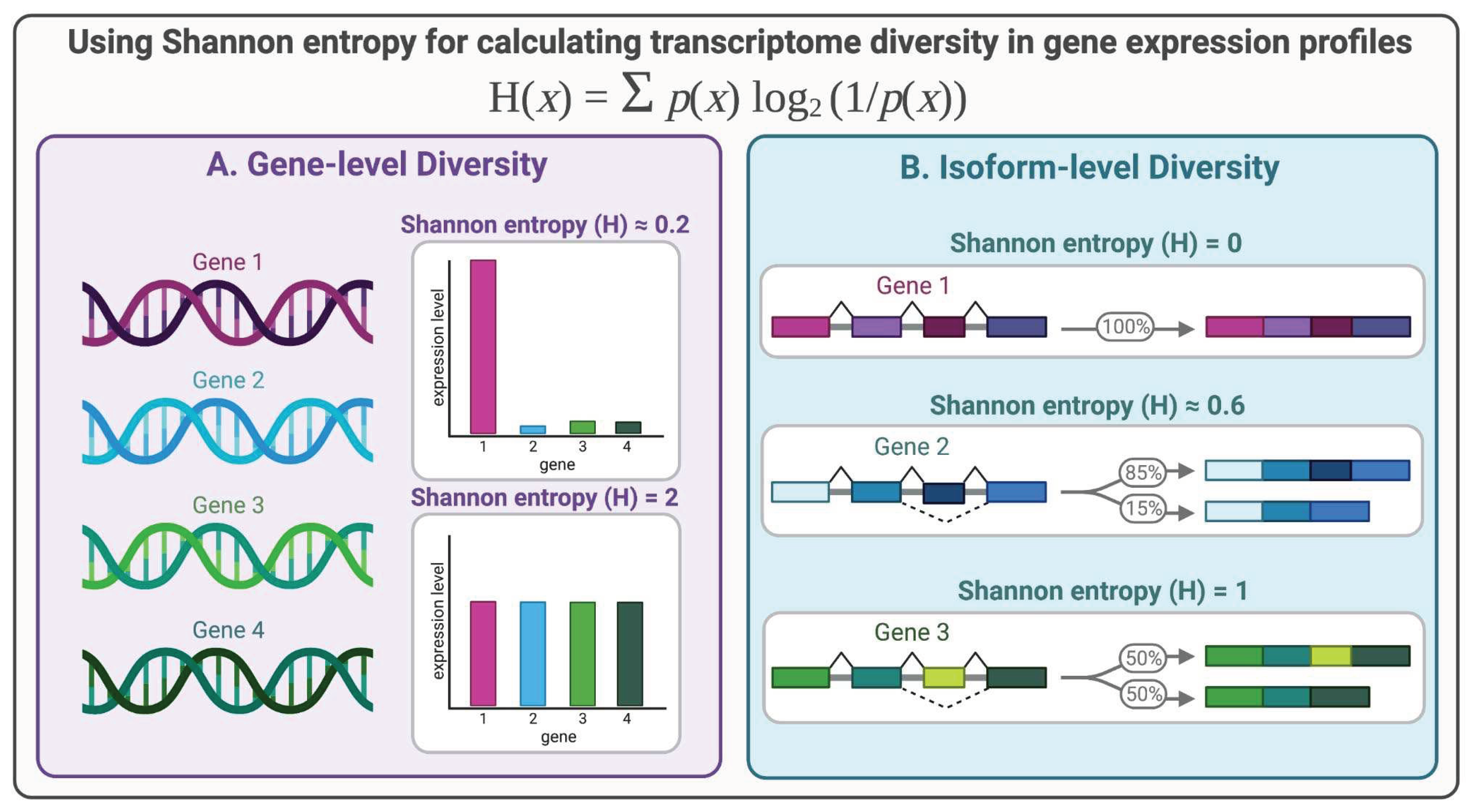
B. Methods for Quantifying Gene-level Transcriptome Diversity
II. Isoform-level Diversity in Gene Expression Profiles
A. Biological Processes that Lead to Isoform-level Diversity
B. Methods for Quantifying Isoform-level Diversity
| Package Name | Year | Bulk or Single Cell | Analysis Type: Exon/Transcript or Other | Citation count | Language |
| Insplico [141] | 2023 | Both | Other - Splicing Order | 0 | Perl |
| acorde [142] | 2022 | Single-cell | DTU and coDTU | 2 | R |
| SpliZ [143] | 2022 | Single-cell | DEU (PSI) | 4 | Python |
| DTUrtle [144] | 2021 | Both | DTU | 3 | R |
| NanoCount [145] | 2021 | Bulk | DTU | 11 | R |
| SplicingFactory [140] | 2021 | Bulk | Other - Diversity | 0 | R |
| scisorseqr [146] | 2021 | Single-cell | DTU (modified) | 39 | R |
| satuRn [147] | 2021 | Both | DTU | 0 | R |
| ASCOT [112] | 2020 | Single-cell | DEU (PSI) | 24 | Python |
| BANDITS [148] | 2020 | Bulk | DTU | 10 | R |
| Sierra [149] | 2020 | Single-cell | DTU | 28 | R |
| RATs [150] | 2019 | Bulk | DTU | 10 | R |
| SUPPA2 [151] | 2018 | Bulk | DEU (PSI) | 193 | Python |
| LeafCutter [152] | 2018 | Bulk | Other - Intron Excision | 246 | R/Python |
| Whippet [139] | 2018 | Bulk | DTU | 61 | Julia |
| GSReg/SEVA [122] | 2018 | Bulk | Other - Variability | 6 | R |
| IsoformSwitchAnalyzeR [153] | 2017 | Bulk | DTU | 104 | R |
| Census/Monocle [154] | 2017 | Single-cell | DEU (PSI) | 610 | R |
| BRIE [155] | 2017 | Single-cell | DEU (PSI) | 50 | Python |
| DRIM-Seq [156] | 2016 | Bulk | DTU | 49 | R |
| JunctionSeq [157] | 2016 | Bulk | DEU (PSI) | 81 | R |
| MAJIQ [158] | 2016 | Bulk | DEU (PSI) | 188 | Python/C++ |
| SGSeq [159] | 2016 | Bulk | DEU (PSI) | 63 | R |
| SingleSplice [160] | 2016 | Single-cell | DTU | 36 | R/Perl |
| Limma (diffSplice) [22] | 2015 | Bulk | DEU (PSI) | 15473 | R |
| VAST-TOOLS [161] | 2014 | Bulk | DTU | 339 | R/Perl |
| rMATS [162] | 2014 | Bulk | DEU (PSI) | 982 | Python/C++ |
| CuffDiff2 [163] | 2013 | Bulk | DEU (PSI) | 2341 | C++ |
| SplicingCompass [164] | 2013 | Bulk | DTU | 39 | R |
| DEXSeq [165] | 2012 | Bulk | DEU (PSI) | 874 | R |
| SpliceTrap [123] | 2011 | Bulk | DEU (PSI) | 59 | C++/Perl |
| MISO [166] | 2010 | Bulk | DEU (PSI) | 876 | Python/C |
Conclusion
Acknowledgments
References
- Mantione KJ, Kream RM, Kuzelova H, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [CrossRef]
- Niedringhaus TP, Milanova D, Kerby MB, et al. Landscape of next-generation sequencing technologies. Anal. Chem. 2011, 83, 4327–4341. [CrossRef] [PubMed]
- Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [CrossRef] [PubMed]
- Haque A, Engel J, Teichmann SA, et al. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75.
- Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [CrossRef]
- Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [CrossRef] [PubMed]
- Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [CrossRef]
- Garalde DR, Snell EA, Jachimowicz D, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201–206. [CrossRef]
- Amarasinghe SL, Su S, Dong X, et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30.
- Parker MT, Knop K, Sherwood AV, et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife 2020, 9.
- Tian L, Jabbari JS, Thijssen R, et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 2021, 22, 1–24.
- Singh M, Al-Eryani G, Carswell S, et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019, 10, 3120. [CrossRef] [PubMed]
- Hardwick SA, Hu W, Joglekar A, et al. Single-nuclei isoform RNA sequencing unlocks barcoded exon connectivity in frozen brain tissue. Nat. Biotechnol. 2022, 1–11.
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [CrossRef] [PubMed]
- Melé M, Ferreira PG, Reverter F, et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015, 348, 660–665. [CrossRef]
- Manczak M, Park BS, Jung Y, et al. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004, 5, 147–162. [CrossRef] [PubMed]
- Brown J 3rd, Theisler C, Silberman S, et al. Differential expression of cholesterol hydroxylases in Alzheimer’s disease. J. Biol. Chem. 2004, 279, 34674–34681. [CrossRef]
- Ho JWK, Stefani M, dos Remedios CG, et al. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics 2008, 24, i390–8. [CrossRef]
- Ran D, Daye ZJ. Gene expression variability and the analysis of large-scale RNA-seq studies with the MDSeq. Nucleic Acids Res. 2017, 45, e127. [CrossRef]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550.
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [CrossRef]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015, 43, e47. [CrossRef] [PubMed]
- Wang K, Phillips CA, Rogers GL, et al. Differential Shannon entropy and differential coefficient of variation: alternatives and augmentations to differential expression in the search for disease-related genes. Int. J. Comput. Biol. Drug Des. 2014, 7, 183–194. [CrossRef] [PubMed]
- Porcu E, Sadler MC, Lepik K, et al. Differentially expressed genes reflect disease-induced rather than disease-causing changes in the transcriptome. Nat. Commun. 2021, 12, 5647. [CrossRef] [PubMed]
- Hudson NJ, Dalrymple BP, Reverter A. Beyond differential expression: the quest for causal mutations and effector molecules. BMC Genomics 2012, 13, 356.
- Squair JW, Gautier M, Kathe C, et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 2021, 12, 5692. [CrossRef]
- Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–62. [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021, 49, D325–D334. [CrossRef] [PubMed]
- de la Fuente, A. From ‘differential expression’ to ‘differential networking’ – identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010, 26, 326–333. [Google Scholar] [CrossRef]
- Glass K, Huttenhower C, Quackenbush J, et al. Passing messages between biological networks to refine predicted interactions. PLoS One 2013, 8, e64832.
- de Jong TV, Moshkin YM, Guryev V. Gene expression variability: the other dimension in transcriptome analysis. Physiol. Genomics 2019, 51, 145–158. [CrossRef] [PubMed]
- Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [CrossRef]
- Storey JD, Madeoy J, Strout JL, et al. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007, 80, 502–509. [CrossRef] [PubMed]
- Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science 2005, 309, 2010–2013. [CrossRef] [PubMed]
- Wang Z, Zhang J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, E67–76.
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007, 3, 1724–1735.
- Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 2002, 13, 1977–2000. [CrossRef]
- Zhang R, Lahens NF, Ballance HI, et al. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 16219–16224. [CrossRef]
- Viñuela A, Snoek LB, Riksen JAG, et al. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010, 20, 929–937. [CrossRef]
- Viñuela A, Brown AA, Buil A, et al. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum. Mol. Genet. 2018, 27, 732–741. [CrossRef]
- Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [CrossRef]
- Nott A, Holtman IR, Coufal NG, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 2019, 366, 1134–1139. [CrossRef]
- Butler JEF, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592. [CrossRef]
- Danino YM, Even D, Ideses D, et al. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [CrossRef]
- Duan P, Xu J, Zeng D, et al. Natural Variation in the Promoter of GSE5 Contributes to Grain Size Diversity in Rice. Mol. Plant 2017, 10, 685–694. [CrossRef]
- Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [CrossRef]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [CrossRef]
- Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [CrossRef]
- Lambert SA, Jolma A, Campitelli LF, et al. The Human Transcription Factors. Cell 2018, 172, 650–665.
- Papavassiliou AG. Molecular medicine. Transcription factors. N. Engl. J. Med. 1995, 332, 45–47. [CrossRef]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [CrossRef]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [CrossRef]
- Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [CrossRef]
- Jones, PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Karlić R, Chung H-R, Lasserre J, et al. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 2926–2931. [CrossRef]
- Cheng C, Yan K-K, Yip KY, et al. A statistical framework for modeling gene expression using chromatin features and application to modENCODE datasets. Genome Biol. 2011, 12, R15.
- Araki Y, Wang Z, Zang C, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 2009, 30, 912–925. [CrossRef]
- Feidantsis K, Giantsis IA, Vratsistas A, et al. Correlation between intermediary metabolism, Hsp gene expression, and oxidative stress-related proteins in long-term thermal-stressed Mytilus galloprovincialis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020, 319, R264–R281. [CrossRef]
- Hasthanasombut, Paisarnwipatpong. Expression of OsBADH1 gene in Indica rice (Oryza sativa L.) in correlation with salt, plasmolysis, temperature and light stresses. Plant Omics.
- Zhang TY, Labonté B, Wen XL, et al. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology 2013, 38, 111–123. [CrossRef]
- Mar JC, Matigian NA, Mackay-Sim A, et al. Variance of gene expression identifies altered network constraints in neurological disease. PLoS Genet. 2011, 7, e1002207.
- Komurov K, Ram PT. Patterns of human gene expression variance show strong associations with signaling network hierarchy. BMC Syst. Biol. 2010, 4, 154.
- Alemu EY, Carl JW Jr, Corrada Bravo H, et al. Determinants of expression variability. Nucleic Acids Res. 2014, 42, 3503–3514.
- Bashkeel N, Perkins TJ, Kærn M, et al. Human gene expression variability and its dependence on methylation and aging. BMC Genomics 2019, 20, 941.
- Simonovsky E, Schuster R, Yeger-Lotem E. Large-scale analysis of human gene expression variability associates highly variable drug targets with lower drug effectiveness and safety. Bioinformatics 2019, 35, 3028–3037. [CrossRef]
- Igolkina AA, Armoskus C, Newman JRB, et al. Analysis of Gene Expression Variance in Schizophrenia Using Structural Equation Modeling. Front. Mol. Neurosci. 2018, 11, 192. [CrossRef]
- Sturm G, List M, Zhang JD. Tissue heterogeneity is prevalent in gene expression studies. NAR Genom Bioinform 2021, 3, lqab077.
- Bachtiary B, Boutros PC, Pintilie M, et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin. Cancer Res. 2006, 12, 5632–5640. [CrossRef]
- Wolf S, Melo D, Garske KM, et al. Characterizing the landscape of gene expression variance in humans. bioRxiv 2023. 2022.11.15.516646.
- Pervouchine DD, Djebali S, Breschi A, et al. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat. Commun. 2015, 6, 5903. [CrossRef]
- Breschi A, Djebali S, Gillis J, et al. Gene-specific patterns of expression variation across organs and species. Genome Biol. 2016, 17, 151.
- Chen Y, Davidson NM, Wan YK, et al. A systematic benchmark of Nanopore long read RNA sequencing for transcript level analysis in human cell lines. bioRxiv 2021. 2021.04.21.440736.
- Shannon, CE. A Mathematical Theory of Communication. Bell System Technical Journal 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Martínez O, Reyes-Valdés MH. Defining diversity, specialization, and gene specificity in transcriptomes through information theory. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9709–9714. [CrossRef]
- Ameri AJ, Lewis ZA. Shannon entropy as a metric for conditional gene expression in Neurospora crassa. G3 2021, 11.
- Fuhrman S, Cunningham MJ, Wen X, et al. The application of shannon entropy in the identification of putative drug targets. Biosystems. 2000, 55, 5–14. [CrossRef]
- Schug J, Schuller W-P, Kappen C, et al. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005, 6, R33.
- Machado T, J. A. Shannon Entropy Analysis of the Genome Code. Math. Probl. Eng. 2012, 2012. [Google Scholar]
- Monaco A, Amoroso N, Bellantuono L, et al. Shannon entropy approach reveals relevant genes in Alzheimer’s disease. PLoS One 2019, 14, e0226190.
- Dérian N, Pham H-P, Nehar-Belaid D, et al. The Tsallis generalized entropy enhances the interpretation of transcriptomics datasets. PLoS One 2022, 17, e0266618.
- Kim MC, Gate R, Lee DS, et al. memento: Generalized differential expression analysis of single-cell RNA-seq with method of moments estimation and efficient resampling. bioRxiv 2022. 2022.11.09.515836.
- Zhang JD, Hatje K, Sturm G, et al. Correction to: Detect tissue heterogeneity in gene expression data with BioQC. BMC Genomics 2018, 19, 558.
- Wang K, Phillips CA, Saxton AM, et al. EntropyExplorer: an R package for computing and comparing differential Shannon entropy, differential coefficient of variation and differential expression. BMC Res. Notes 2015, 8, 832.
- Salzberg, SL. Open questions: How many genes do we have? BMC Biol. 2018, 16, 94. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [CrossRef]
- Barbosa-Morais NL, Irimia M, Pan Q, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [CrossRef]
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [CrossRef]
- Wright CJ, Smith CWJ, Jiggins CD. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022.
- Ayoubi TA, Van De Ven WJ. Regulation of gene expression by alternative promoters. FASEB J. 1996, 10, 453–460. [CrossRef]
- Alasoo K, Rodrigues J, Danesh J, et al. Genetic effects on promoter usage are highly context-specific and contribute to complex traits. Elife 2019, 8.
- Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA*. Proceedings of the National Academy of Sciences 1977, 74, 3171–3175. [CrossRef]
- Patrick E, Buckley M, Yang YH. Estimation of data-specific constitutive exons with RNA-Seq data. BMC Bioinformatics 2013, 14, 31.
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [CrossRef] [PubMed]
- Graveley, BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001, 17, 100–107. [Google Scholar] [CrossRef]
- Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000, 25, 381–388. [CrossRef]
- Wan Y, Larson DR. Splicing heterogeneity: separating signal from noise. Genome Biol. 2018, 19, 86.
- Weatheritt RJ, Sterne-Weiler T, Blencowe BJ. The ribosome-engaged landscape of alternative splicing. Nat. Struct. Mol. Biol. 2016, 23, 1117–1123. [CrossRef]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 1989, 56, 997–1010. [CrossRef]
- McIntyre LM, Bono LM, Genissel A, et al. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006, 7, R79.
- Brown JB, Boley N, Eisman R, et al. Diversity and dynamics of the Drosophila transcriptome. Nature 2014, 512, 393–399. [CrossRef]
- Gibilisco L, Zhou Q, Mahajan S, et al. Alternative Splicing within and between Drosophila Species, Sexes, Tissues, and Developmental Stages. PLoS Genet. 2016, 12, e1006464.
- Naftaly AS, Pau S, White MA. Long-read RNA sequencing reveals widespread sex-specific alternative splicing in threespine stickleback fish. Genome Res. 2021, 31, 1486–1497. [CrossRef] [PubMed]
- Rogers TF, Palmer DH, Wright AE. Sex-Specific Selection Drives the Evolution of Alternative Splicing in Birds. Mol. Biol. Evol. 2021, 38, 519–530. [CrossRef] [PubMed]
- Blekhman R, Marioni JC, Zumbo P, et al. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010, 20, 180–189. [CrossRef] [PubMed]
- Trabzuni D, Ramasamy A, Imran S, et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 2013, 4, 2771. [CrossRef] [PubMed]
- Karlebach G, Veiga DFT, Mays AD, et al. The impact of biological sex on alternative splicing. bioRxiv 2020, 490904.
- Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002, 30, 3754–3766. [CrossRef]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, et al. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008, 36, 4823–4832. [CrossRef]
- Zhang C, Zhang Z, Castle J, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008, 22, 2550–2563. [CrossRef]
- Buljan M, Chalancon G, Eustermann S, et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 2012, 46, 871–883. [CrossRef]
- Zhang X, Chen MH, Wu X, et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 2016, 166, 1147–1162.e15. [CrossRef]
- Ling JP, Wilks C, Charles R, et al. ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat. Commun. 2020, 11, 137. [CrossRef]
- Song Y, Botvinnik OB, Lovci MT, et al. Single-Cell Alternative Splicing Analysis with Expedition Reveals Splicing Dynamics during Neuron Differentiation. Mol. Cell 2017, 67, 148–161.e5. [CrossRef] [PubMed]
- Shalek AK, Satija R, Adiconis X, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [CrossRef] [PubMed]
- Kim HK, Pham MHC, Ko KS, et al. Alternative splicing isoforms in health and disease. Pflugers Arch. 2018, 470, 995–1016. [CrossRef]
- Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [CrossRef] [PubMed]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992, 90, 41–54.
- Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 2015, 347, 1254806. [CrossRef]
- Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [CrossRef]
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [CrossRef]
- Steijger T, Abril JF, Engström PG, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [CrossRef]
- Afsari B, Guo T, Considine M, et al. Splice Expression Variation Analysis (SEVA) for inter-tumor heterogeneity of gene isoform usage in cancer. Bioinformatics 2018, 34, 1859–1867. [CrossRef]
- Wu J, Akerman M, Sun S, et al. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics 2011, 27, 3010–3016. [CrossRef]
- Venables JP, Klinck R, Bramard A, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008, 68, 9525–9531. [CrossRef]
- Merino GA, Conesa A, Fernández EA. A benchmarking of workflows for detecting differential splicing and differential expression at isoform level in human RNA-seq studies. Brief. Bioinform. 2019, 20, 471–481. [CrossRef]
- Soneson C, Matthes KL, Nowicka M, et al. Isoform prefiltering improves performance of count-based methods for analysis of differential transcript usage. Genome Biol. 2016, 17, 12.
- Dougherty ML, Underwood JG, Nelson BJ, et al. Transcriptional fates of human-specific segmental duplications in brain. Genome Res. 2018, 28, 1566–1576. [CrossRef] [PubMed]
- Sharon D, Tilgner H, Grubert F, et al. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013, 31, 1009–1014. [CrossRef] [PubMed]
- ang AD, Soulette CM, van Baren MJ, et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [CrossRef]
- Chen Y, Sim A, Wan Y, Goeke J. bambu: Reference-guided isoform reconstruction and quantification for long read RNA-Seq data. 2022.
- Leung SK, Jeffries AR, Castanho I, et al. Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing. Cell Rep. 2021, 37, 110022. [CrossRef]
- Palmer CR, Liu CS, Romanow WJ, et al. Altered cell and RNA isoform diversity in aging Down syndrome brains. Proc. Natl. Acad. Sci. U. S. A. 2021, 118.
- Ritchie W, Granjeaud S, Puthier D, et al. Entropy measures quantify global splicing disorders in cancer. PLoS Comput. Biol. 2008, 4, e1000011.
- Oguchi Y, Ozaki Y, Abdelmoez MN, et al. NanoSINC-seq dissects the isoform diversity in subcellular compartments of single cells. Sci Adv 2021, 7.
- Padonou F, Gonzalez V, Provin N, et al. Aire-dependent transcripts escape Raver2-induced splice-event inclusion in the thymic epithelium. EMBO Rep. 2022, 23, e53576. [CrossRef] [PubMed]
- Roberts A, Trapnell C, Donaghey J, et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [CrossRef] [PubMed]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [CrossRef] [PubMed]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [CrossRef] [PubMed]
- Sterne-Weiler T, Weatheritt RJ, Best AJ, et al. Efficient and Accurate Quantitative Profiling of Alternative Splicing Patterns of Any Complexity on a Laptop. Mol. Cell 2018, 72, 187–200.e6. [CrossRef]
- Dankó B, Szikora P, Pór T, et al. SplicingFactory-splicing diversity analysis for transcriptome data. Bioinformatics 2021.
- Gohr A, Iñiguez LP, Torres-Méndez A, et al. Insplico: effective computational tool for studying splicing order of adjacent introns genome-wide with short and long RNA-seq reads. Nucleic Acids Res. 2023.
- Arzalluz-Luque A, Salguero P, Tarazona S, et al. acorde unravels functionally interpretable networks of isoform co-usage from single cell data. Nat. Commun. 2022, 13, 1828. [CrossRef] [PubMed]
- Olivieri JE, Dehghannasiri R, Salzman J. The SpliZ generalizes ‘percent spliced in’ to reveal regulated splicing at single-cell resolution. Nat. Methods 2022, 19, 307–310. [CrossRef]
- Tekath T, Dugas M. Differential transcript usage analysis of bulk and single-cell RNA-seq data with DTUrtle. Bioinformatics 2021.
- Gleeson J, Leger A, Prawer YDJ, et al. Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 2022, 50, e19. [CrossRef] [PubMed]
- Joglekar A, Prjibelski A, Mahfouz A, et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 2021, 12, 463. [CrossRef]
- Gilis J, Vitting-Seerup K, Van den Berge K, et al. satuRn: Scalable analysis of differential transcript usage for bulk and single-cell RNA-sequencing applications. F1000Res. 2021, 10, 374. [CrossRef]
- Tiberi S, Robinson MD. BANDITS: Bayesian differential splicing accounting for sample-to-sample variability and mapping uncertainty. Genome Biol. 2020, 21, 69.
- Patrick R, Humphreys DT, Janbandhu V, et al. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 2020, 21, 167.
- Froussios K, Mourão K, Simpson G, et al. Relative Abundance of Transcripts ( RATs): Identifying differential isoform abundance from RNA-seq. F1000Res. 2019, 8, 213. [CrossRef]
- Trincado JL, Entizne JC, Hysenaj G, et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40.
- Li YI, Knowles DA, Humphrey J, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018, 50, 151–158. [CrossRef]
- Vitting-Seerup K, Sandelin A. The Landscape of Isoform Switches in Human Cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [CrossRef] [PubMed]
- Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [CrossRef] [PubMed]
- Huang Y, Sanguinetti G. BRIE: transcriptome-wide splicing quantification in single cells. Genome Biol. 2017, 18, 123.
- Nowicka M, Robinson MD. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Res. 2016, 5, 1356. [CrossRef] [PubMed]
- Hartley SW, Mullikin JC. Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res. 2016, 44, e127.
- Vaquero-Garcia J, Barrera A, Gazzara MR, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 2016, 5, e11752. [CrossRef] [PubMed]
- Goldstein LD, Cao Y, Pau G, et al. Prediction and Quantification of Splice Events from RNA-Seq Data. PLoS One 2016, 11, e0156132. [CrossRef]
- Welch JD, Hu Y, Prins JF. Robust detection of alternative splicing in a population of single cells. Nucleic Acids Res. 2016, 44, e73. [CrossRef]
- Irimia M, Weatheritt RJ, Ellis JD, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 2014, 159, 1511–1523. [CrossRef]
- Shen S, Park JW, Lu Z-X, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E5593–601.
- Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [CrossRef]
- Aschoff M, Hotz-Wagenblatt A, Glatting K-H, et al. SplicingCompass: differential splicing detection using RNA-seq data. Bioinformatics 2013, 29, 1141–1148. [CrossRef]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [CrossRef]
- Katz Y, Wang ET, Airoldi EM, et al. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 2010, 7, 1009–1015. [CrossRef]
- Zhao S, Ye Z, Stanton R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020, 26, 903–909. [CrossRef] [PubMed]
- Van den Berge K, Hembach KM, Soneson C, et al. RNA Sequencing Data: Hitchhiker’s Guide to Expression Analysis. Annu. Rev. Biomed. Data Sci. 2019, 2, 139–173. [CrossRef]
- Jiang R, Sun T, Song D, et al. Statistics or biology: the zero-inflation controversy about scRNA-seq data. Genome Biol. 2022, 23, 31.
- Lun ATL, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016, 17, 75.
- Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019, 15, e8746. [CrossRef] [PubMed]
- Archer N, Walsh MD, Shahrezaei V, et al. Modeling Enzyme Processivity Reveals that RNA-Seq Libraries Are Biased in Characteristic and Correctable Ways. Cell Syst 2016, 3, 467–479.e12. [CrossRef] [PubMed]
| Name of Package | Year | Bulk or Single Cell | Gene-level Transcriptome Diversity Metrics | Citation Count | Language |
| memento [81] | 2022 | Single-cell | Variation | 0 | Python |
| BioQC [82] | 2017 | Bulk | Shannon Entropy | 26 | R |
| MDSeq [19] | 2017 | Bulk | Variation | 15 | R |
| EntropyExplorer [83] | 2015 | Bulk | Differential Shannon Entropy | 9 | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





