1. Introduction
Iron is an essential nutrient and it is well-known that it is an integral constituent of many metalloproteins, primarily as part of heme groups and iron-sulfur clusters. As such, iron is essential for oxygen transport, electron transfer (redox) and catalytic reactions (Papanikolaou and Pantopoulos, 2005). The biological versatility of iron is based on its capacity to be coordinated by proteins and to act as electron donor and acceptor. Thus, iron can readily convert between its two common oxidation states Fe2+ and Fe3+, by the loss or gain of one electron. However, iron is also potentially toxic due to its redox reactivity. Indeed, free iron acts as a catalyst of oxidative stress via Fenton reactions, which yields hazardous radicals with the capacity to attack cellular macromolecules and cause tissue damage. Consequently, a tight control of iron homeostasis is imperative to satisfy metabolic needs for iron, and to prevent the accumulation of toxic iron concentrations. Iron homeostasis involves all the processes that regulate the balance between iron uptake, its intracellular storage and utilization (Collins and Anderson, 2012).
In soil Fe2+ undergoes spontaneous aerobic oxidation to Fe3+, which is virtually insoluble at physiological pH. This makes acquisition of iron by cells and organisms challenging, despite its high abundance. The mechanism of iron uptake in the roots of Arabidopsis thaliana is now well-described and it involves an acidification reduction-transport mechanism (Curie et al., 2009). Under iron deficiency, ferric chelates are solubilized by local rhizosphere acidification caused by the release of protons by the ARABIDOPSIS PLASMA MEMBRANE H+-ATPase2 (AHA2; Santi and Schmidt, 2009). Solubilized Fe3+ ions are then reduced to Fe2+ by the REDUCTASE FERRIC REDUCTION OXIDASE2 (FRO2) (Robinson et al., 1999) and, finally, transported into the cell by the IRON TRANSPORTER IRON REGULATED TRANSPORTER1 (IRT1; Eide et al., 1996; Vert et al., 2002). The mechanisms governing the distribution of iron to specific organs, cells and organelles are still very poorly understood.
In plants, in addition to its role in the mitochondrial electron transport chain, common to eukaryotes, iron is essential for chloroplast photosynthesis, as shown by chlorosis of plants grown under iron-deficient conditions (Guerinot and Yi, 1994; Yi and Guerinot, 1996; Briat et al., 2015). A total of twenty-two iron atoms are required per photosynthetic electron transport chain (Schmidt et al., 2020). Iron import into the chloroplast was proposed to be performed by the PERMEASE IN CHLOROPLAST1 (PIC1) localized in the inner envelope of this organelle (Duy et al., 2007). PIC1 knockout mutations result in dwarf plants with altered iron homeostasis. Before being transported into the chloroplast, iron is thought to be first reduced by the FERRIC REDUCTASE7 (FRO7), also localized in the chloroplast envelope (Jeong et al., 2008). Indeed, chloroplasts isolated from fro7 loss-of-function mutants have significantly reduced iron content and altered photosynthetic complexes (Jeong et al., 2008). Iron remobilization from leaves chloroplasts seems to be mediated by YSL4 and YSL6 (Divol et al., 2013). Recently, it has been suggested that FPN3 has a role in the iron export from Arabidopsis chloroplast and mitochondria (Kim et al., 2021).
In plant mitochondria, iron is more abundant than other transition metals such as Cu, Zn and Mn, consistent with its crucial role as component in electron transfer reactions (Tan et al., 2010). It has been suggested that iron is transported to the mitochondria through the outer membrane by Voltage Dependent Anion protein Channels (VDACs), and then to the mitochondrial matrix by the Mitochondrial Carrier Family (MCF) transporters. MCF gene family, with more than fifty members in Arabidopsis thaliana, encodes membrane proteins containing six transmembrane domains (Haferkamp and Schmitz-Esser, 2012). Mitochondrial Iron Transporter (MIT), a member of the MCF in Oryza sativa, was the first mitochondrial iron transporter identified in plants (Bashir et al., 2011). Complementation studies using OsMIT demonstrated that it is able to transport iron into yeast mitochondria and its function is essential in Oryza sativa. Knockdown plants for OsMIT showed a decrease of iron content in mitochondria and in the aconitase activity, an iron-sulfur protein (Bashir et al., 2011). In Arabidopsis thaliana it has been described that two genes encode MIT proteins (Jain et al., 2019). Fusions with fluorescent proteins demonstrated that AtMIT1 and AtMIT2 localized to mitochondria, and plants knockout for MIT1 and knockdown for MIT2 showed mitochondrial defects when plants were grown in iron deficiency conditions (Jain et al., 2019). Both mitochondrial iron deficiency or excess seems to provoke oxidative stress in a mammalian model (Walter et al., 2002). In plants, mitochondrial iron deficiency or excess also affects mitochondrial function (Vigani et al., 2009; Kim et al., 2021), indicating that plant mitochondria have a crucial role in cellular metal homeostasis (Vigani et al., 2019).
In this article, we characterize Atmit1 Atmit2 double mutant plants (knockout for MIT1 and knockdown for MIT2) grown in iron sufficient conditions. These plants showed pleiotropic developmental defects, some of which strikingly resemble those found in auxin transport and sensing mutants. Transcriptomic data revealed a misregulation of genes involved in iron acquisition, synthesis of coumarins, formation of the Casparian strip, suberization and root hair development. Furthermore, we demonstrate unambiguously, by crossing knockout mutants for MIT1 and MIT2, that MIT function is essential in Arabidopsis.
2. Results
Isolation of mutants in the two Arabidopsis genes encoding mitochondrial iron transporters (MIT)
In Arabidopsis two genes (At1g07030 and At2g30160) encode proteins with high similarity to the rice
Mitochondrial
Iron
Transporter (MIT) (Bashir et al., 2011, Vigani et al., 2016) and have recently been characterised (Jain et al. 2019). The Arabidopsis MIT isoforms share 82% peptide sequence identity and similarity, including the putative mitochondrial targeting peptide. Both are 66-67 % identical (77-78 % similar) to the rice MIT protein (excluding the putative mitochondrial targeting peptides). To evaluate the potential role of the Arabidopsis proteins as mitochondrial iron transporters, At1g07030 (
MIT2) and At2g30160 (
MIT1) were used to transform the
MRS3-MRS4 knockout yeast (Δ
mrs3Δ
mrs4).
Saccharomyces cerevisiae Mrs3 and Mrs4 are members of the MCF responsible for transporting Fe into mitochondria under low-Fe conditions, and the double knockout Δ
mrs3Δ
mrs4 mutant grows poorly when Fe availability is low (Foury and Roganti, 2002; Mühlenhoff et al., 2003). Each Arabidopsis MIT isoform was able to complement the growth defect of Δ
mrs3Δ
mrs4 yeast cells (Supplemental
Figure S1), indicating that they can act as mitochondrial iron carriers. Our results are largely in agreement with previous observations by Jain et al. (2019), with the exception of the lack of evidence for a significant difference in the efficiency of complementation by MIT1 and MIT2 (Supplemental
Figure S1).
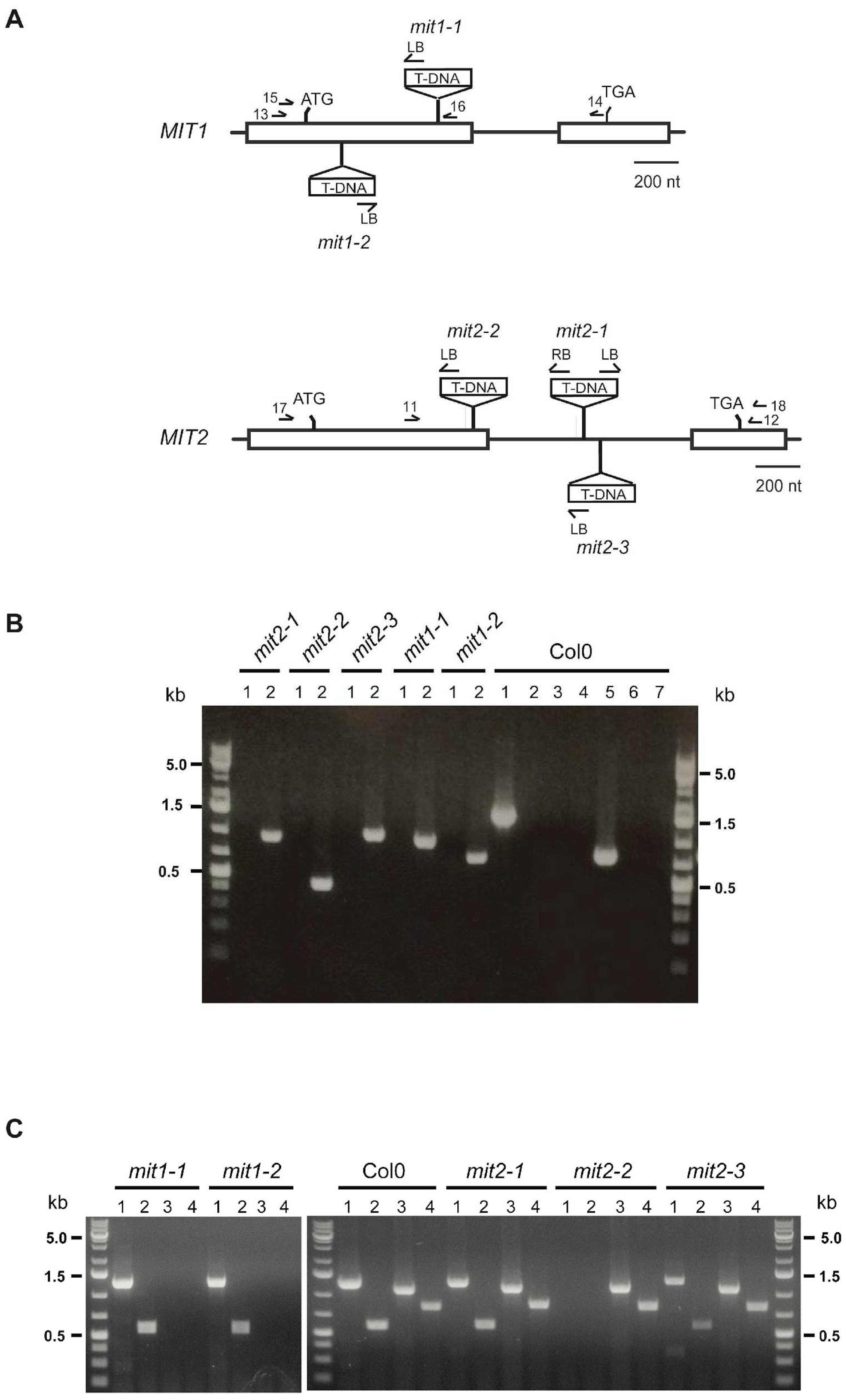
To explore MIT1 and MIT2 function in Arabidopsis we identified two and three T-DNA insertion mutant lines for
MIT1 and
MIT2, respectively (
Figure 1A, Supplemental
Figures S2 and S3). The T-DNA insertion is located in exon 1 for both
mit1 mutants, causing an interruption in gene expression 200 bp and 53 bp downstream of the start codon in
mit1-1 and
mit1-2 respectively. In
mit2-1 and
mit2-3 mutants, the T-DNA is located in the intron (419 and 500 bp downstream of the 5’ splice site), while
mit2-2 contains an insertion in the first exon (239 bp downstream of the start codon). We analyzed the progeny of selfed heterozygous
mit1-1,
mit1-2 and
mit2-1 plants and found that the progeny did not deviate significantly from 1:2:1 (wild type: heterozygous: homozygous). Furthermore, homozygous mutant plants for each of the
mit1 alleles and
mit2 alleles did not show any phenotypic alteration when compared to wild type plants (data not shown).
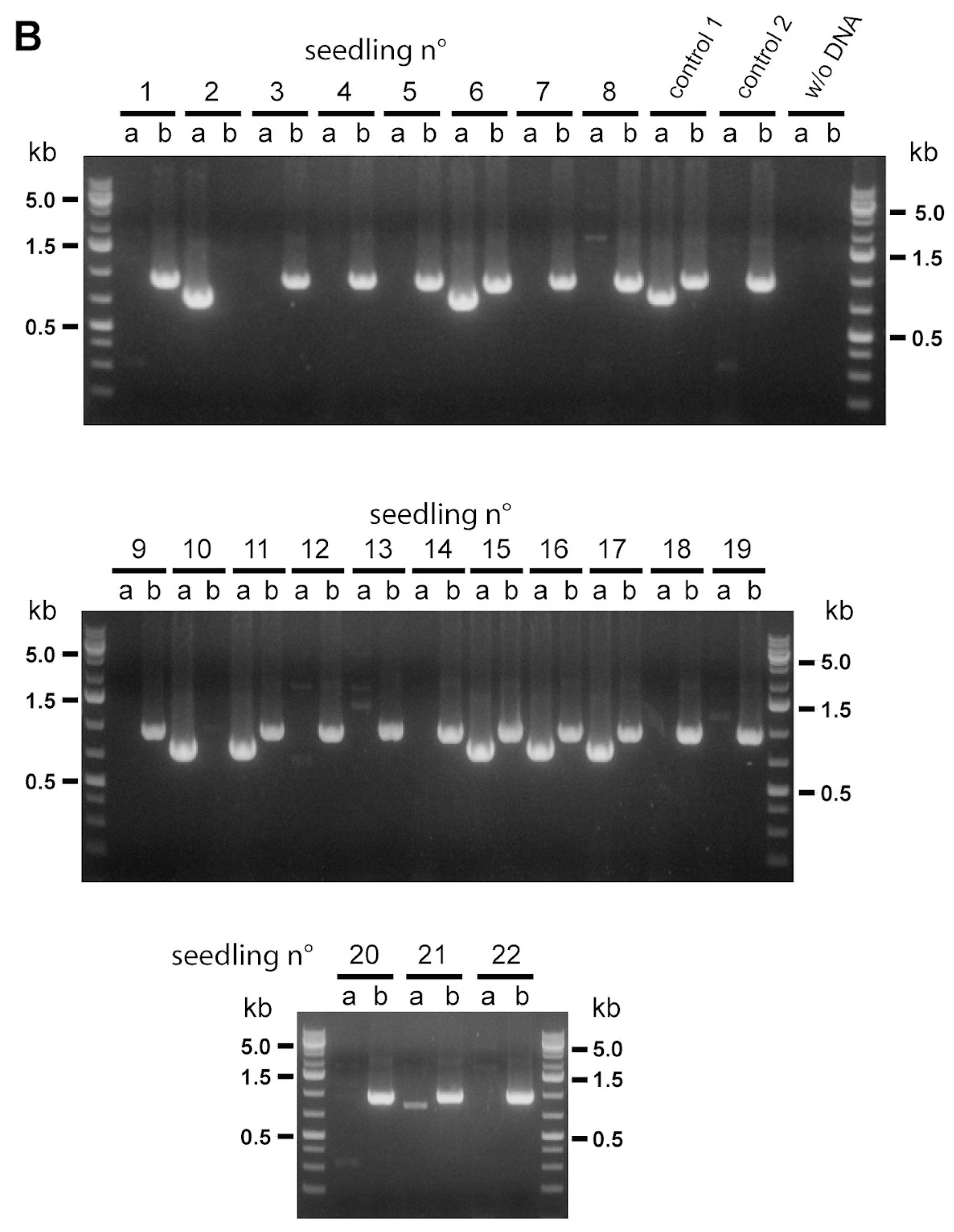
Next, RT-PCR analysis of
MIT1 and
MIT2 expression was carried out to ascertain homozygous mutant plants obtained for all five mutants (
Figure 1B) were truly null mutants (
Figure 1C). Results show clearly that
mit1-1,
mit1-2 and
mit2-2 plants are knockout mutants, expressing only the non-mutated
MIT gene. Unexpectedly,
mit2-1 and
mit2-3 expressed
MIT2, in addition to
MIT1, and thus are not knockout mutants. Characterization of the two T-DNA/
MIT2 junctions in
mit2-1 (
Supplementary Figure S3) showed that no major deletions or chromosomal rearrangements occurred during the T-DNA insertion, which is 4080 bp long (with deletions of 303 bp in the right border and 102 bp in the left border), and thus this insertion converts a 913 nt intron into one of 4993 nt. Sequencing of the two
MIT2 RT-PCR products obtained from
mit2-1 RNA demonstrated that the intron is correctly spliced. Splicing of long introns is very unusual in plants but is not without precedent (e.g. Chang et al., 2017). However,
MIT2 transcript level as determined by RT-qPCR is significantly decreased (
Supplementary Figure S6), confirming that both
mit2-1 and likely
mit2-3 are knockdown mutants.
Given that mit1-1, mit1-2 and mit2-2 are knockout mutants their normal growth showed that neither MIT1 nor MIT2 are essential per se, and that they may be redundant. These results led us to perform mit1 x mit2 crosses.
2.1. MIT Function Is Essential in Arabidopsis
Given that mit1-1, mit1-2 and mit2-2 are knockout mutants their normal growth showed that neither MIT1 nor MIT2 are essential per se. To determine whether MIT function is essential, we crossed the knockout mutants mit1-1 and mit2-2. F2 seeds from selfed double heterozygous plants (MIT1mit1-1 MIT2mit2-2) were sown on 0.5X MS plates and a plant carrying three mutated alleles was identified (MIT1mit1-1 mit2-2mit2-2). Visual inspection of F3 seeds in three siliques from this selfed plant showed that they contain 77.5 ± 7.0 % normal seeds and 22.5 ± 7.0 % aborted seeds. Furthermore, when F3 seeds were allowed to developed on soil under iron-sufficient conditions, we were unable to identify a plant carrying a double homozygous mutation (65 plants analyzed). Altogether, these results confirm that MIT function is essential, that the absence of MIT1 and MIT2 is embryo-lethal, and that MIT1 and MIT2 genes are redundant.
2.2. Isolation of mit1-1 x mit2-1, a MIT Knockdown Mutant
Next, we crossed homozygous
mit1-1 with homozygous
mit2-1 plants. One hundred and seventy seven F2 plants grown from seeds of three selfed F1 double heterozygous plants (genotype
MIT1mit1 MIT2mit2) were genotyped (
Supplementary Table S2). No double homozygous mutants were identified, and we noted a bias against plants homozygous for
mit1 and heterozygous for
mit2 (4 plants
mit1-1mit1-1 MIT2mit2-1) is apparent and not observed for plants heterozygous for
mit1 and homozygous for
mit2 (22 plants
mit1-1MIT1 mit2-1mit2-1). This may be due to
mit1-1 being a knockout mutation and
mit2-1 a knockdown mutation (see below). The plants carrying three mutated alleles did not show visible phenotypic alterations when compared to wild type plants, at least under standard growth conditions.
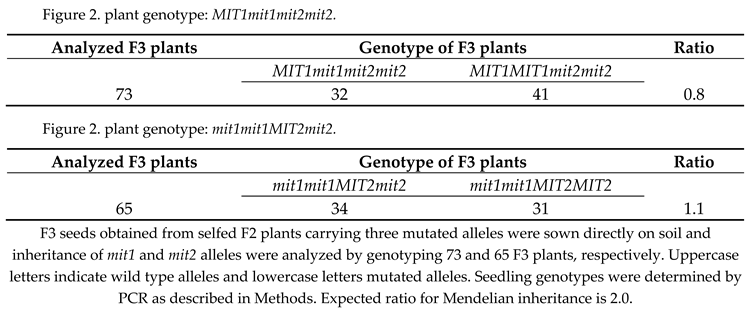
Then F3 seeds from selfed F2 plants carrying three mutated alleles and one wild type allele (either
MIT1 or
MIT2) were sown directly on soil and grown plants were genotyped. Again, no double homozygous mutant plants were obtained (73 and 65 plants analyzed,
Table 1). These results suggest that MIT function is essential and that
MIT1 and
MIT2 genes are redundant. Furthermore, instead of the expected ratio of 2:1 for heterozygous: wild type plants,
MIT1mit1 :
MIT1MIT1 in the
mit2mit2 background, and
MIT2mit2 :
MIT2MIT2 in the
mit1mit1 background, ratios of 0.8 and 1.1 were observed. These ratios suggest a gametophytic defect, i.e. a defect in gametes carrying only mutated alleles of the mitochondrial iron transporters (
mit1mit2 gametes).
F3 seeds obtained from selfed F2 plants carrying three mutated alleles were sown directly on soil and inheritance of mit1 and mit2 alleles were analyzed by genotyping 73 and 65 F3 plants, respectively. Uppercase letters indicate wild type alleles and lowercase letters mutated alleles. Seedling genotypes were determined by PCR as described in Methods. Expected ratio for Mendelian inheritance is 2.0.
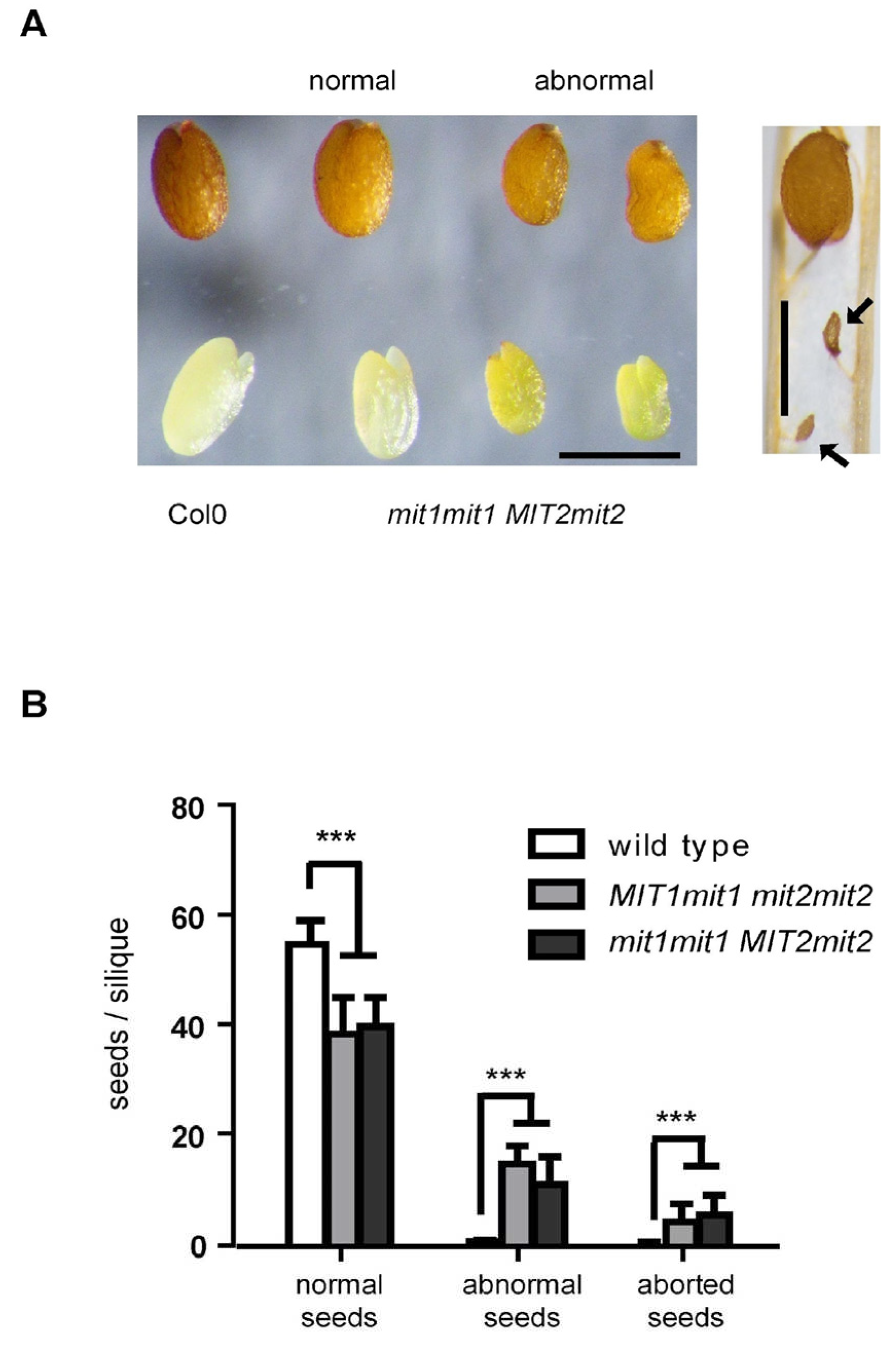
Visual inspection of F4 seeds from F3 plants carrying three mutated alleles showed that in addition to “normal” seeds that resemble those from wild type plants, plants carrying three mutated alleles produced seeds with altered phenotypes: (i) smaller, irregular seeds (“abnormal” seeds), and (ii) shrunken, collapsed seeds (“aborted” seeds) (
Figure 2A). Sets of normal, abnormal and aborted seeds per silique were quantitatively determined, using four
MIT1mit1 mit2mit2 plants, four
mit1mit1 MIT2mit2 plants and two wild type plants as control (
Figure 2B). In siliques from plants with three mutated alleles, 26 % (plants with one
MIT1 wild type allele) and 20% (plants with one
MIT2 wild type allele) of total seeds were considered “abnormal”. Only a minor proportion of seeds were aborted (7 % and 9 %, respectively). There are no significant differences in the total number of seeds per silique between wild type plants and plants with three mutated alleles, nor are significant numbers of non-fertilized ovules. Furthermore, these numbers were evenly distributed along the inflorescence axis, indicating that seed phenotypes are not due to flower heterogeneity (
Supplementary Figure S4).
When these “abnormal” seeds were sown on plates containing half-concentrated MS medium, almost all were able to germinate (
Table 2) and expand their green cotyledons, although at a lower rate than “normal” and wild type seeds (
Supplementary Figure S5). However, only around 50 % of seedlings were established with true leaves and elongated roots (
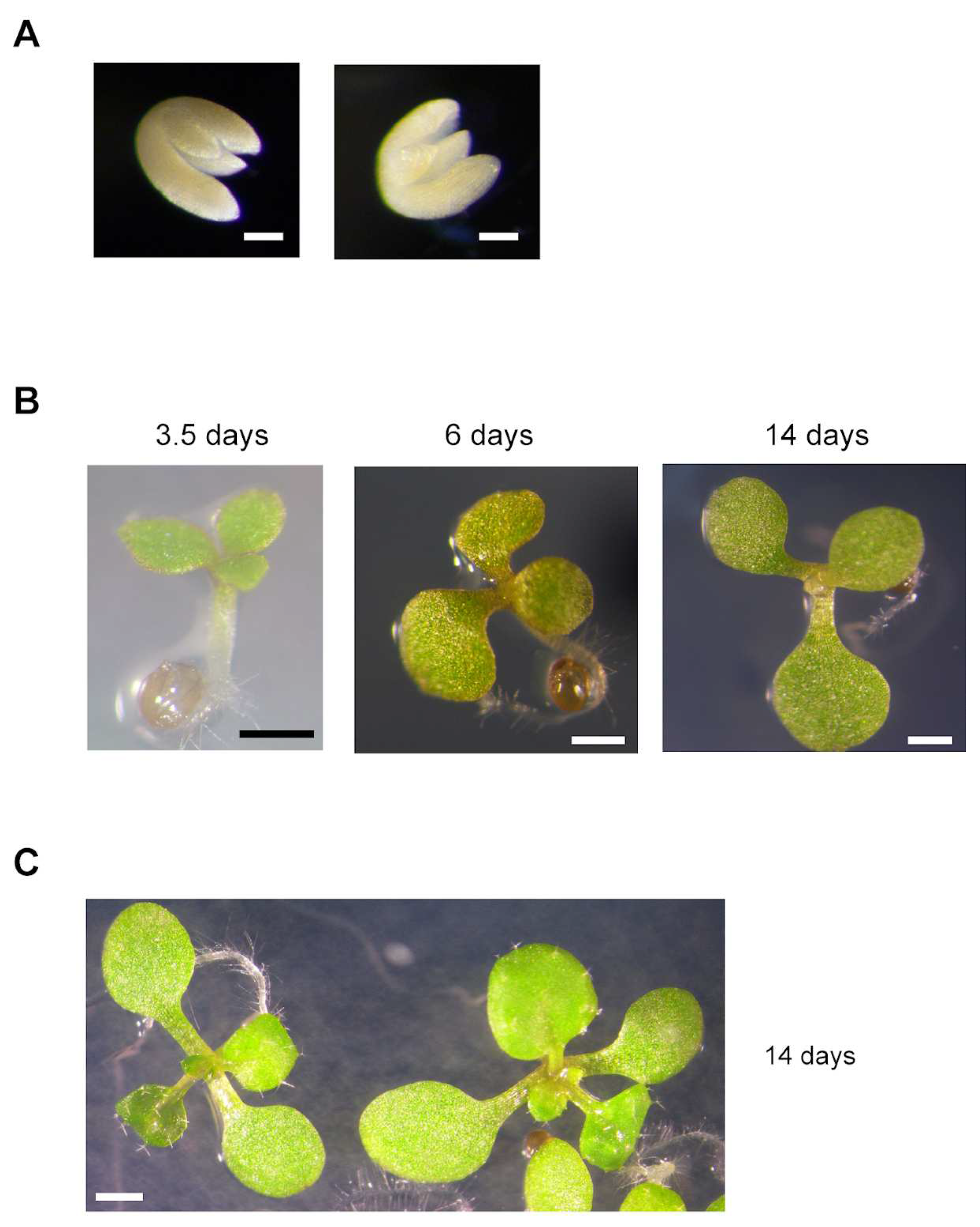
Table 2). A significant proportion of these abnormal seeds possessed embryos with three cotyledons (
Table 2,
Figure 3). We were able to identify double homozygous mutants (
mit1-1mit1-1 mit2-1mit2-1) among plants grown from “abnormal” seeds, in addition to plants with three mutated alleles (
Figure 4). Furthermore, there is a strong correlation between growth rate and genotype: all double homozygous mutants presented a delayed growth rate when compared with plants with either 1 or 2 wild type
MIT1 alleles. These results showed on one hand that seed morphology was not a clear-cut criterion to identify genotype, and on the other hand that double homozygous mutants are viable. This unexpected result led us to determine that
mit2-1 is not a knockout mutant (see above,
Figure 1C) and that
MIT2 is expressed, at a lower level (12.2%), in the double homozygous mutants (
Supplementary Figure S6).
Five individual siliques from MIT1mit1 mit2mit2 plants in two experiments were used to genotype all seedlings (established or not) grown from normal and abnormal seeds: 16.0 ± 3.5 % of total seeds were double homozygous mutants (43 out of 264 total seeds in the 5 siliques), and 27.7 ± 8.9 % of the double homozygous mutants possessed three cotyledons (11 plants out of 43). It is important to point out that all genotyped 3-cotyledon plants in this and other experiments were double homozygous mutants.
2.3. Growth of Double Homozygous Mutant Plants Is Severely Affected
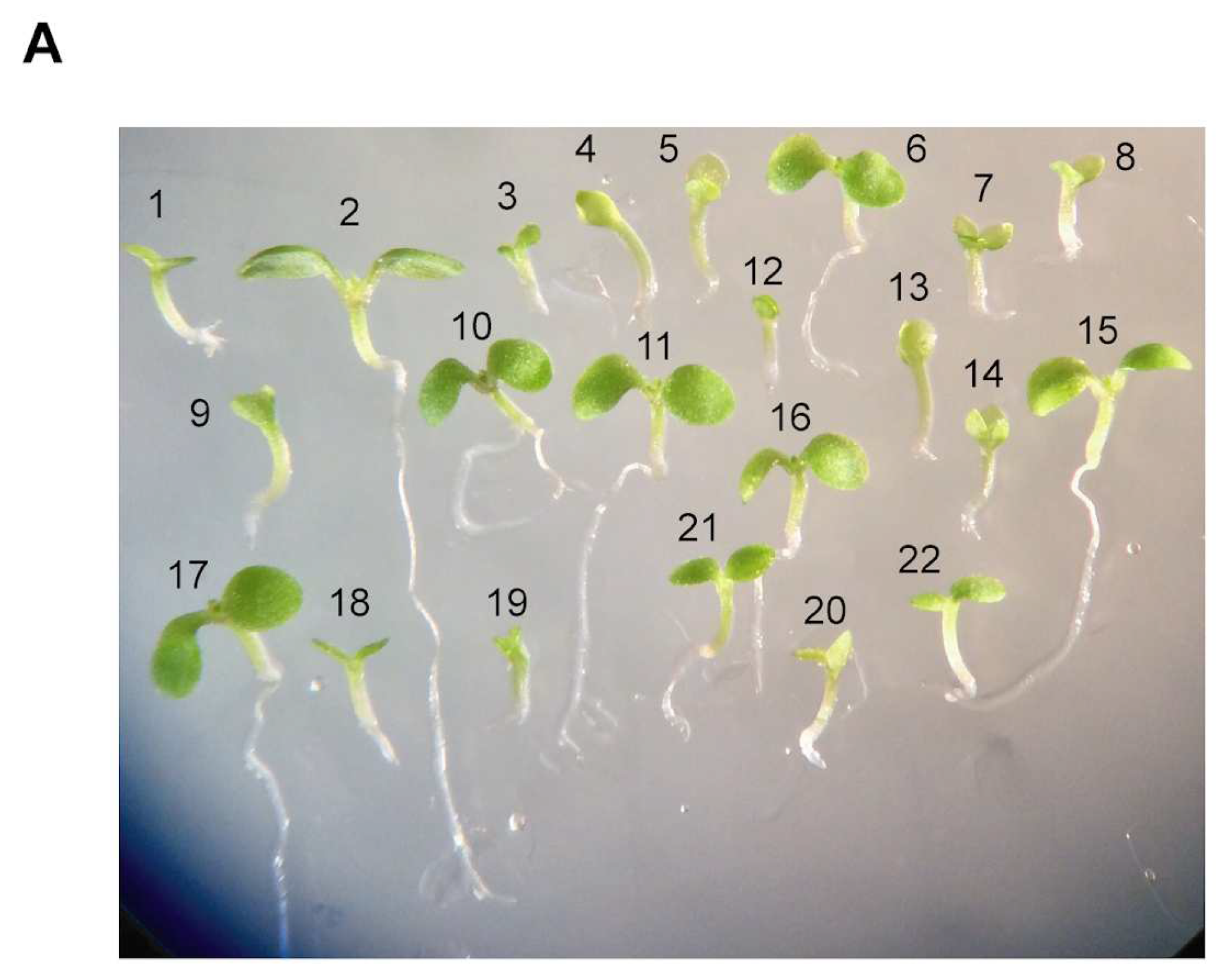
Viable double homozygous
mit1-1mit1-1 mit2-1mit2-1 plants were easily identified by their severe phenotype. First, as already mentioned, germination and early post-germinative growth were slower than that of wild type plants and plants with three mutated alleles (
Figure 4,
Supplementary Figure S5). For instance, in one experiment, at 13 days when all Col0 seedlings (90 out of 90) were at least at stage 1.02 on day 13 according to Boyes et al. (2001), only one out of 59 (1.9%) of the double homozygous mutant seedlings attained this stage. Three weeks after germination, only 1.7 % (1/59) and 10 % (6/59) of these seedlings were at stages 1.06 and 1.04, respectively.
Growth of double homozygous mutant plants on soil was severely affected throughout the entire life cycle, and senescence was delayed by 1.5-2 months (
Supplementary Figure S7). Drastic reduction of MIT expression has pleiotropic effects on double homozygous plants (
Figure 5), including pinoid stems (
Figure 5A and B) similar to those observed for mutants of auxin efflux carriers (PIN) (39 out of the 46 plants, 85 %), stems terminated at either cauline leaves (25 plants, 54%,
Figure 5B), a unique flower (21 plants, 46%,
Figure 5C) or multiple floral buds and cauline leaves (18 plants, 39%,
Figure 5D). Furthermore, phyllotaxis in the appearance of cauline leaves was altered: there were a higher number of these leaves in some stems of 15 plants (33%,
Figure 5E), their position was less regular (
Figure 5D to F), and in some cases three cauline leaves were found at the same position (in 7 plants, 15%,
Figure 5F). Additionally, some enlarged stems were found in 5 (11%) of the plants, as if two stems had been fused (
Figure 5G, also visible in the plant shown in
Figure 5F).
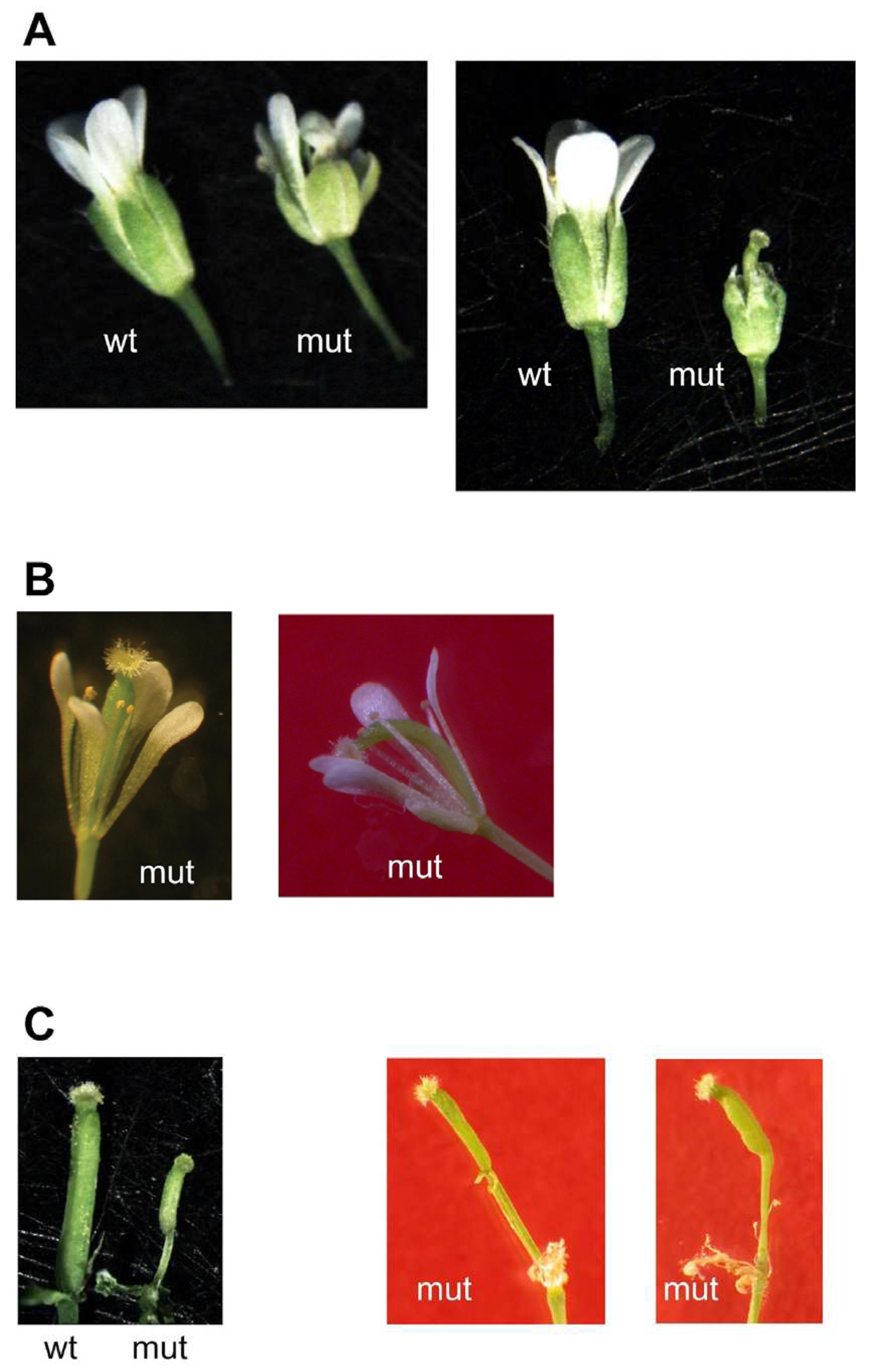
All double mutant plants showed, alongside with some normal flowers, flowers with all their structures (sepals, petals, anthers, pistils) altered (
Figure 6). This resulted in 17 out of 46 plants (37 %) unable to give seeds, and remaining plants showed a reduced seed set (less than 50 seeds in 1 to 6 siliques for 17 plants, between 50 and 250 seeds in 6 to 16 siliques for 10 plants, and more than 600 seeds for 2 plants with at least 80 siliques).
2.4. The Next Generation of Double Homozygous mit1-1 mit2-1 Mutant Plants Showed a Normal Phenotype
When seeds obtained from mit1-1mit1-1 mit2-1mit2-1 plants were sown, almost all germinated (97.2 ± 5.0 %) and plant establishment was variable (55.5 ± 17.9 %). Most importantly, plant growth was similar to that of wild type plants, for instance all established seedlings were at stage 1.0 at 7 days and at stage 1.02-1.03 at 2 weeks, and this similarity extends to vegetative and reproductive growth. From now these plants have been designated as “compensated” double homozygous mutant plants. Their genotype was verified by PCR to be mit1-1mit1-1 mit2-1mit2-1.
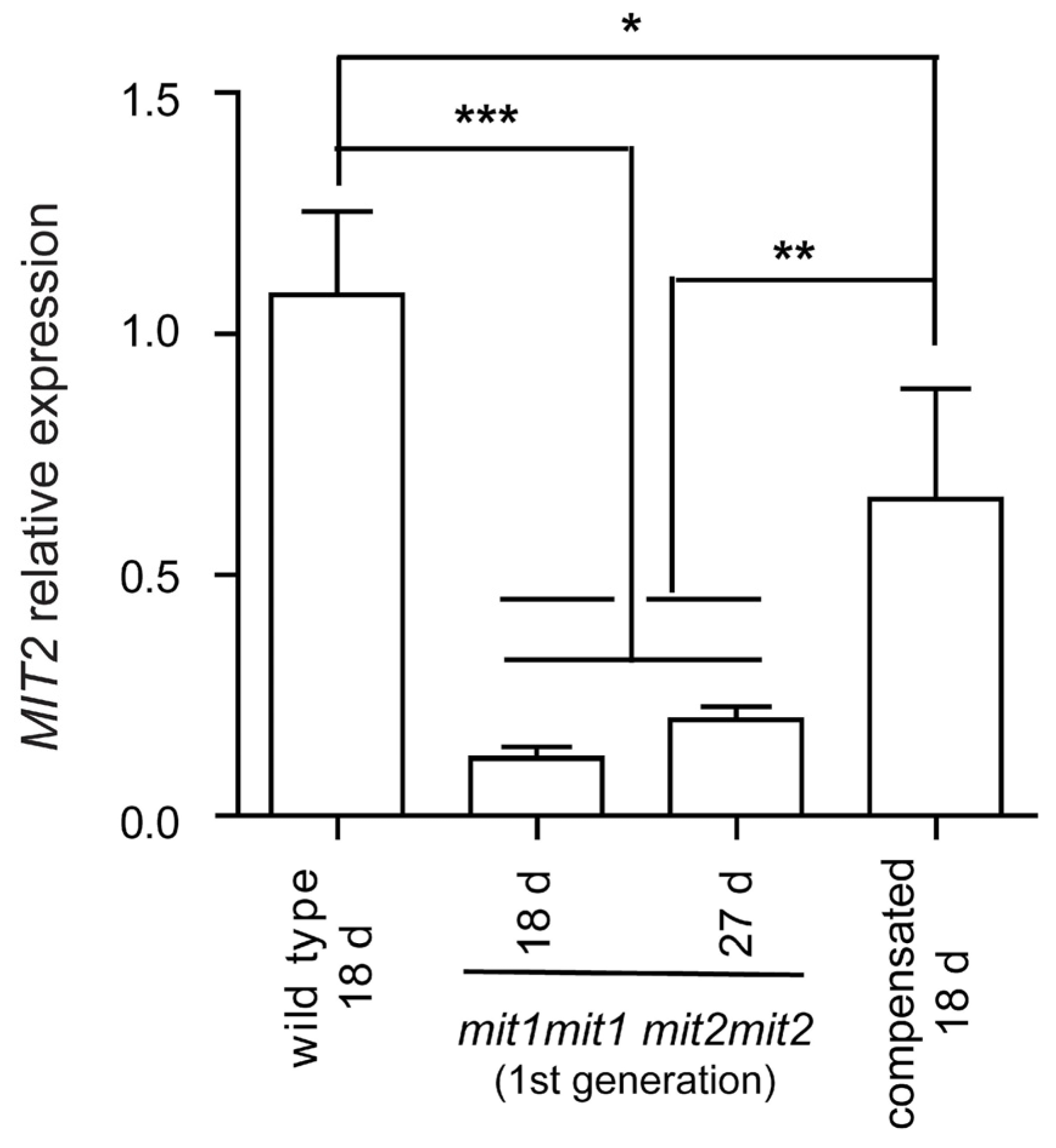
This intriguing result led us to analyze
MIT2 expression by RT-qPCR in these “compensated” double homozygous mutant plants and compare it to that observed in plants showing an affected phenotype (first generation of double homozygous mutant plants, arising from seeds obtained from plants carrying three mutated alleles) (
Figure 7). Interestingly,
MIT2 expression is significantly higher in compensated plants (60.9 % that of wild type plants) compared to affected plants (11.0 % - 18.5 % that of wild type).
These results are consistent with the view that increased splicing of the MIT2 intron containing the T-DNA is responsible for phenotypic recovery of the double homozygous mutant plants, and may be related to a relatively recently described phenomenon called “T-DNA suppression” (see Discussion).
2.5. Analysis of Mitochondrial Function
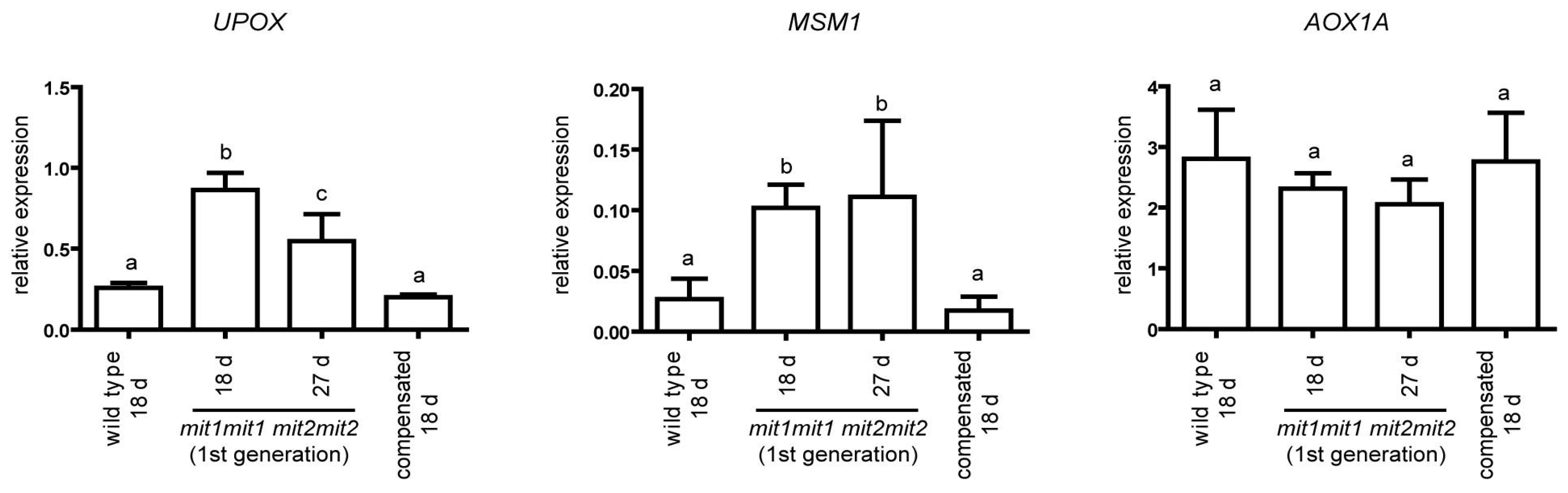
Marker genes for mitochondrial response to stress have been identified (Van Aken et al., 2009), and included the genes encoding mitochondrial proteins alternative oxidase 1A and UPOX (
up-regulated by
oxidative stress) (Clifton et al., 2005; Ho et al., 2008). On the other hand, Van Aken and Whelan (2012) were able to identify marker genes that respond to mitochondrial and chloroplast dysfunction (e.g.
UPOX) or are specific for mitochondrial dysfunction (e.g.
MSM1, for Mitochondrial Stress Marker 1, also designated
At12cys-2, Wang et al., 2016). We evaluated the expression of these three genes,
AOX1a,
UPOX and
MSM1, and found that
UPOX and
MSM1 are significantly up-regulated in the first generation of double homozygous mutant plants (
mit1-1mit1-1 mit2-1mit2-1) but return to wild type levels in the “compensated” second generation plants (
Figure 8). In contrast,
AOX1a transcript levels were not significantly altered in any genotype. These results suggest some degree of mitochondrial perturbation in the first generation of double homozygous mutant plants with drastic reduction of
MIT expression.
Unfortunately, we were unable to purify mitochondria from first generation double homozygous mutant plants (mit1-1mit1-1 mit2-1mit2-1) with an affected phenotype. To do this it would be necessary to grow plants with three mutated alleles, collect seeds, manually separate “abnormal” seeds and grow plants from these seeds; so it was unfeasible to obtain enough biological material. Thus, mitochondria were purified from “compensated” double homozygous mutant (mit1-1mit1-1 mit2-1mit2-1) and wild type seedlings as described in Supplementary Methods.
Targetted proteomic analysis was performed on 4 biological replicates of both compensated and wild-type mitochondria by Selective Reaction Monitoring (SRM) mass spectrometry. In this way more than one hundred proteins (listed in
Supplementary Table S3) were quantified allowing a focused dissection of responses in the TCA cylce, electron transport chain, mitochondrial localised iron-related proteins, and MIT1/2 proteins (
Supplementary Figure S8 A). Significant differences in protein levels between compensated and wild type plants were found only for MIT1 and MIT2. The specific quantified peptide for MIT1 was found in wild-type, but was below the limit of detection in mitochondria from “compensated” double homozygous plants (as expected for a knockout mutation). The specific peptide for MIT2 in compensated plants was 30.9 ± 12.4 (SD) % the level found in wild type mitochondria, confirming that
mit2-1 is a knockdown allele.
Despite this large reduction in MIT abundance (absence of MIT1, and 30 % of MIT2), no differences were observed in the oxygen consumption rate of isolated mitochondria (
Supplementary Figure S8 B). Furthermore, no differences were detected in the abundance or native size of respiratory complexes or in complex I activity when analyzed by BN gel electrophoresis (
Supplementary Figure S8C). Complex I activity was assessed as it is the respiratory complex having the higher number of iron ions in its structure present as iron-sulfur centers. These results show that, at least when plants are grown under standard conditions, mitochondria with a drastic reduction in MIT are not functionally impaired.
2.6. RNA-seq Analysis of Gene Expression in Double Homozygous Mutant Plants
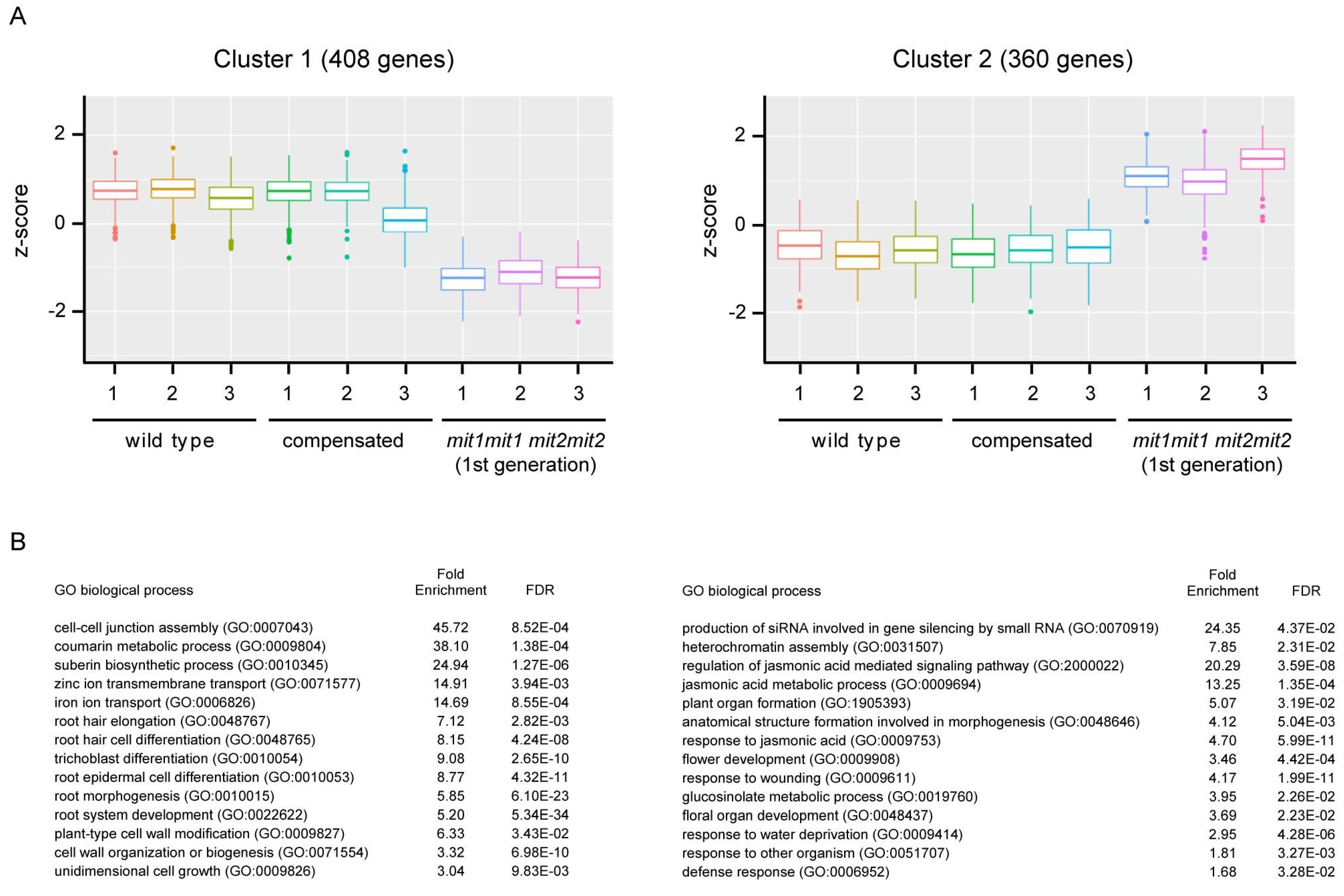
Since RNA-seq analysis required less biological material, we were able to perform this analysis with the first generation of double homozygous mutant plants (mit1-1mit1-1 mit2-1mit2-1) which showed an affected phenotype, and compare this transcriptome to that of “compensated” plants having the same genotype, and that of wild type plants. Total RNA was prepared from three biological replicates of 18 days-old wild type seedlings and compensated double homozygous mutant seedlings. For double homozygous plants of the first generation (thus, presenting a severe phenotype), 27 days-old seedlings were considered in order to compare plants at the same developmental stage, in this case 1.04 of Boyes et al. (2001). Poly A-enriched RNA fractions were employed to construct libraries for Illumina sequencing (see Methods).
Differentially expressed genes between genotypes were identified (padj < 0.05, log
2 [fold change] > 1 in any condition, LRT test) and grouped in two clusters by k-means (
Figure 9). Interestingly, no significant differences (except for
MIT expression) were found between wild type plants and compensated plants (Wald test, padj = 3.08E-18, log
2 fold change = -4.2), further supporting the conclusion that partial expression (around 30 %) of one of the two
MIT genes is sufficient for normal plant growth and development. In cluster 1 (408 genes, listed in
Supplementary Table S4) expression is down regulated in double homozygous mutant plants (1
st generation) when compared to wild type and compensated plants. In cluster 2 (360 genes, listed in
Supplementary Table S5) higher levels of transcripts are found in the double homozygous mutant plants (1
st generation).
Cluster 1 shows an overrepresentation of genes belonging to the Gene Ontology annotation categories of iron ion transport (GO:0006826; FDR 8.6 x 10
-4, 14.7 fold enrichment) and coumarin metabolic process (GO: 0009804; FDR 1.4 x 10
-4, 38.1 fold enrichment); coumarins being involved in iron chelation in the rhizosphere for incorporation into the plant (Rajniak et al., 2018; Tsai et al., 2018; Riaz and Guerinot, 2021). For instance,
FRO2 (At1g01580, ferric reduction oxidase 2),
IRT1 (At4g19690, iron-regulated transporter 1),
IRT2 (At4g19680, iron-regulated transporter 2),
FIT1/FRU/bHLH29 (At2g28160, FER-like Iron deficiency Induced-Transcription factor),
IREG2/FPN2 (At5g03570, iron-regulated transporter 2, ferroportin 2),
BTSL2 (At1g74770, Zinc finger BRUTUS-like protein 2) and
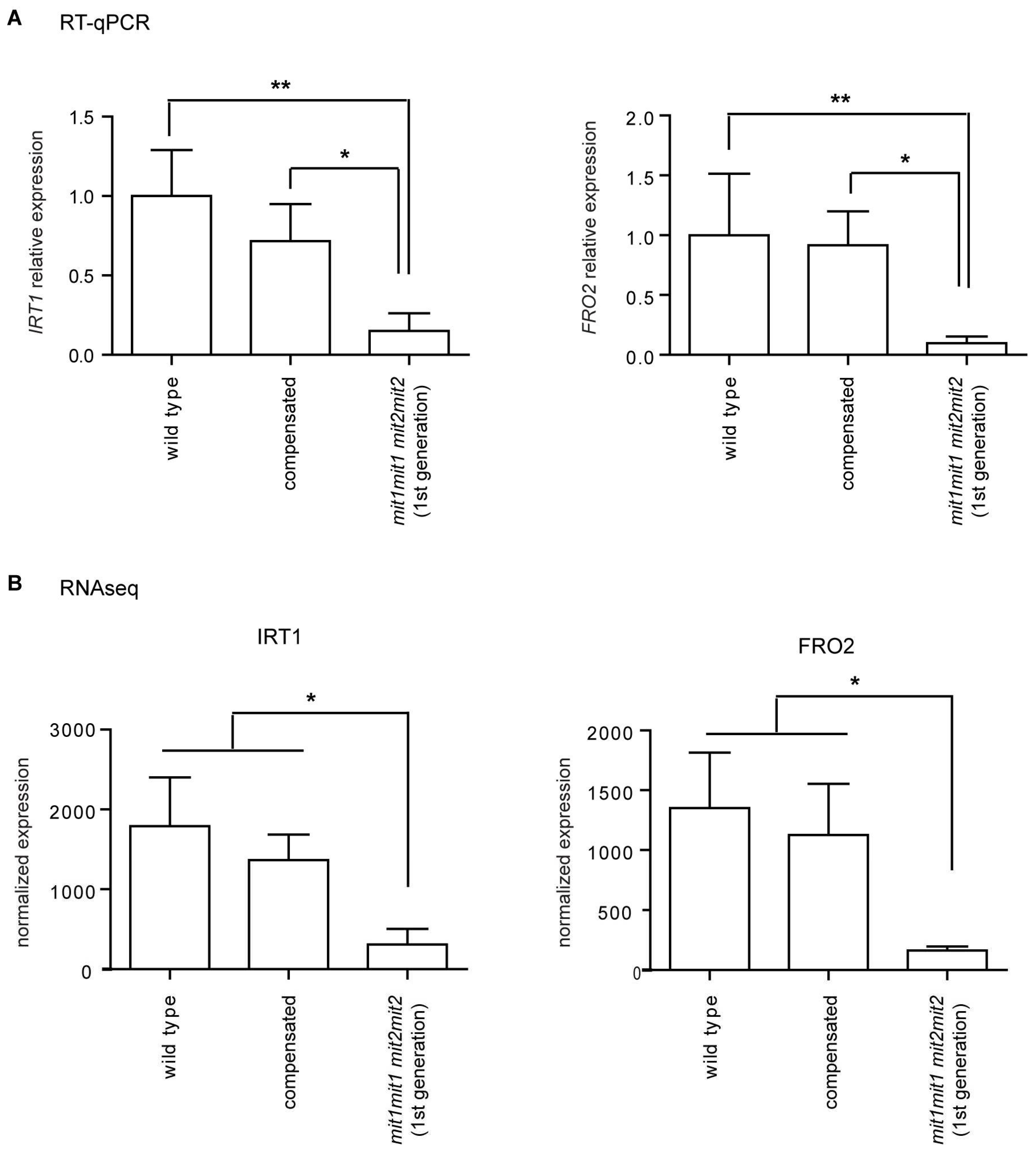
NAS2 (At5g56080, nicotianamine synthase 2) are in cluster 1. Down regulation of
FRO2 and
IRT1 were also independently verified by RT-qPCR (
Figure 10). Other relevant overrepresented biological processes in Cluster 1 are related to growth processes, including root hair elongation, plant-type cell wall modification and unidimensional cell growth, consistent with the growth deficiency phenotypes presented in the double homozygous mutants (
Supplementary Table S6).
Cluster 2, containing genes with an increased expression relative to wild-type and compensated plants, shows an overrepresentation of biological processes that can be related to plant defense, including different terms related to jasmonic acid (regulation of jasmonic acid mediated signaling pathway, jasmonic acid metabolic process, response to jasmonic acid), regulation of defense response, response to other organism and glucosinolate metabolic process. As well, processes related to wounding and response to water deprivation were found in cluster 2. This suggests that double homozygous plants have a basal activation of stress-related responses, which might impact plant growth in these plants.
3. Discussion
Arabidopsis
MIT1 and
MIT2 encode proteins highly similar to rice MIT and with significant similarity to yeast mitochondrial iron transporters MRS3 and MRS4 (38 % identity). Mitochondrial localization of the plant proteins has been demonstrated by either fusion to GFP or YFP (Bashir et al., 2011; Jain et al., 2019), subcellular fractionation (Jain et al., 2019), or proteomic analysis of purified mitochondria (Supplemental
Figure 8). Expression of plant
MIT genes appears to be ubiquitous (Bashir et al., 2011; Jain et al., 2019;
http://bar.utoronto.ca). MIT1 and MIT2, with their own targeting peptides (
Supplementary Figure S1) or that of MRS3 (Jain et al., 2019) were able to complement the defect of the yeast Δ
mrs3Δ
mrs4 mutant, demonstrating that they function as high affinity iron transporters in yeast mitochondria, and likely also in plant mitochondria.
The rice MIT1 gene is essential since its absence is embryo-lethal (Bashir et al., 2011). In contrast, in Arabidopsis MIT1 and MIT2 appear to be redundant since individual mutants, including the knockout mutants mit1-1 and mit2-2 were indistinguishable from wild type plants. However, MIT function is essential in Arabidopsis since when crossing these two null mutants no double homozygous plants could be obtained, and almost 25 % of the seeds from plants with three mutated alleles (MIT1mit1-1 mit2-2mit2-2) aborted.
3.1. Phenotypic Alterations of the mit1-1 mit2-1 Double Homozygous Mutants
When crossing the knockout
mit1-1 mutant with the knockdown
mit2-1 mutant we were able to obtain double homozygous mutant plants from the so-called “abnormal” seeds plated on MS x 0.5 (
Figure 2 and
Figure 4). The 1
st generation of these plants expressed only low levels of
MIT2 (10-20 %,
Figure 7 and
Supplementary Figure S6) and showed striking phenotypes, highlighting the importance of MIT function. Pleiotropic defects include polycotyly (three cotyledon-embryos,
Figure 3 and
Table 2), retarded germination and early post-germinative growth with reduced establishment (
Supplementary Figure S5,
Figure 4,
Table 2), delayed and altered reproductive development including
pin stems, abnormalities in phyllotaxy and in all organs of the flowers (
Figure 5 and
Figure 6), and reduced seed set.
Some of these phenotypes are clearly indicative of defects in auxin signaling. For instance, it is well known that polar auxin transport is involved in cotyledon emergence (Liu et al., 1993; Benková et al., 2003), and both
pin and
pid mutants show polycotyly (Bennett et al., 1995; Benjamins et al., 2001; Aida et al., 2002; Furutani et al., 2004). However, whereas the presence of one cotyledon is more frequent in
pin1 mutants (auxin efflux carrier, PINFORMED1), mutants in the ser/thr protein kinase PID (PINOID) which phosphorylates PIN1, show higher frequency of three-cotyledon embryos, and thus resembles double homozygous
mit1-1 mit2-1 embryos. In all these cases phenotype penetrance is incomplete. Furthermore, striking similarities are found between
pin,
pid and
mit1-1mit2-1 mutants during reproductive development: inflorescence stems without flowers and cauline leaves (
pin stems), alterations in cauline leaves phyllotaxis, abnormal flowers, reduced fertility (
Figure 5 and
Figure 6; Okada et al., 1991; Bennett et al., 1995; Christensen et al., 2000).
Although somewhat surprising at first sight, an interplay between mitochondrial function and auxin signaling is well documented. For instance, an antagonistic relationship has been described between mitochondrial stress signaling (MRR, mitochondrial retrograde regulation) and auxin signaling (Kerchev et al., 2014; Ivanova et al., 2014). Furthermore, a knockdown mutant in a mitochondrial Pyruvate dehydrogenase subunit (mab1-1, macchi-bou1), which has reduced respiration and pyruvate dehydrogenase activity, is also defective in polar auxin transport and shows auxin-related phenotypes (Ohbayashi et al., 2019). Knockdown of the succinate dehydrogenase assembly factor SDHAF2 is also known to induce ROS-mediated auxin hypersensitivity, causing pH-dependent root elongation in Arabidopsis (Tivendale et al 2021). The exact mechanisms linking in each case mitochondrial dysfunction and auxin signaling are not known but several competing or complementary hypotheses have been proposed based on the deep connections between auxin synthesis, conjugation and post-translational regulation of auxin signalling pathways (Tivendale and Millar 2022). These assembled examples and our results suggest that at least some of the phenotypes observed in the mit1-1mit2-1 double homozygous mutants are mediated by defects in auxin homeostasis.
On the other hand, phenotypes like reduced plant establishment, delayed germination and slow early post-germinative growth are characteristic of mitochondrial deficiency. Seed germination and early post-germinative growth are crucial for seedling establishment and involved the mobilization of seed nutrient reserves to fuel growth until the seedling becomes photoautotrophic. Mitochondria are expected to play a crucial role at these early stages, supplying energy and carbon skeletons for growth, and a role for respiratory complexes I, II and IV has been described (e.g. de Longevialle et al., 2007; Meyer et al., 2009; Roschzttardtz et al., 2009; Kühn et al., 2015; Restovic et al., 2017; Kolli et al., 2019). For instance, Kühn et al. (2015) found a correlation between the severity of complex I mutants and the delay in germination, cotyledon emergence, and seedling establishment. Unfortunately, as already mentioned, we were unable to obtain mitochondria from these seedlings to assess mitochondrial function, and only indirect evidence for mitochondrial dysfunction, i.e. higher transcript levels of
MSM1 and
UPOX, was documented (
Figure 8). However, Bashir et al. (2011) characterized a knockdown mutant of the unique rice
MIT gene (
mit-2, T-DNA insertion in the gene promoter) which displays a less severe phenotype, allowing mitochondrial preparation from homozygous mutant plants. Those rice mitochondria contain less Fe and have less aconitase (an iron-sulfur protein) activity, thus supporting a mitochondrial dysfunction due to iron deficiency also in our Arabidopsis double mutant.
3.2. Phenotypic Recovery of Double Homozygous mit1-1 mit2-1 in Next Generations
When the few seeds obtained from the affected
mit1-1mit1-1 mit2-1mit2-1 plants were sown, plant growth was similar to that of wild type plants. In these “compensated” double homozygous mutant plants,
MIT2 expression is enhanced with respect to parent plants (
Figure 7), and targetted proteomic analysis of purified mitochondria showed they differ significantly from wild type mitochondria only in MIT1 (undetectable) and MIT2 (30 %) content (
Supplementary Figure S8). Furthermore, no differences were observed in mitochondrial respiratory rate, respiratory complex size and abundance or complex I activity. Thus, increased splicing of the
MIT2 intron containing the T-DNA is likely responsible for phenotypic recovery of the double homozygous mutant plants. In the past years a phenomenon called “T-DNA suppression” has been described, occurring when crossing two mutants with similar T-DNA insertions (e.g. two SALK lines). At least one of the T-DNA insertions must be intronic, and the “suppressed” phenotype is then caused by the T-DNA (Xue et al., 2012; Gao and Zhao, 2013; Sandhu et al., 2013; Osabe et al., 2017). In all cases a significant proportion of the large intron containing the T-DNA is correctly spliced, enhancing expression of the mutated gene and suppressing the phenotype. Although the mechanism is not well known, T-DNA hypermethylation and heterochromatinisation are necessary, and the RdDM (RNA-dependent DNA methylation) pathway is involved (Osabe et al., 2017). In our experiments, the altered phenotypes observed in the first generation of double homozygous mutant plants are “suppressed” by an increase of
MIT2 intron splicing.
3.3. Transcriptome of Double Homozygous mit1-1 mit2-1 Mutant Plants
No significant differences (except for
MIT expression) were found between wild type plants and “compensated”
mit1-1mit2-1 double homozygous mutant plants, supporting the conclusion that partial expression (around 30 %) of MIT2 is sufficient for normal plant growth and development, at least under standard growth conditions. In contrast, in the first generation of
mit1-1mit2-1 double homozygous mutant plants, which expressed 10-20 % of
MIT2 (at the transcript level), 408 genes are down-regulated (cluster 1) and 360 genes up-regulated (cluster 2) compared to wild type and compensated plants (
Figure 9,
Supplementary Tables S4 and S5).
Interestingly, data suggest down-regulation of the iron acquisition system. Besides FRO2, IRT1 and IRT2, genes encoding either proteins involved in coumarin biosynthesis (F6’H1, feruloyl CoA ortho-hydroxylase 1; S8H, Scopoletin 8-hydroxylase; CYP82C4, fraxetin 5-hydroxylase) or coumarin export (ABCG37/PDR9) are included in cluster 1. These genes are part of the Fe deficiency response (Kobayashi and Nishizawa, 2012) and are regulated by the master transcription factor FIT1/FRU (Fe-deficiency induced transcription factor) which is also found in cluster 1 and known to control expression of additional cluster 1 genes involved in Fe homeostasis: IREG2, iron-regulated transporter 2/ferroportin 2; BTSL2, E3 ubiquitin ligase BRUTUS-like protein 2; and MTPA2, metal tolerance protein A2. Therefore, the response to a deficiency in iron uptake by mitochondria may be opposite to the response observed under Fe deficiency (reviewed in Kobayashi and Nishizawa, 2012; Riaz and Guerinot, 2021) and reminiscent of the root Fe exclusion strategy described in rice (Aung and Masuda, 2020).
Other enriched GO categories in cluster 1 related to root system may be indirectly relevant to iron acquisition (
Supplementary Table S6): e.g. “root system development” (GO:0022622; FDR 5.2 x 10
-34, 3.7 fold enrichment), “root morphogenesis” (GO:0010015; FDR 6.1 x 10
-23, 5.9 fold enrichment), “root epidermal cell differentiation” (GO:0010053; FDR 4.3 x 10
-11, 8.8 fold enrichment) and “trichoblast differentiation” (GO:0010054; FDR 2.7 x 10
-10, 9.1 fold enrichment). Given the role of root epidermis and in particular of root hairs in water and nutrient uptake (Gilroy and Jones, 2000; Müller and Schmidt, 2004; Grierson et al., 2014), future work will be necessary to analyze root development in the mutant plants.
Relevant enriched GO terms in cluster 2 (up-regulated in mutant plants) may be involved in the observed pleiotropic phenotypes (
Supplementary Table S7), for instance “plant organ formation” (GO:1905393; FDR 3.2 x 10
-2, 5.1 fold enrichment), “anatomical structure formation involved in morphogenesis” (GO:0048646; FDR 5.0 x 10
-3, 4.1 fold-enrichment), “flower development” (GO:0009908; FDR 4.4 x 10
-4, 3.5 fold enrichment) and “floral organ development” (GO:0048437; FDR 2.2 x 10
-2, 3.7 fold enrichment). Furthermore, the enriched GO categories “production of siRNA involved in gene silencing by small RNA” (GO:0070919; FDR 4.4 x 10
-2, 24.4 fold enrichment), which has three RNA-dependent RNA polymerases (RDR1, RDR2 and RDR3), and “heterochromatin assembly” (GO:0031507; FDR 2.3 x 10
-2, 7.9 fold enrichment), which contains the same three RDR, nucleolin 2 and two chromatin remodeling factors (chr31/SNF2 domain-containing protein CLASSY 3 and chr42/ SNF2 domain-containing protein CLASSY 2), may be relevant to explain the T-DNA suppression phenomenon discussed above.
Although enriched GO terms related to auxin were not found, careful examination of the genes in both clusters (
Supplementary Tables S4 and S5) highlighted a number of genes related to either auxin metabolism, transport, signaling or response (9 in cluster 1, 13 in cluster 2). For instance, PIN2 (At5g57090), ABCG37/PDR9 (At3g53480), ABCB11 (At1g02520), ERULUS (At5g61350), MYB93 (At1g34670) and YUCCA3 (At1g04610) are down regulated in mutant plants (cluster 1). Other genes were found to be up-regulated (cluster 2), for example AIL7/PLT7 (At5g65510), LRP1 (At5g12330), SHI (At5g66350), SGR5 (At2g09140), SKP2A (At1g21410), ENP/MACCHI-BOU4/NPY1 (At4g31820), TCP18 (At3g18530) and GH3.5 (At4g27260). Whether these changes are related to the observed phenotypes similar to those of polar auxin transport mutants remains to be explored.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
All A. thaliana plants used were in the Columbia (Col-0) background. Seeds were sown on half-concentrated MS agar medium and stratified for 48 h at 4 °C in the dark. After two weeks in a 16 h/8 h day/night cycle at 22 °C, seedlings were transferred to soil and grown under long day conditions (16 h light/ 8 h dark).
Seeds from five T-DNA insertion mutants were obtained from the ABRC stock center:
mit1-1 (SALK_013388),
mit1-2 (SALK_208340C),
mit2-1 (SALK_096697),
mit2-2 (SAIL_653_B10, CS828300) and
mit2-3 (SALK_095187). For genotyping DNA was extracted from either 15 days-old seedlings or leaves of 4 weeks-old plants and analyzed as described (León et al. 2007). Primers used to amplify wild type and mutant alleles are described in
Figure 1 and
Supplementary Figures S2 and S3. To further characterize the T-DNA insertion in
mit2-1, located in the intron spliced out in spite of its size, both T-DNA/
MIT2 junctions were amplified (
Supplementary Figure S3). All amplified junctions were characterized by DNA sequencing.
Homozygous mutant mit1-1 plants were crossed with either mit2-1 or mit2-2 homozygous mutant plants. F1 seeds were germinated and F1 plants verified to be double heterozygous plants (genotype MIT1mit1 MIT2mit2). F2 seeds obtained from these selfed F1 plants were used to characterize F2 generation and obtain next generations (F3, F4 and so on).
4.2. Complementation of Mutant Δmrs3Δmrs4 Yeast Cells
The
Δmrs3Δmrs4 strain (
MATa,
ura3-52,
leu2-3,
112,
trp1-1,
his3-11,
ade2-1,
can1-100(
oc),
Δmrs3::kanMax Δmrs4::kanMax) was kindly provided by Liangtao Li and Diane Ward (Department of Pathology, School of Medicine, University of Utah), and is described in Li and Kaplan (2004). The constructs containing the
ADH1 promoter,
MIT1 (At2g30160) or
MIT2 (At1g07030) cDNAs, and the
ADH2 terminator were generated using
in vivo assembly yeast recombinational cloning (Oldenburg et al. 1997, Gibson et al. 2008).
ADH1 promoter and
ADH2 terminator were amplified by PCR using as template a plasmid kindly provided by Dr. Luis Larrondo, and Phusion High-Fidelity DNA polymerase (Thermo Scientific,
https://www.thermofisher.com). For the promoter either primers 1 and 2 (with an overlap in its 5’ end with the beginning of
MIT2 cds) or primers 1 and 3 (with an overlap in its 5’ end with the beginning of
MIT1 cds) were employed, for the terminator either primers 8 (with an overlap in its 5’ end with the end of
MIT2 cds) and 9, or primers 10 (with an overlap in its 5’ end with the end of
MIT1 cds) and 9 were used.
MIT1 and
MIT2 cDNAs were obtained by RT-PCR. Total RNA was prepared from 15 day-old seedlings with the Spectrum Plant Total RNA kit (Sigma-Aldrich). cDNAs were synthesized with the Superscript First Strand synthesis system for RT-PCR (Invitrogen, Life Technologies), and PCR amplifications were performed with primers 4 and 5 for
MIT2, and primers 6 and 7 for
MIT1. PCR products were co-transformed with linear pRS426 plasmid into the BY4741 yeast strain (
MATa,
his3Δ
1,
leu2Δ
0,
LYS2,
met15Δ
0,
ura3Δ
0), and circular plasmids obtained from several ura
+ colonies were transferred to E. coli DH5α. Positive colonies were identified by PCR, plasmids prepared (AxyPrep Plasmid Miniprep kit) and construct integrity verified by DNA sequencing. These plasmids were used to transform the
Δmrs3Δmrs4 strain and transformants were selected by plating on synthetic defined (SD) medium without uracyl. Complementation assays were performed growing yeast cells in liquid SD medium (without uracyl) to DO
600nm of 1.5, concentrating 5 times and plating serial dilutions onto agarose-SD plates with or without 50 µM of the impermeable iron chelator bathophenanthroline disulfonate (BPDS).
4.3. Expression Analysis by RT-PCR and RT-qPCR
Total RNA was obtained from frozen seedlings with the TRIzol reagent, treated with DNase I (Promega,
http://www.promega.com/) and quantified using a Nanodrop spectrophotometer (Thermo Scientific). cDNA synthesis was carried out on 1-2 µg of RNA, using oligodT as primer and SuperScript II Reverse transcriptase (Thermo Scientific).
To analyze
MIT expression by RT-PCR in individual mutants, total RNA was prepared from 15 days-old seedlings. Then PCR reactions were performed with 1/10 of the cDNA and the following primer pairs (
Supplementary Table S1): for
MIT1, either primers 13 and 14 or 15 and 16; for
MIT2, either primers 17 and 18 or 11 and 12.
For RT-qPCR total RNA was obtained from seedlings at stages 1.02 to 1.04 (Boyes et al., 2001). RT-qPCR experiments were performed on 1/10 of the cDNA using the StepOne Plus Real-Time PCR System (Applied Biosystems,
http://www.appliedbiosystems.com/) according to the manufacturer’s instructions and the Brilliant III Ultra-fast SYBR GREEN QPCR reagents (Agilent). RNA levels were estimated considering the amplification efficiency of each primer pair, and were normalized relative to either the clathrin adaptor (At4g24550) or the TIP41L (At4g34270) transcripts as internal controls. The primer pairs (
Supplementary Table S1) used for clathrin adaptor were At4g24550F and At4g24550R, and for TIP41L At4g34270F and At4g34270R. Those for
MIT2 are indicated in figure legends. Primer sequences for
UPOX,
MSM1,
AOX1A,
FRO2 and
IRT1 are also shown in
Supplementary Table S1.
4.4. Transcriptome Analysis by RNA Sequencing
Total RNA was prepared from seedlings (stage 1.04) with the Spectrum Plant Total RNA Mini kit (Sigma-Aldrich). Following DNase I (Invitrogen, Life Technologies) treatment, RNA integrity was evaluated by capillary electrophoresis on a Qsep100 Bio-fragment analyzer (BiOptic,
https://www.bioptic.com.tw). Libraries for RNA-Seq were prepared with the Illumina TruSeq Stranded mRNA kit, quantified by qPCR with KAPA Library Quantification Kit (Universal) (Roche,
https://sequencing.roche.com), and sequenced using the NextSeq 500 System (Illumina,
https://illumina.com), considering 150 bp paired-end reads. Raw sequences were processed with Trimmomatic v.0.39 (Bolger et al., 2014;
http://www.usadellab.org), using the following settings: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:2:keepBothReads LEADING:30 TRAILING:30 SLIDINGWINDOW:10:30 MINLEN:36. Filtered sequences were mapped to the Arabidopsis genome (The Arabidopsis Information Resource TAIR v.10,
www.arabidopsis.org) using HISAT2 v.2.1.0 with standard settings (Kim et al., 2019;
http://daehwankimlab.github.io/hisat2/). Count tables were generated using the featureCounts function from the Rsubread (v.2.10.4) library from R (Liao et al., 2019), and the Araport11 GTF gene annotation (Cheng et al., 2017).
Differential gene expression between genotypes was analyzed with the DESeq2 package (v. 1.24) (Love et al., 2014), using the Likelihood ratio test (LRT). We considered differentially expressed genes those with log2 > 1 or log2 < 1, and adjusted p-value < 0.01. Clustering of differentially expressed genes was performed with the R library pheatmap v. 1.0.12 (kmeans_k = 2).
Overrepresentation of Gene Ontology (GO) terms (biological processes) was analyzed for each cluster using the PANTHER Overrepresentation Test tool (http://geneontology.org; Mi et al., 2013).
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary Figure S1. Arabidopsis MIT1 and MIT2 complement the growth defect of yeast Δmrs3Δmrs4 in Fe-deficient medium. Serial dilutions of the yeast Δmrs3Δmrs4 strain transformed with a construct expressing either Arabidopsis MIT1 (AT2G30160) or MIT2 (AT1G07030) or with the pRS426 vector alone as a control were plated onto SD medium (solified with agarose to avoid Fe contamination and lacking uracyl) with or without the iron chelator BPDS. Supplementary Figure S2. Characterization of mutant mit1 alleles. (A) Exon-intron structure of the MIT1 gene with exons represented by boxes. Horizontal arrows indicate the position of primers and the T-DNA insertion sites are shown (T-DNA inserts are not drawn to scale). (B) Sequence of the MIT1/T-DNA junction in the mit1-1 mutant allele. PCR amplification was performed using primers 13 and LBb1.3, and the product was sequenced. T-DNA is inserted 595 bp downstream of the ATG initiation codon, interrupting codon 200. In the alignment the first row corresponds to the left border T-DNA sequence (in blue), the third row to the wild type MIT1 allele sequence (in red), and the second row to the junction between MIT1 and T-DNA in the mutant allele (inserted nucleotides of unknown origin in green). (C) Sequence of the T-DNA/MIT1junction in the mit1-2 allele. PCR amplification was performed using primers LBb1.3 and 14. T-DNA is inserted 154 bp downstream of the ATG initiation codon, interrupting codon 53. In the alignment the first row corresponds to the left border T-DNA sequence (in blue), the third row to the wild type MIT1 sequence (in red), and the second row to the junction between T-DNA and MIT1 in the mutant allele (inserted nucleotides of unknown origin in green). Supplementary Figure S3. Characterization of mutant mit2 alleles. (A) Exon-intron structure of the MIT2 gene with exons represented by boxes. Horizontal arrows indicate the position of primers and the T-DNA insertion sites are shown (T-DNA inserts are not drawn to scale). (B) Sequences of the T-DNA/MIT2 junctions in the mit2-1 mutant allele. PCR amplifications were performed with either primers LBb1.3 and 12 (3’ side of insertion) or primers 11 and RB (5’ side of insertion), and products were sequenced. T-DNA is inserted 419 bp downstream of the exon-intron junction, 18 nucleotides from the intron have been deleted (not shown) and 15 nucleotides of unknown origin inserted (in green). In the alignments first rows correspond to the left or right border T-DNA sequences (in blue), the third row to the wild type MIT2 sequence (in red), and the second row to the junctions between MIT2 and T-DNA in mit2-1. (C) Sequence of the MIT2/T-DNA junction in the mit2-2 allele. PCR amplification was performed using primers 11 and LB1sail. T-DNA is inserted 712 bp downstream of the ATG initiation codon, interrupting codon 239. In the alignment the first row corresponds to the left border T-DNA sequence (in blue), the third row to the wild type MIT2 sequence (in red), and the second row to the junction between MIT2 and T-DNA in the mutant allele. (D) Sequence of the MIT2/T-DNA junction in the mit2-3 allele. PCR amplification was performed using primers 11 and LBb1.3. T-DNA is inserted 500 bp downstream of exon-intron junction. In the alignment the first row corresponds to the left border T-DNA sequence (in blue), the third row to the wild type MIT2 sequence (in red), and the second row to the junction between MIT2 and T-DNA in the mutant allele. Supplementary Figure S4. A significant proportion of seeds in plants with three mit mutated alleles have an abnormal phenotype. Seed set per silique was scored along the main inflorescence axis for four F3 MIT1mit1 mit2mit2 plants, four F3 mit1mit1MIT2mit2 plants and two wild type plants. Silique 5 (older) to silique 16 (younger) were grouped in three groups of four siliques for each plant, and normal, abnormal and aborted seeds were counted in each of the 16 siliques of plants carrying three mutated alleles and the 8 siliques of Col-0 plants. Error bars are SD. Supplementary Figure S5. Abnormal seeds from plants with only one wild type MIT allele germinate slowly. Wild type seeds and both normal and abnormal seeds from MIT1mit1-1 mit2-1mit2-1 plants were sown on 0.5 x MS, stratified for 48 h and grown for 3 and 4 days before being photographed. Supplementary Figure S6. MIT2 expression in double homozygous mutant plants. MIT2 expression levels were determined by RT-qPCR using primers 19 and 21 (Supplementary Table S1) and normalized to clathrin adaptor transcript level. Means ± SD of three biological replicates are shown. RNAs were prepared from 6-12 seedlings at stage 1.02 (Boyes et al., 2001), i.e. 12 days-old seedlings for wild type and 19 days-old seedlings for the first generation of the double homozygous mutant plants (mit1-1mit1-1 mit2-1mit2-1). Three asterisks indicate a value that was determined by the t-test to be significantly different from wild type (p < 0.001). To evaluate MIT2 gene expression, primers 19 and 21 were used. Supplementary Figure S7. Growth of double homozygous mutant plants was retarded throughout the entire life cycle. Wild type Col-0 plants and double homozygous mutant plants (mit1-1mit1-1 mit2-1mit2-1, 1st generation) were grown as described in Materials and methods and three representative plants of each genotype were photographed at 2 months (A) and 3.5 months (B). Supplementary Figure S8. Analysis of purified mitochondria from “compensated” double homozygous plants (mit1-1mit1-1 mit2-1mit2-1) and wild type plants. (A) Proteomic analysis of the purified mitochondria (four mitochondrial preparations for each genotype). Heat map showing the relative abundance in both genotypes of 274 peptides corresponding to 108 polypeptides (Supplementary Table S3). Only the abundances of MIT1 (not detected in the mutant) and of MIT2 were significantly different (p < 0.001, t-test) between wild type and compensated plants. (B) Oxygen consumption by purified mitochondria using pyruvate, malate and ADP as substrates. Four mitochondrial preparations per genotype were tested and means ± SD are shown. Difference between genotypes is not significant (t-test). (C) BN-PAGE separation of respiratory chain complexes and supercomplexes followed by staining for Complex I activity (NADH:UQ oxidoreductase; right panel) and Coomassie blue staining (left panel). Three replicates (each 0.3 mg mitochondrial protein) of purified mitochondria from wild-type and “compensated” double homozygous seedlings were loaded. Mobility of respiratory complexes I, II, III, IV and V and supercomplexes are indicated.
Acknowledgments
This work was supported by a grant from FONDECYT-Chile 1160334 to HR, Facultad de Ciencias Biológicas-PUC grant to HR, and CONICYT-Chile grant 21151344 to JVP, and the Australian Research Council to AHM (CE140100008, FL200100057). We thank Ricarda Fenske for helping with the samples preparation and analysis of proteomics studies.
Conflicts of Interest
The authors have no conflict of interest to declare
Abbreviations
BN-PAGE: blue native polyacrylamide gel electrophoresis; bp, base pair; BPDS, bathophenanthroline disulfonate; nt, nucleotide; GO, gene ontology; MIT, mitochondrial iron transporter; qRT-PCR: quantitative reverse transcription-PCR; RNA-seq, RNA sequencing.
References
- Aida, M. , Vernoux, X., Furutani, M., Traas, J. and Tsaka, M. Roles. of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 2002, 129, 3965–3974. [Google Scholar] [CrossRef]
- Anders, S. and Huber, W. Differential. expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Aung, M. S. and Masuda, H. (2020). How does rice defend against excess iron?: Physiological and molecular mechanisms. Front. Plant Sci. 2020, 11, 1102. [Google Scholar] [CrossRef]
- Bashir, K. , Ishimaru, Y., Shimo, H., Nagasaka, S., Fujimoto, M., Takanashi, H., Tsutsumi, N., An, G. and Nishizawa, N. K. The. rice mitochondrial iron transporter is essential for plant growth. Nature Commun 2011, 2, 322. [Google Scholar] [CrossRef] [PubMed]
- Benjamins, R. , Quint, A., Weijers, D., Hooykaas, P. and Offringa, R. The. PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 2001, 128, 4057–4067. [Google Scholar] [CrossRef]
- Benková, E. , Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G. and Friml, J. Local,. efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S. R. M. , Alvarez, J., Bossinger, G. and Smith, D.R. Morphogenesis. in pinoid mutants of Arabidopsis thaliana. Plant J. 1995, 8, 505–520. [Google Scholar] [CrossRef]
- Bolger, A. M. , Lohse, M. and Usadel, B. Trimmomatic:. a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Boyes, D.C. , Zayed, A.M., Ascenzi, R., McCaskill, A.J., Hoffman, N.E., Davis, K.R., et al. Growth. stage-based phenotypic analysis of Arabidopsis: a model for hgh throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar]
- Briat, J.F. Briat, J.F., Dubos, C., Gaymard, F. Iron nutrition, biomass production, and plant product quality. TRENDS in Plant Sci. 2015, 20: 33-40.
- Chang, N. , Sun, Q., Hu, J., An, C. and Gao, H. Large. introns of 5 to 10 kilo base pairs can be spliced out in Arabidopsis. Genes 2017, 8, 200. [Google Scholar] [CrossRef]
- Cheng, C. Y. , Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S. and Town, C. D. Araport11:. a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S. K. , Dagenais, N., Chory, J. and Weigel, D. Regulation. of auxin response by the protein kinase PINOID. Cell 2000, 100, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Clifton, R. , Lister, R., Parker, K. L, Sappl, P. G., Elhafez, D., Millar, A. H., Day, D. A. and Whelan, J. Stress-induced. co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant. Mol. Biol. 2005, 58, 193–212. [Google Scholar] [CrossRef] [PubMed]
- Collins, J. , Anderson G. Physiology. of the Gastrointestinal Tract (Fifth Edition) 2012, Elsevier, New York.
- Curie, C. , Cassin, G., Couch, D., Divol, F., Higuchi, K., Le Jean, M., Misson, J., Schikora, A., Czernic, P., Mari, S. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Annals of botany 2009, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- De Longevialle, A. F. , Meyer, E. H., Andrés, C., Taylor, N. L., Lurin, C., Millar, A. H. and Small, I. D. The. pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 2007, 19, 3256–3265. [Google Scholar] [CrossRef] [PubMed]
- 18. Divol F, Couch D, Conéjéro G, Roschzttardtz H, Mari S, Curie C. The. Arabidopsis YELLOW STRIPE LIKE4 and 6 transporters control iron release from the chloroplast. Plant Cell. 25(3):1040-55.
- Duy, D. , Wanner, G., Meda, A.R., Wiren, N. von, Soll, J. Philippar, K. PIC1, an Ancient Permease in Arabidopsis Chloroplasts, Mediates Iron Transport. Plant Cell 2007, 19, 986–1006. [Google Scholar] [CrossRef] [PubMed]
- Eide, D. , Broderius, M., Fett, J., Guerinot, M. L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences of the United States of America 1996, 93, 5624–5628. [Google Scholar] [CrossRef]
- Eubel, H. , Braun, H. P. and Millar, A. H. Blue-native. PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 2005, 1, 11. [Google Scholar]
- Eubel, H. , Jänsch, L. and Braun, H. P. New. insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 2003, 133, 274–286. [Google Scholar]
- Foury, F. and Roganti, T. Deletion. of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 2002, 277, 24475–24483. [Google Scholar]
- Furutani, M. , Vernoux, T., Traas, J., Kato, T., Tasaka, M. and Aida, M. PIN-FORMED1. and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 2004, 131, 5021–5030. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y. and Zhao, Y. Epigenetic suppression of T-DNA insertion mutants in arabidopsis. Mol. Plant 2013, 6, 539–545. [Google Scholar] [CrossRef]
- Guerinot, M.L. , Yi Y. Iron:. nutritious, noxious, and not readily available. Plant Physiol. 1994, 104, 815–820. [Google Scholar]
- Gibson, D. G. , Benders, G. A., Axelrod, K. C., Zaveri, J., Algire, M. A., Moodie, M., Montague, M. G., Craig Venter, J., Smith, H. O. and Hutchison III, C. A. One-step. assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Nat. Acad. Sci. USA 2008, 105, 20404–20409. [Google Scholar]
- Gilroy, S. and Jones, D. L. Through. form to function: root hair development and nutrient uptake. Tr. Plant Sci. 2000, 5, 56–60. [Google Scholar] [CrossRef]
- Grierson, C. , Nielsen, E., Ketelaarc, T. and Schiefelbein, J. Root. Hairs. The Arabidopsis Book 2014, 12, e0172. [Google Scholar] [CrossRef]
- Haferkamp I, Schmitz-Esser S. (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci. 8;3:2.
- Ho 2012, L. H. , Giraud, E., Uggalla, V., Lister, R., Clifton, R., Glen, A., Thirkettle-Watts, D., Van Aken, O. and Whelan, J. Identification. of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 2008, 147, 1858–1873. [Google Scholar]
- Ivanova, A. , Law, S. R., Narsai, R., Duncan, O., Lee, J-H., Zhang, B., Van Aken, O., Radomiljac, J. D., van der Merwe, M., Yi, K. K. and Whelan, J. A. functional relationship between auxin and mitochondrial retrograde signaling regulates Alternative Oxidase1a expression in Arabidopsis. Plant Physiol. 2014, 165, 1233–1254. [Google Scholar]
- Jain, A. , Dashner, Z. S. and Connolly, E. L. Mitochondrial. iron transporters (MIT1 and MIT2) are essential for iron homeostasis and embryogenesis in Arabidopsis thaliana. Front. Plant Sci 2019, 10, 1449. [Google Scholar] [CrossRef]
- Jeong, J. , Cohu, C., Kerkeb, L., Pilon, M., Connolly, E.L. Guerinot, M. Lou Chloroplast. Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10619–10624. [Google Scholar] [CrossRef]
- Kerchev, P. I. , De Clercq, I., Denecker, J., Mühlenbock, P., Kumpf, R., Nguyen, L., Audenaert, D., Dejonghe, W. and Van Breusegem, F. Mitochondrial. perturbation negatively affects auxin signaling. Mol. Plant 2014, 7, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Kim, D. , Paggi, J. M., Park, C., Bennett, C. and Salzberg, S. L. Graph-based. genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- 37. Kim LJ, Tsuyuki KM, Hu F, Park EY, Zhang J, Iraheta JG, Chia JC, Huang R, Tucker AE, Clyne M, Castellano C, Kim A, Chung DD, DaVeiga CT, Parsons EM, Vatamaniuk OK, Jeong J. Ferroportin. 3 is a dual-targeted mitochondrial/chloroplast iron exporter necessary for iron homeostasis in Arabidopsis. Plant J.
- Kobayashi, T. and Nishizawa, N. K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Kolli, R. , Soll, J. and Carrie, C. OXA2b. is crucial for proper membrane insertion of COX2 during biogenesis of Complex IV in Plant Mitochondria. Plant Physiol. 2019, 179, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K. , Obata, T. , Feher, K., Bock, R., Fernie, A. R. and Meyer, E. H. Complete. mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015, 168, 1537–1549. [Google Scholar] [PubMed]
- León, G. , Holuigue, L. and Jordana, X. Mitochondrial. complex II is essential for gametophyte development in Arabidopsis. Plant Physiol. 2007, 143, 1534–1546. [Google Scholar] [CrossRef] [PubMed]
- Li, L. and Kaplan, J. A. mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem. 2004, 279, 33653–33661. [Google Scholar]
- Liao, Y. , Smyth G. , Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads, Nucleic Acids Research 2019, 47, 8. [Google Scholar]
- Liu, C. , Xu, A. and Chua, N. Auxin. polar transport is essential for the establishment of bilateral simmetry during early plant embryogenesis. Plant Cell 1993, 5, 621–630. [Google Scholar] [CrossRef]
- Love, M.I. , Huber, W. & Anders, S. Moderated. estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar]
- Meyer, E. H. , Tomaz, T., Carroll, A. J., Estavillo, G., Delannoy, E., Tanz, S. K., Small, I. D., Pogson, B. J. and Millar, A. H. Remodeled. respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009, 151, 603–619. [Google Scholar] [PubMed]
- Mi, H. , Muruganujan, A., Casagrande, J. T. and Thomas, P. D. Large-scale. gene function analysis with the panther classification system. Nature Prot. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Mühlenhoff, U. , Stadler, J. A., Richhardt, N., Seubert, A., Eickhorst, T., Schweyen, R. J., Lill, R. and Wiesenberger, G. A. specific role of the yeast mitochondrial carriers Mrs3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 2003, 278, 40612–40620. [Google Scholar] [CrossRef] [PubMed]
- Müller, M. and Schmidt, W. Environmentally. induced plasticity of root hair development in Arabidopsis. Plant Physiol. 2004, 134, 409–419. [Google Scholar] [CrossRef]
- Ohbayashi, I. , Huang, S., Fukaki, H., Song, X., Sun, S., Morita, M. T., Tasaka, M., Millar, A. H. and Furutani, M. Mitochondrial. pyruvate dehydrogenase contributes to auxin-regulated organ development. Plant Physiol. 2019, 180, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Okada, K. , Ueda, J., Komaki, M. K., Bell, C. J. and Shimura, Y. Requirement. of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K. R. , Vo, K. T., Michaelis, S. and Paddon, C. Recombination-mediated. PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997, 25, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Osabe, K. , Harukawa, Y. , Miura, S. and Saze, H. Epigenetic Regulation of Intronic Transgenes in Arabidopsis. Sci. Rep. 2017, 7, 45166. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, G. , Pantopoulos K. Iron. metabolism and toxicity. Toxicol. Appl. Pharmacol. 2005, 202, 199–211. [Google Scholar] [CrossRef]
- Rajniak, J. , Giehl, R. F. H., Chang, E., Murgia, I., von Wirén, N. and Sattely, E. S. Biosynthesis. of redox-active metabolites in response to iron deficiency in plants. Nature Chem. Biol. 2018, 14, 442–450. [Google Scholar] [CrossRef]
- Restovic, F. , Espinoza-Corral, R., Gómez, I., Vicente-Carbajosa, J. and Jordana, X. An. active mitochondrial complex II present in mature seeds contains an embryo-specific iron–sulfur subunit regulated by ABA and bZIP53 and is involved in germination and seedling establishment. Front. Plant Sci. 2017, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N. and Guerinot, M. L. All. together now: regulation of the iron deficiency response. J. Exp. Bot. 2021, 72, 2045–2055. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N. J. , Procter, C. M., Connolly, E. L., Guerinot, M. L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Roschzttardtz, H. , Fuentes, I., Vásquez, M., Corvalán, C., León, G., Gómez, I., Araya, A., Holuigue, L., Vicente-Carbajosa, J. and Jordana, X. A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol. 2009, 150, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K. S. , Koirala, P. S. and Neff, M. M. The. ben1-1 brassinosteroid-catabolism mutation is unstable due to epigenetic modifications of the intronic T-DNA insertion. G3 (Bethesda) 2013, 3, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Santi, S. , Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. The New phytologist 2009, 183, 1072–1084. [Google Scholar] [PubMed]
- Schmidt SB, Eisenhut M, Schneider A. Chloroplast Transition Metal Regulation for Efficient Photosynthesis. Trends Plant Sci. 2020, 25, 817–828. [Google Scholar] [CrossRef]
- Tan YF, O’Toole N, Taylor NL, Millar AH Divalent. metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol. 2010, 152, 747–761.
- Tivendale ND, Belt K, Berkowitz O, Whelan J, Millar AH, Huang S. (2021) Knockdown of Succinate Dehydrogenase Assembly Factor 2 Induces Reactive Oxygen Species-Mediated Auxin Hypersensitivity Causing pH-dependent Root Elongation. Plant Cell Physiol. 62:1185-1198.
- Tivendale ND, Millar AH. (2022) How is auxin linked with cellular energy pathways to promote growth? New Phytol. 2022 233:2397-2404.
- Tsai, H. H. , Rodríguez-Celma, J. , Lan, P., Wu, Y. C., Vélez-Bermúdez, I. C. and Schmidt, W. Scopoletin. 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol. 2018, 177, 194–207. [Google Scholar]
- Van Aken, O. , Zhang, B., Carrie, C., Uggalla, V., Paynter, E., Giraud, E. and Whelan, J. Defining. the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2009, 2, 1310–1324. [Google Scholar] [CrossRef]
- Van Aken, O. and Whelan, J. Comparison. of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front. Plant Sci. 2012, 3, 281. [Google Scholar] [CrossRef] [PubMed]
- Vert, G. , Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M.L., Briat, J.F., Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Vigani, G. , Bashir, K., Ishimaru, Y., Lehmann, M., Casiraghi, F. M., Nakanishi, H., Seki, M., Geigenberger, P., Zocchi, G. and Nishizawa, N. K. Knocking. down mitochondrial iron transporter (MIT) reprogramsprimary and secondary metabolism in rice plants. J. Exp. Bot. 2016, 67, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Vigani G, Maffi D, Zocchi G. Iron. availability affects the function of mitochondria in cucumber roots. New Phytol. 2009,182:127-136.
- Vigani G, Solti A, Thomine SB, Philippar K. Essential. and Detrimental - an Update on Intracellular Iron Trafficking and Homeostasis. Plant Cell Physiol. 2019, 60, 1420–1439. [Google Scholar] [CrossRef] [PubMed]
- Walter PB, Knutson MD, Paler-Martinez A, Lee S, Xu Y, Viteri FE, Ames BN. Iron. deficiency and iron excess damage mitochondria and mitochondrial DNA in rats. Proc Natl Acad Sci USA 2002, 99, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. , Lyu, W., Berkowitz, O., Radomiljac, J. D., Law, S. R., Murcha, M. W., Carrie, C., Teixera, P. F., Kmiec, B., Duncan, O., Van Aken, O., Narsai, R., Glaser, E., Huang, S., Roessner, U., Millar, A. H. and Whelan, J. Inactiavtion of complex I induces the expression of a twin cysteine protein that targets and affects cytosolic, chloroplastidic and mitochondrial function. Mol. Plant 2016, 9, 696–710. [Google Scholar] [PubMed]
- Xue, W. , Ruprecht, C., Street, N., Hematy, K., Chang, C., Frommer, W. B., Persson, S. and Niittylä, T. Paramutation-like. Interaction of T-DNA loci in Arabidopsis. PLoS ONE 2012, 7, e51651. [Google Scholar] [CrossRef]
- Yi, Y. , & Guerinot, M. L. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary foriron uptake under iron deficiency. The Plant journal: for cell and molecular biology 1996, 10, 835–844. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).