Submitted:
05 January 2023
Posted:
06 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- health data scientists, companies, or institutions that need a general workflow for routine - possibly automated - clustering projects on data of various types and volumes (e.g. for preliminary data mining), or
- health data scientists with limited experience, who are looking for both an overview of the literature and an accessible and reusable workflow with concrete practical recommendations to quickly implement a complete unsupervised clustering approach adapted to various projects.
2. Statistical rationale and literature review on data clustering
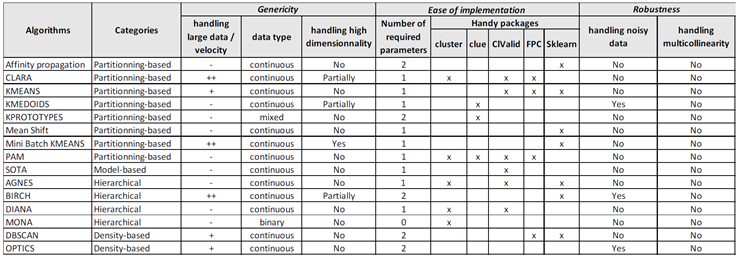
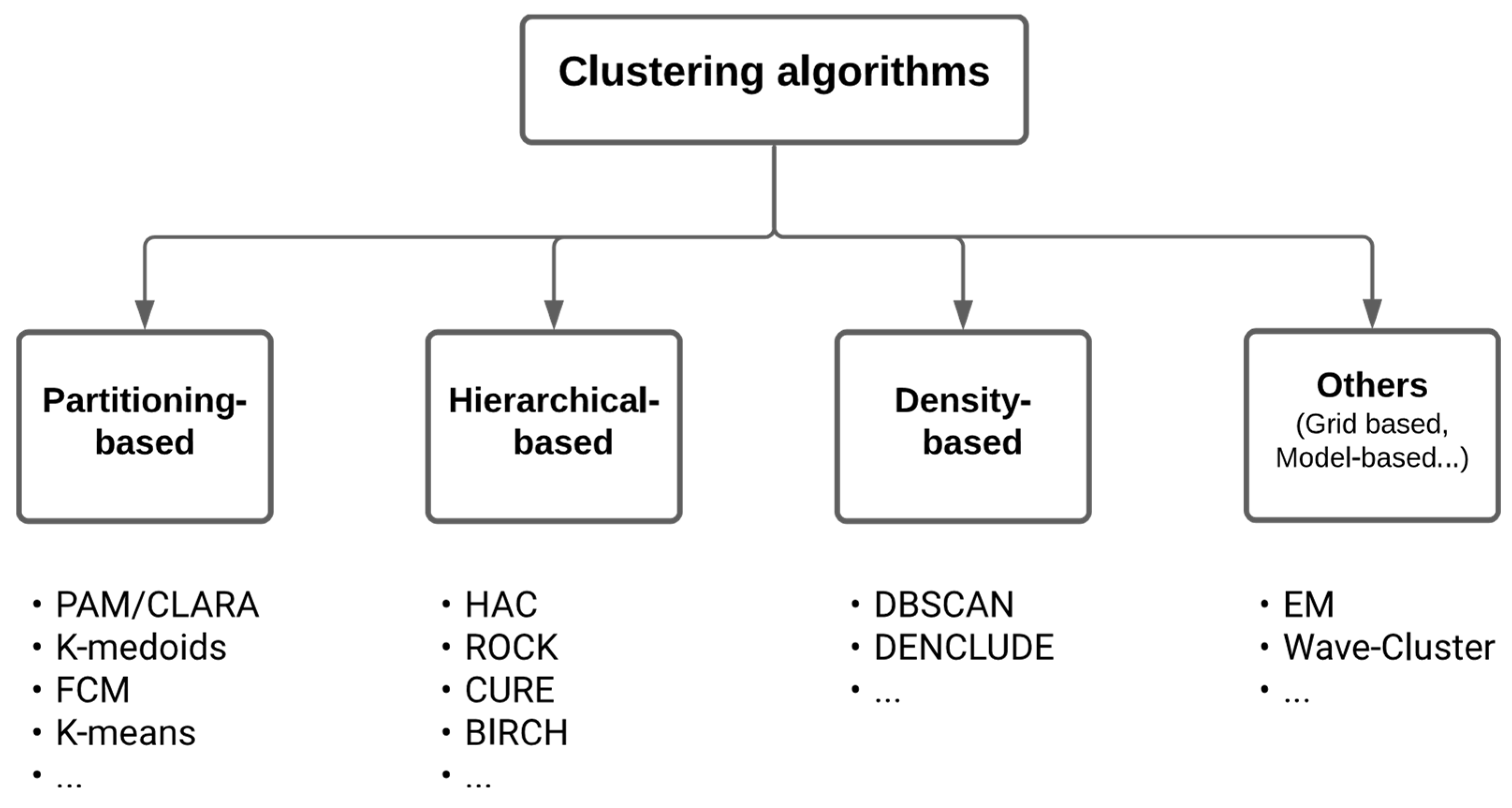
2.1. Overview of unsupervised clustering methods
- Partitioning-based methods (e.g. K-means (MacQueen 1967), K-medoid (Jin et al. 2010), PAM (Ng et al. 1994), K-modes (Huang 1997), K-prototype (Huang 1998), CLARA (Kaufman et al. 2009), FCM (Bezdek, Ehrlich, et al. 1984), etc 2.),
- Hierarchical-based methods (e.g. BIRCH (Zhang et al. 1996), CURE (Guha et al. 1998), ROCK (Guha et al. 2000), etc.),
- Density-based methods (e.g. DBSCAN (Ester et al. 1996), DENCLUDE (Keim et al. 1998), etc.).
2.2. Choosing an appropriate clustering approach
- its ability to handle the desired type of data (binary / nominal / ordinal / numerical),
- the dimensionality of the data (see e.g. Mittal et al. (2019)),
- the size of the data (small to large data),
- the availability of reliable implementations in software (e.g. R and Python - the 2 most used statistical software by data scientists).
2.3. Methods for clusters description
2.4. Methods for clustering validity and stability assessment
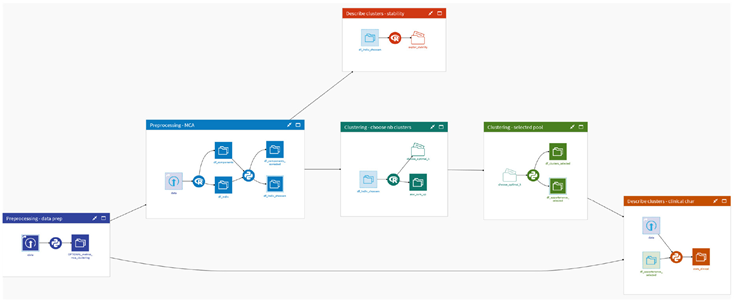
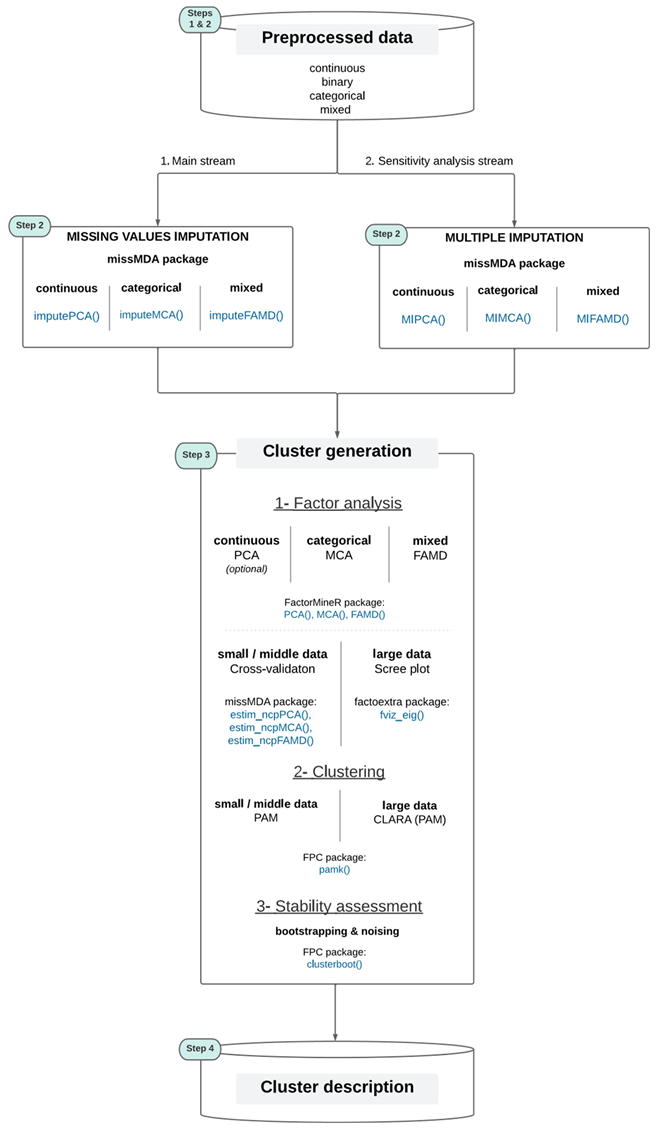
3. The Qluster workflow
3.1. Research objective
3.2. Method
- Criteria for achieving genericity: applicability to small and big data, applicability to continuous or categorical or mixt data, and management of high dimensionality.
- Criteria for achieving ease of implementation and use: number of packages used, of algorithms used, of parameters to tune, use of “all-inclusive” packages covering at best the general clustering process
- Criteria for achieving robustness and reliability: management of noise data, of multicollinearity, methods considered for clusters’ stability assessment, reliability of packages used (e.g. hosting site, renown, …), reliability of algorithms used (e.g. renown, literature, …).
3.3. Preliminary work
3.4. Qluster
- Adapted to variables of any nature, be it categorical only, continuous only, or a mix of both. This is made possible by transforming all data in the continuous setting (that is both more mature in the literature and simpler to process) using methods for factor analysis (MCA for categorical data only or FAMD for mixed data (Pages` 2004)). As mentioned in section 2.2, the latter also allows for dealing with collinearity, high-dimensionality and noise. It also makes the work for a clustering algorithm easier, as there are both fewer variables to deal with and greater clarity in information to cluster (factor analysis methods are themselves meant to uncover profiles into components of richer information).
- Adapted to datasets of any volume, be it small or large data. Indeed, the same partitioning-based algorithm is used (PAM), either applied entirely on a dataset of reasonable size, or on samples of a large 18 dataset (CLARA algorithm), using the same pamk() function from the FPC R package. In addition to this practical aspect, PAM was chosen over the (widely used) K-means algorithm based on its ability to be deterministic and to deal with Manhattan distance which is less sensitive to outliers than the Euclidean distance (Jin et al. 2010). Moreover, PAM is known for its simplicity of use (fewer parameters e.g. than with DBSCAN or BIRCH (Fahad et al. 2014)) and is also implemented in an easy-to-use all-inclusive package (not the case for e.g. CLARANS, KAMILA, Mini batch K-means, DENCLUE and STING that are suited for large datasets but where assessing clusters stability would require extensive code development by data scientists). More details on the choice of the clustering algorithm can be found in section 5.
- Both the tasks of clustering and clusters stability assessment are handled using the FPC R package (functions pamk() and clusterboot() respectively). R has been chosen over Python because the former offers all the clustering methods desired, and no package including all the steps of interest to clustering was found for the latter (one would have to code some steps by him/herself, see section 2 for more details). The clusterboot() function offers many ways to assess clusters’ stability, but one selects the two followings for routine practice and for their complementarity as mentioned in Hennig (2008): bootstrapping and noising.
- The factor analysis part is handled using the FactoMineR R package (function PCA(), MCA(), FAMD(), for continuous, categorical, and mixed data respectively - the latter function generalizing the others). This step is optional in the case where only continuous variables are in input 19. To select the optimal number of components to keep, one recommends for small data to use the deterministic cross-validation technique implemented in the missMDA package (function estim ncpPCA(), estim ncpMCA(), estim ncpFAMD() (Josse, Chavent, et al. 2012)). As this method requires high computing time, the standard “elbow” method in a scree plot is recommended for large data, using the factoextra R package (function fviz eig()).
| Algorithm 1: The Qluster pseudo-code |
 |
4. Detailed workflow methodology
4.1. The Cardiovascular Disease dataset and the objective
- Age (days, converted into years) - age
- Height (cm) - height
- Weight (kg) - weight
- Gender (M/F) - gender
- Systolic blood pressure (SBP) (mmHg) - ap hi
- Diastolic blood pressure (DBP) (mmHg) - ap lo
- Cholesterol (categories 1: normal, 2: above normal, 3: well above normal) - cholesterol
- Glucose (categories 1: normal, 2: above normal, 3: well above normal) - gluc
- Smoking (Y/N) - smoke
- Alcohol intake (Y/N) - alco
- Physical activity (Y/N) - active
- Presence or absence of cardiovascular disease (Y/N) - cardio
4.2. Step-by-step application of the Qluster workflow
4.2.1. Data preparation
- Glucose (gluc): modalities 2 (above normal, 8.8%) and 3 (well above normal, 9.5%) were grouped into one modality 2 (above normal)
- BMI (BMI): modalities “underweight” (0.5%) and “normal” were grouped into one modality “underweight & normal”.
4.2.2. Perform Multiple Correspondence Analysis
4.2.3. Clustering of the data
- Distance measure: dissimilarity matrix was computed using the Manhattan distance. The latter is more robust and less sensitive to outliers than the standard Euclidean distance (Jin et al. 2010).
-
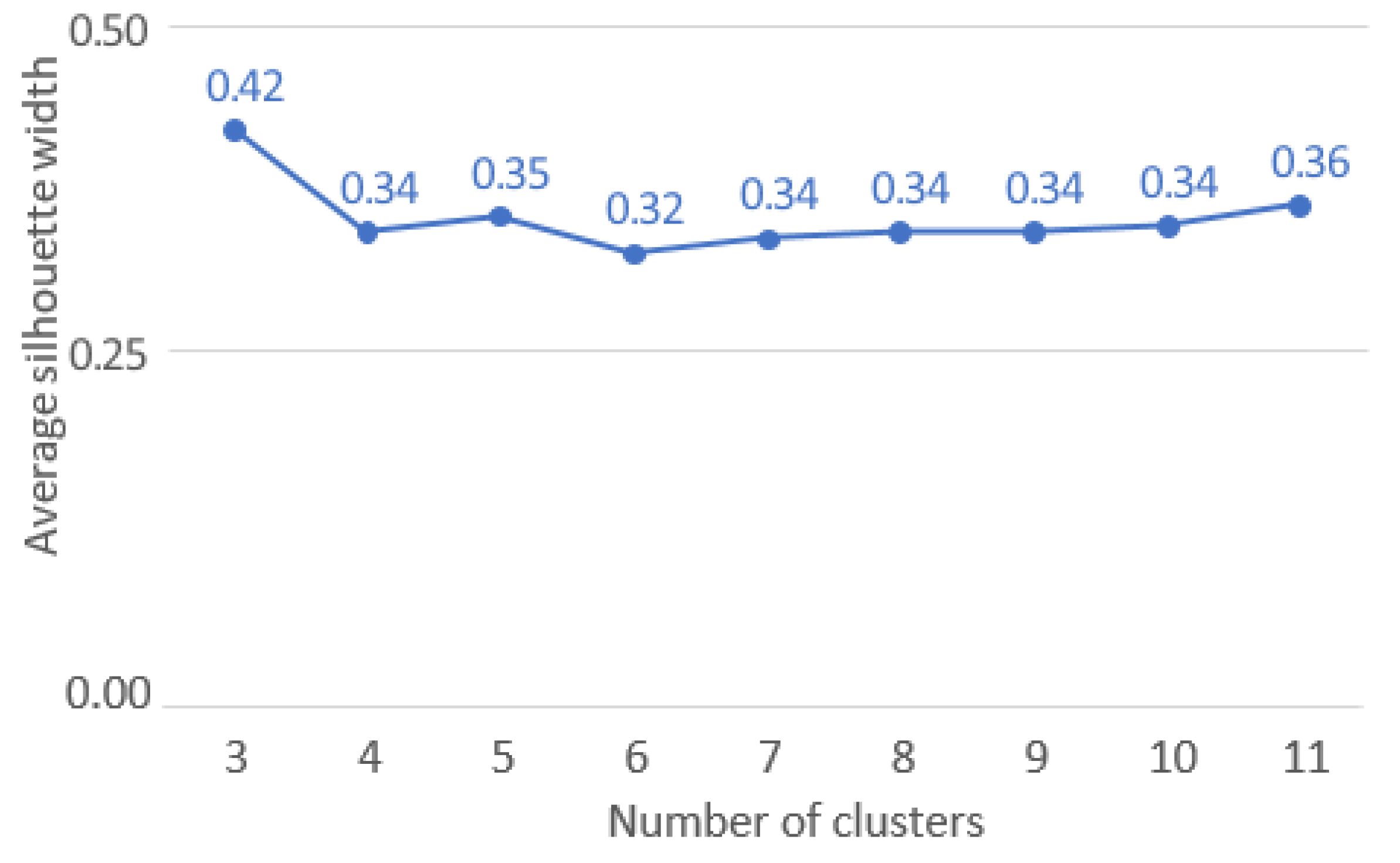
Number K of clusters: From 3 to 11.
- -
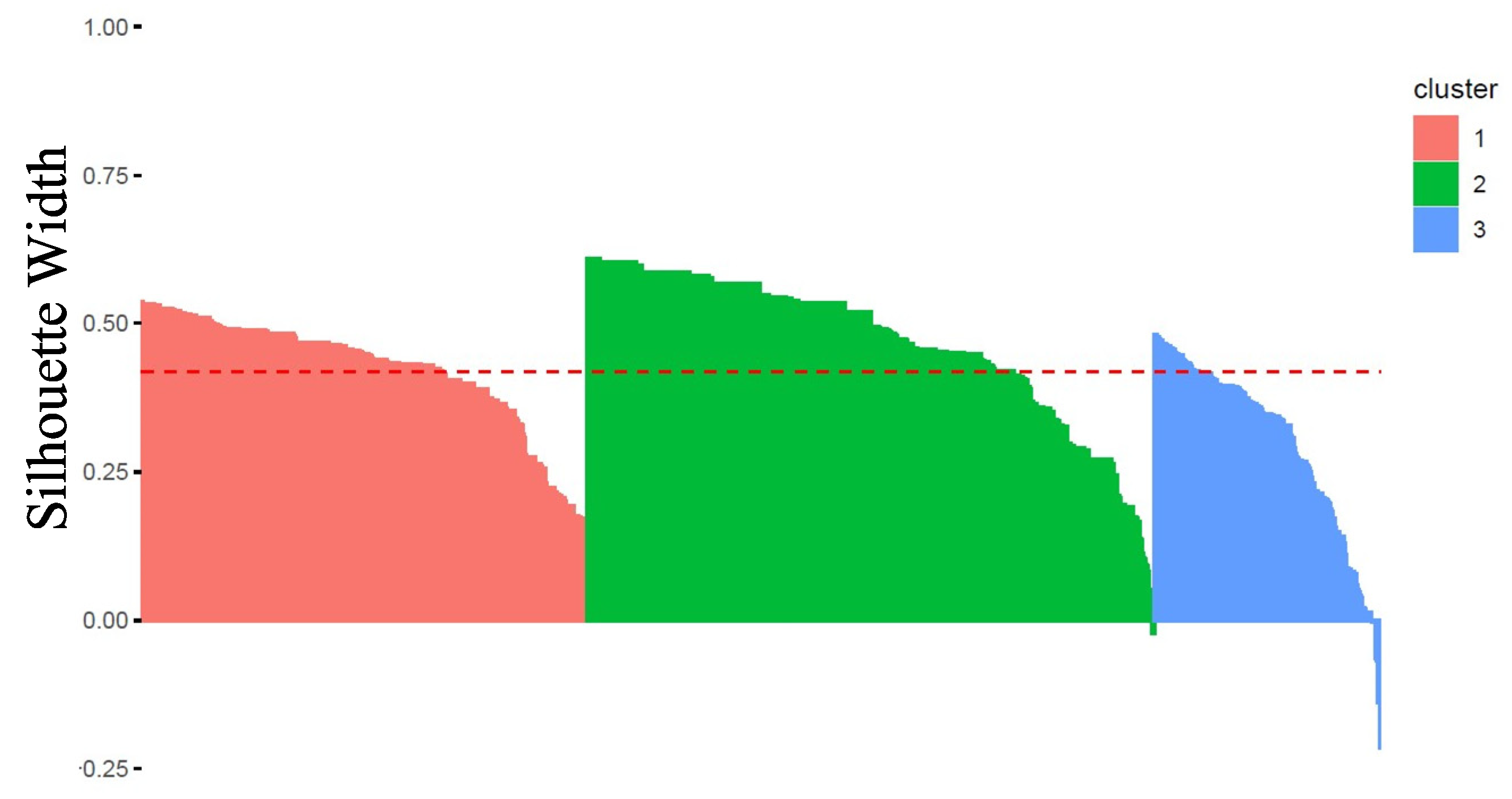
- The number of clusters was optimized on the Average Silhouette Width (ASW) quality measure, that is an internal validity metric reflecting the compactness and separation of the clusters. The ASW is based on Silhouettes Width that were calculated for all patients in the best sample, i.e., the one used to obtain cluster medoids and generate clusters (Rousseeuw 1987).
- -
- The range of clusters to be tested was determined to enable the identification of phenotypically similar subgroups while not generating an excessive number of subgroups for interpretation.
-
Number of samples and sample size: 100 samples of 5% study population size (1,706 patients).
- -
- Experiments have shown that 5 samples of size 40+2C (with C the number of variables in input) give satisfactory results (Kaufman et al. 1990). However, increasing these numbers is recommended (if feasible), to limit sampling biases and favor converging toward the best solution. Equally, the higher the sample size is, the higher it is representative of the entire dataset. We therefore recommends to pretest on his own material up to what parameters values the computation times are acceptable considering the size of the input dataset and the other steps of the workflow (including the clusters stability evaluation step, the most time consuming).
4.2.4. Clusters stability assessment
-
Bootstrap approach:
- -
- This approach consists in performing the clustering as described in section 4.2.3 on B = 50 bootstrapped data (i.e. random sampling with replacement (Efron 1979; Efron and R. J. Tibshirani 1994)), using the clusterboot() function in the FPC R package (version 2.2-5).
- -
- The Jaccard similarity metric is used to compute, for each cluster, the proximity between the clusters of patients obtained on the modified population and the original clusters. It is given by the number of patients in common between the new cluster (modified population) and the original cluster divided by the total number of distinct patients considered (i.e. present in either the new or the original cluster).
- -
-
For each cluster, the following results are provided:
- ■
- the mean of the Jaccard similarity statistic
- ■
- the number of times the cluster has been “dissolved”, defined as a Jaccard similarity value ≤ 0.5. This value indicates instability.
- ■
- the number of times the cluster has been “recovered”, defined as a Jaccard similarity value ≥ 0.75. This value indicates stability.
-
Noise approach:
- -
-
This approach consists in performing the clustering as described in section 4.2.3 on B = 50 noisy data and for different values of noise, using the clusterboot() function in the FPC R package.
- ■
- Level of noise values: from 1% to 10%
- -
- The number of times each cluster was “dissolved” and “recovered” is provided, as well as the mean of the Jaccard similarity statistic, according to the noise values.
- For the bootstrap approach, clusters 1, 2 and 3 have all 100% of Jaccard similarity statistics over 50 iterations. The 3 clusters were recovered for 100% of bootstrapped iterations, which characterizes a very high stability to resampling with replacement.
- For the noise approach, clusters 1, 2 and 3 have at worst (for 2% of noise) 100%, 98%, 96% of Jaccard similarity statistics respectively over 50 iterations. The 3 clusters were recovered for 100% of iterations and regardless of the level of noise (from 1% to 10%), which characterizes a very high stability to noise.
4.2.5. Clusters interpretation
5. Discussion
5.1. Limitations and proposition for enhancing this workflow
- The packages used cannot handle the ordinal nature of the variables. The latter must be treated as categorical or continuous.
- The observations x components matrix is continuous, although some raw variables could be categorical. This prevents the user from favoring (respectively, not favoring) positive cooccurrence over negative co-occurrence via the Jaccard (respectively, Hamming) distance
- one of the best known, studied and used algorithm by the community, for general purpose, - adapted to the continuous setting (i.e., the most mature in the literature).
- meant for the most frequent use case of clustering (i.e., hard partitioning),
- suitable to the Manhattan distance, a less sensitive to outliers distance, unlike its counterpart on the euclidean distance (K-means),
- deterministic, thanks to its internal medoid initialization procedure, unlike the basic K-means algorithm that may lead to inconsistent or non-reproducible clusters
- requiring few parameters to set up (e.g. conversely to BIRCH and DBSCAN, see Fahad et al.(2014)),
- very well implemented within a recognized reference R package (the FPC package) facilitating its use within a complete and robust clustering approach,
- usable within the same R function (pamk()) regardless of the volume of data.
5.2. Discussion and recommendation on the practical use of the workflow
6. Conclusion
Supplementary Material
Author Contributions
Acknowledgment
Conflict of Interest
Appendix
 |
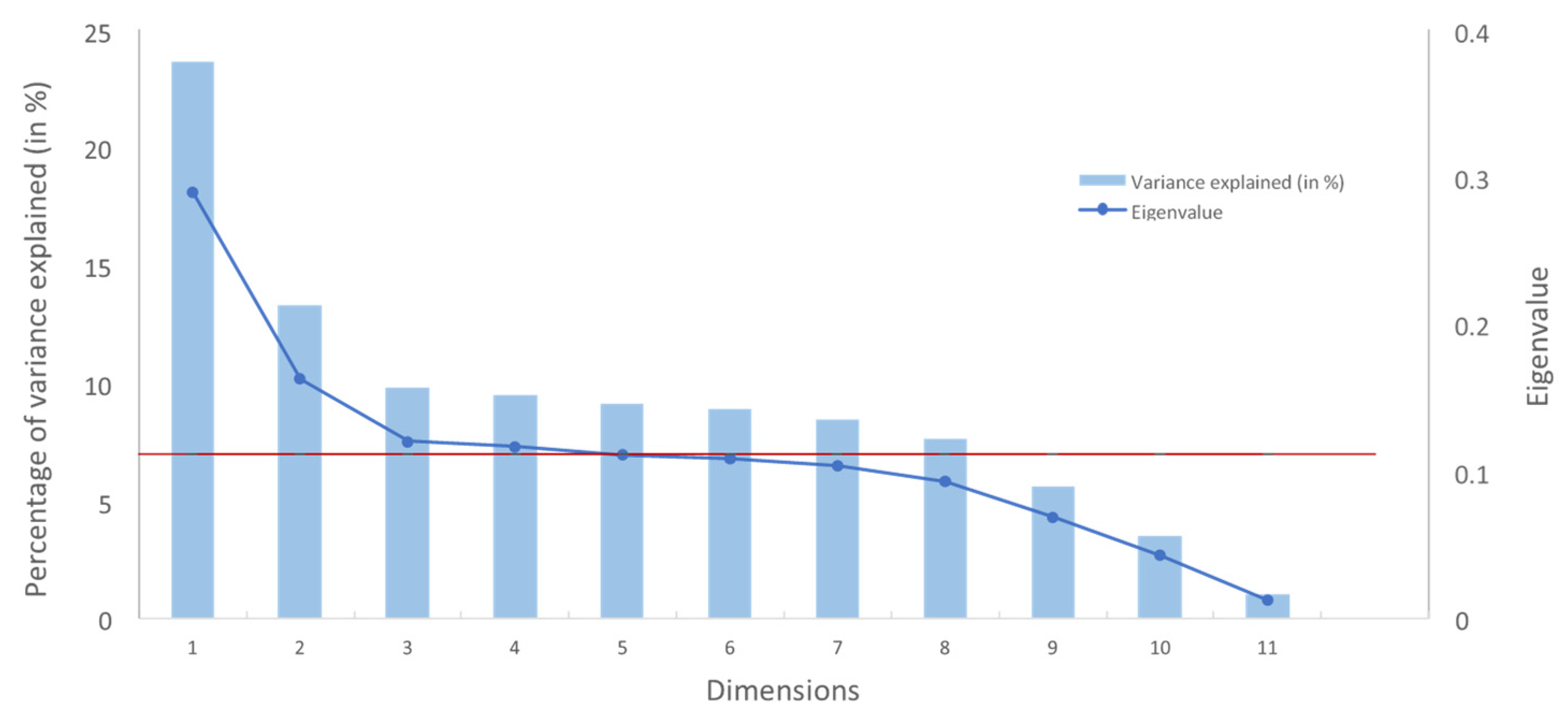
| Dimension | Eigenvalue | Variance explained (in %) | Cumulative variances explained (in %) |
Eigenvalue corrected by Benzecri |
Variances explained corrected by Benzecri (in %) |
Cumulative variances explained corrected by Benzecri (in %) |
| 1 | 0.29 | 23.6 | 23.6 | 0.04 | 92.1 | 92.1 |
| 2 | 0.16 | 13.3 | 36.9 | < 0.01 | 7.7 | 99.7 |
| 3 | 0.12 | 9.8 | 46.7 | < 0.01 | 0.2 | 99.9 |
| 4 | 0.12 | 9.5 | 56.1 | < 0.01 | 0.1 | 100.0 |
| 5 | 0.11 | 9.0 | 65.2 | − | − | 100.0 |
| 6 | 0.11 | 8.8 | 74.0 | − | − | 100.0 |
| 7 | 0.10 | 8.4 | 82.4 | − | − | 100.0 |
| 8 | 0.09 | 7.6 | 90.0 | − | − | 100.0 |
| 9 | 0.07 | 5.6 | 95.6 | − | − | 100.0 |
| 10 | 0.04 | 3.5 | 99.0 | − | − | 100.0 |
| 11 | 0.01 | 1.0 | 100.0 | − | − | 100.0 |
| 1 | |
| 2 | For further details, see Celebi (2014). |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 | |
| 10 | |
| 11 | |
| 12 | |
| 13 |
https://github.com/sandysa/Interpretable Clustering |
| 14 | |
| 15 | A discussion on other possible criteria and ways to integrate them into the workflow is presented in section 5.1 (e.g. management of missing data and outliers). |
| 16 | e.g. CRAN Task View for cluster analysis: https://cran.r-project.org/web/views/Cluster.html
|
| 17 | e.g. of handy R packages: https://towardsdatascience.com/a-comprehensive-list-of-handy-r-packages-e85dad294b3d
|
| 18 | The notion of large data, as well as how to fix the hyperparameters samples and sampsize in CLARA algorithm, may vary according to the computing capabilities of the user’s system. One recommends users to pre-tests different scenarios to adapt these thresholds to their own settings. For guidance, this workflow applied on the case study in section 4 (34,134 observations and 9 variables) took 5 hour and 30 minutes with 8 CPUs and 10 GB RAM |
| 19 | It is worth noting that standardization of continuous data is recommended before using PCA, to not give excessive importance to the variables with the largest variances. |
| 20 | |
| 21 | |
| 22 | |
| 23 | Prise en charge des patients adultes atteints d’hypertension arterielle essentielle - Actualisation 2005´ https://www.has-sante.fr/upload/docs/application/pdf/2011-09/hta 2005 - recommandations.pdf |
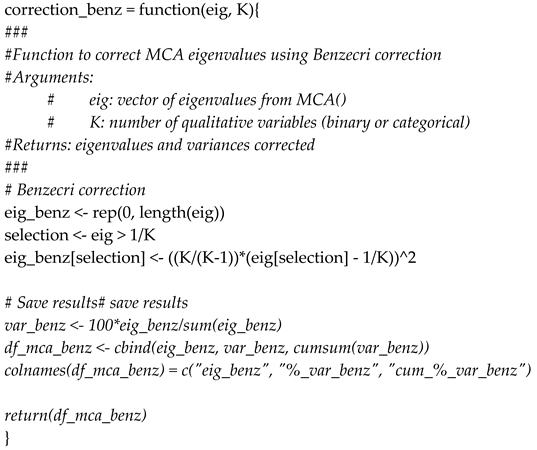
| 24 | One may prefer the Greenacre adjustment to Benzecri correction, which tends to be less optimistic than the Benzecri correction. Please note that both methods are not currently implemented in the proposed R packages and must therefore be implemented by data scientists if desired. R code for Benzecri correction is provided in Appendix in section C
|
| 25 | Similarly, the user may want to compare several methods for selecting the optimal number of clusters, including other direct methods (e.g. elbow method on the total within-cluster sum of square) or methods based on statistical testing (e.g. gap statistic) |
References
- Achtert, E., S. Goldhofer, H-P. Kriegel, E. Schubert and A. Zimek, "Evaluation of Clusterings -- Metrics and Visual Support," 2012 IEEE 28th International Conference on Data Engineering, 2012, pp. 1285-1288. [CrossRef]
- Ahmad, Amir and Shehroz S. Khan. “Survey of state-of-the-art mixed data clustering algorithms”. In: IEEE Access 7 (2019), pp. 31883–31902. ISSN: 2169-3536. [CrossRef]
- Aljalbout, Elie, Vladimir Golkov, Yawar Siddiqui, Maximilian Strobel, and Daniel Cremers. “Clustering with Deep Learning: Taxonomy and New Methods”. In: arXiv:1801.07648 [cs, stat] (Sept. 13, 2018). arXiv: 1801.07648. URL: http://arxiv.org/abs/1801.07648 (visited on 09/09/2021).
- Alqurashi, T. and W., Wang. Clustering ensemble method. Int. J. Mach. Learn. & Cyber. 10, 1227–1246 (2019). [CrossRef]
- Altman, Naomi and Martin Krzywinski. “Clustering”. In: Nature Methods 14.6 (June 1, 2017). Bandiera abtest: a Cg type: Nature Research Journals Number: 6 Primary atype: News Publisher: Nature Publishing Group Subject term: Data mining;Data processing Subject term id: data-mining;data-processing, pp. 545–546. ISSN: 1548-7105. [CrossRef]
- Arabie, P. and L. Hubert. “Cluster analysis in marketing research”. In: Advanced Methods of Marketing Research (1994), pp. 160–189.
- Arbelaitz, Olatz, Ibai Gurrutxaga, Javier Muguerza, Jesus M.´ Perez´, and Inigo˜ Perona. “An extensive comparative study of cluster validity indices”. en. In: Pattern Recognition 46.1 (Jan. 2013), pp. 243–256. ISSN: 0031-3203. [CrossRef]
- Arthur, D. , and Vassilvitskii, S. "k-means++: the advantages of careful seeding". Proceedings of the eighteenth annual ACM-SIAM symposium on Discrete algorithms. Society for Industrial and Applied Mathematics Philadelphia, PA, USA. (2007). pp. 1027-1035.
- Audigier, Vincent, Franc¸ois Husson, and Julie Josse. “A principal components method to impute missing values for mixed data”. In: arXiv:1301.4797 [stat] (Feb. 19, 2013). arXiv: 1301.4797. URL: http://arxiv.org/abs/1301.4797 (visited on 09/09/2021).
- Bandalos, Deborah L. and Meggen R. Boehm-Kaufman. “Four common misconceptions in exploratory factor analysis”. In: Statistical and methodological myths and urban legends: Doctrine, verity and fable in the organizational and social sciences. New York, NY, US: Routledge/Taylor & Francis Group, 2009, pp. 61–87. ISBN: 978-0-8058-6238-6 978-0-8058-6237-9.
- Bertsimas, Dimitris, Agni Orfanoudaki, and Holly Wiberg. “Interpretable clustering: an optimization approach”. In: Machine Learning 110.1 (Jan. 1, 2021), pp. 89–138. ISSN: 1573-0565. [CrossRef]
- Bezdek, J.C. , Robert Ehrlich, and William Full. “FCM: The fuzzy c-means clustering algorithm”. en. In: Computers & Geosciences 10.2 (Jan. 1984), pp. 191–203. ISSN: 0098-3004. [CrossRef]
- Bezdek, J.C. and N.R. Pal. “Some new indexes of cluster validity”. In: IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics) 28.3 (June 1998). Conference Name: IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), pp. 301–315. ISSN: 1941-0492. [CrossRef]
- Bock, H. H. “On the Interface between Cluster Analysis, Principal Component Analysis, and Multidimensional Scaling”. en. In: Multivariate Statistical Modeling and Data Analysis: Proceedings of the Advanced Symposium on Multivariate Modeling and Data Analysis May 15–16, 1986. Ed. by H. Bozdogan and A. K. Gupta. Theory and Decision Library. Dordrecht: Springer Netherlands,1987, pp. 17–34. ISBN: 978-94-009-3977-6. [CrossRef]
- Bousquet, Philippe Jean, Philippe Devillier, Abir Tadmouri, Kamal Mesbah, Pascal Demoly, and Jean Bousquet. “Clinical Relevance of Cluster Analysis in Phenotyping Allergic Rhinitis in a Real-Life Study”. In: International Archives of Allergy and Immunology 166.3 (2015). Publisher: Karger Publishers, pp. 231–240. ISSN: 1018-2438, 1423-0097. [CrossRef]
- Bro, R., K. Kjeldahl, A. K. Smilde, and H. A. L. Kiers. “Cross-validation of component models: A critical look at current methods”. en. In: Analytical and Bioanalytical Chemistry 390.5 (Mar. 2008), pp. 1241–1251. ISSN: 1618-2650. [CrossRef]
- Brock, Guy, Vasyl Pihur, Susmita Datta, and Somnath Datta. “clValid: An R Package for Cluster Validation”. en-US. In: (Mar. 2008). URL: https://www.jstatsoft.org/article/ view/v025i04 (visited on 12/16/2021). Buuren, Stef van and Willem J. Heiser. “Clusteringn objects intok groups under optimal scaling of variables”. In: Psychometrika 54.4 (Sept. 1, 1989), pp. 699–706. ISSN: 1860-0980. [CrossRef]
- Calinski´, T. and J Harabasz. “A dendrite method for cluster analysis”. In: Communications in Statistics 3.1 (Jan. 1, 1974). Publisher: Taylor & Francis eprint: https://www.tandfonline.com/doi/pdf/10.1080/03610927408827101, pp. 1–27. ISSN: 0090-3272. [CrossRef]
- Caruana, R. , Elhawary, M., Nguyen, N. and C. Smith, "Meta Clustering," Sixth International Conference on Data Mining (ICDM'06), 2006, pp. 107-118. [CrossRef]
- Cattell, Raymond B. “The Scree Test For The Number Of Factors”. In: Multivariate Behavioral Research 1.2 (Apr. 1, 1966). Publisher: Routledge eprint: https://doi.org/10.1207/s15327906mbr0102 10, pp. 245–276. ISSN: 0027-3171. [CrossRef]
- Celebi, M Emre. Partitional clustering algorithms. Springer, 2014.
- Ciampi, Antonio and Yves Lechevallier. “Clustering Large, Multi-level Data Sets: An Approach Based on Kohonen Self Organizing Maps”. In: Principles of Data Mining and Knowledge Discovery. Ed. by Djamel A. Zighed, Jan Komorowski, and Jan Zytkow˙. Lecture Notes in Computer Science. Berlin, Heidelberg: Springer, 2000, pp. 353–358. ISBN: 978-3-540-45372-7. [CrossRef]
- Clausen, Sten Erik. Applied Correspondence Analysis: An Introduction. es. Google-Books-ID: FsCXR3uk 0gC. SAGE, June 1998. ISBN: 978-0-7619-1115-9. “Cluster analysis”. In: Wikipedia. July 18, 2021. URL: https://en.wikipedia.org/w/ index.php?title=Cluster_analysis&oldid=1034255231.
- Costa, Patr´ıcio Soares and Nuno Sousa. The Use of Multiple Correspondence Analysis to Explore Associations between Categories of Qualitative Variables in Healthy Ageing. 2013. URL: https://www.hindawi.com/journals/jar/2013/302163/ (visited on 12/16/2021).
- Datta, Susmita and Somnath Datta. “Methods for evaluating clustering algorithms for gene expression data using a reference set of functional classes”. In: BMC Bioinformatics 7.1 (Aug. 2006), p. 397. ISSN: 1471-2105. [CrossRef]
- De Soete, Geert and J. Douglas Carroll. “K-means clustering in a low-dimensional Euclidean space”. en. In: New Approaches in Classification and Data Analysis. Ed. by Edwin Diday, Yves Lechevallier, Martin Schader, Patrice Bertrand, and Bernard Burtschy. Studies in Classification, Data Analysis, and Knowledge Organization. Berlin, Heidelberg: Springer, 1994, pp. 212–219. ISBN: 978-3-642-51175-2. [CrossRef]
- DeSarbo, Wayne S., Daniel J. Howard, and Kamel Jedidi. “Multiclus: A new method for simultaneously performing multidimensional scaling and cluster analysis”. en. In: Psychometrika 56.1 (Mar. 1991), pp. 121–136. ISSN: 1860-0980. [CrossRef]
- Desormais, Ileana and Williams Bryan. “2018 ESC/ESH Guidelines for the management of arterial hypertension”. In: Journal of Hypertension (). URL: https://journals.lww.com/ jhypertension/Fulltext/2018/10000/2018_ESC_ESH_Guidelines_for_ the_management_of.2.aspx (visited on 12/16/2021).
- Di Franco, Giovanni. “Multiple correspondence analysis: one only or several techniques?” In: Quality & Quantity 50.3 (May 1, 2016), pp. 1299–1315. ISSN: 1573-7845. [CrossRef]
- Do, Chuong B. and Serafim Batzoglou. “What is the expectation maximization algorithm?” In: Nature Biotechnology 26.8 (Aug. 2008). Bandiera abtest: a Cg type: Nature Research Journals Number: 8 Primary atype: Reviews Publisher: Nature Publishing Group, pp. 897–899. ISSN: 1546-1696. [CrossRef]
- Drennan, Robert D. Statistics for Archaeologists: A Common Sense Approach. en. Google-BooksID: wZEmzgEACAAJ. Springer US, Mar. 2010. ISBN: 978-1-4419-6071-9.
- Efron, B. “Bootstrap Methods: Another Look at the Jackknife”. In: The Annals of Statistics 7.1 (Jan. 1979). Publisher: Institute of Mathematical Statistics, pp. 1–26. ISSN: 0090-5364, 2168-8966. [CrossRef]
- Efron, B. and R. J. Tibshirani. An Introduction to the Bootstrap. Google-Books-ID: gLlpIUxRntoC. CRC Press, May 15, 1994. 456 pp. ISBN: 978-0-412-04231-7.
- Esnault, Cyril, May-Line Gadonna, Maxence Queyrel, Alexandre Templier, and Jean-daniel Zucker. “Q-Finder: An Algorithm for Credible Subgroup Discovery in Clinical Data Analysis - An Application to the International Diabetes Management Practice Study.” In: Frontiers in Artificial Intelligence 3 (2020), p. 559927. [CrossRef]
- Ester, Martin, Hans-Peter Kriegel, Jorg¨ Sander, and Xiaowei Xu. “A density-based algorithm for discovering clusters in large spatial databases with noise”. In: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining. KDD’96. Portland, Oregon: AAAI Press, Aug. 1996, pp. 226–231. (Visited on 12/05/2021).
- Estivill-Castro, Vladimir. “Why so many clustering algorithms: a position paper”. In: ACM SIGKDD Explorations Newsletter 4.1 (June 1, 2002), pp. 65–75. ISSN: 1931-0145. [CrossRef]
- Fahad, Adil, Najlaa Alshatri, Zahir Tari, Abdullah Alamri, Ibrahim Khalil, Albert Y. Zomaya, Sebti Foufou, and Abdelaziz Bouras. “A Survey of Clustering Algorithms for Big Data: Taxonomy and Empirical Analysis”. In: IEEE Transactions on Emerging Topics in Computing 2.3 (Sept. 2014). Conference Name: IEEE Transactions on Emerging Topics in Computing, pp. 267–279. ISSN: 2168-6750. [CrossRef]
- Fahrmeir, Ludwig, Thomas Kneib, Stefan Lang, and Brian Marx. “Categorical Regression Models”. In: Regression: Models, Methods and Applications. Ed. by Ludwig Fahrmeir, Thomas Kneib, Stefan Lang, and Brian Marx. Berlin, Heidelberg: Springer, 2013, pp. 325–347. ISBN: 978-3642-34333-9. [CrossRef]
- Fisher, Douglas H. “Knowledge Acquisition Via Incremental Conceptual Clustering”. In: Machine Learning 2.2 (Sept. 1, 1987), pp. 139–172. ISSN: 1573-0565. [CrossRef]
- Foss, Alexander H. and Marianthi Markatou. “kamila: Clustering Mixed-Type Data in R and Hadoop”. en. In: Journal of Statistical Software 83 (Feb. 2018), pp. 1–44. ISSN: 1548-7660. [CrossRef]
- Franti¨ , Pasi, Sami Sieranoja, Katja Wikstrom¨ , and Tiina Laatikainen. “Clustering Diagnoses From 58 Million Patient Visits in Finland Between 2015 and 2018”. EN. In: JMIR Medical Informatics 10.5 (May 2022). Company: JMIR Medical Informatics Distributor: JMIR Medical Informatics Institution: JMIR Medical Informatics Label: JMIR Medical Informatics Publisher: JMIR Publications Inc., Toronto, Canada, e35422. [CrossRef]
- Gordon. Classification - 2nd Edition - A.D. Gordon - Routledge Book. 1999. URL: https://www. routledge.com/Classification/Gordon/p/book/9780367399665 (visited on 12/15/2021).
- Green, Paul E. and Abba M. Krieger. “A Comparison of Alternative Approaches to Cluster-Based Market Segmentation”. en. In: Market Research Society. Journal. 37.3 (May 1995). Publisher: SAGE Publications, pp. 1–19. ISSN: 0025-3618. [CrossRef]
- Greenacre, Michael. Correspondence Analysis in Practice. 2nd ed. New York: Chapman and Hall/CRC, May 2007. ISBN: 978-0-429-14614-5. [CrossRef]
- Theory and Applications of Correspondence Analysis. en. Google-Books-ID: LsPaAAAAMAAJ. Academic Press, 1984. ISBN: 978-0-12-299050-2.
- Greenacre, Michael and Jorg Blasius, eds. Multiple Correspondence Analysis and Related Methods. New York: Chapman and Hall/CRC, June 2006. ISBN: 978-0-429-14196-6. [CrossRef]
- Greene, D., A. Tsymbal, N. Bolshakova and P. Cunningham, "Ensemble clustering in medical diagnostics," Proceedings. 17th IEEE Symposium on Computer-Based Medical Systems, 2004, pp. 576-581. [CrossRef]
- Guha, Sudipto, Rajeev Rastogi, and Kyuseok Shim. “CURE: an efficient clustering algorithm for large databases”. In: ACM SIGMOD Record 27.2 (June 1998), pp. 73–84. ISSN: 0163-5808. [CrossRef]
- Rock: A robust clustering algorithm for categorical attributes - ScienceDirect. URL: https:// www.sciencedirect.com/science/article/abs/pii/S0306437900000223 (visited on 12/15/2000).
- Halkidi, Maria, Yannis Batistakis, and Michalis Vazirgiannis. “On Clustering Validation Techniques”. In: Journal of Intelligent Information Systems 17.2 (Dec. 1, 2001), pp. 107–145. ISSN: 1573-7675. [CrossRef]
- Handl, Julia, Joshua Knowles, and Douglas B. Kell. “Computational cluster validation in postgenomic data analysis”. In: Bioinformatics 21.15 (Aug. 2005), pp. 3201–3212. ISSN: 1367-4803. [CrossRef]
- Hennig, Christian. “Cluster validation by measurement of clustering characteristics relevant to the user”. In arXiv:1703.09282 [stat] (Sept. 8, 2020). arXiv: 1703.09282. URL: http:// arxiv.org/abs/1703.09282 (visited on 09/09/2021).
- Cluster-wise assessment of cluster stability - ScienceDirect. 2007. URL: https://www. sciencedirect.com/science/article/abs/pii/S0167947306004622?via%3Dihub (visited on 09/09/2021).
- “Dissolution point and isolation robustness: Robustness criteria for general cluster analysis meth-ods”. In: Journal of Multivariate Analysis 99.6 (July 1, 2008), pp. 1154–1176. ISSN: 0047-259X. [CrossRef]
- “How Many Bee Species? A Case Study in Determining the Number of Clusters”. In: Data Analysis, Machine Learning and Knowledge Discovery. Ed. by Myra Spiliopoulou, Lars SchmidtThieme, and Ruth Janning. Studies in Classification, Data Analysis, and Knowledge Organization. Cham: Springer International Publishing, 2014, pp. 41–49. ISBN: 978-3-319-01595-8. [CrossRef]
- Hennig, Christian. How to find an appropriate clustering for mixed-type variables with application to socio-economic stratification - Hennig - 2013 - Journal of the Royal Statistical Society: Series C (Applied Statistics) - Wiley Online Library. 2013. URL: https://rss.onlinelibrary.wiley.com/doi/10.1111/j.1467-9876.2012.01066.x (visited on 09/09/2021).
- Herawan, Tutut and Ali Seyed Shirkhorshidi. Big Data Clustering: A Review — SpringerLink. Springer Link. 2014. URL: https://link.springer.com/chapter/10.1007/9783-319-09156-3_49 (visited on 03/16/2021).
- Huang, Zhexue. “A Fast Clustering Algorithm to Cluster Very Large Categorical Data Sets in Data Mining,” in: In Research Issues on Data Mining and Knowledge Discovery, 1997, pp. 1–8.
- “Extensions to the k-Means Algorithm for Clustering Large Data Sets with Categorical Val-ues”. en. In: Data Mining and Knowledge Discovery 2.3 (Sept. 1998), pp. 283–304. ISSN: 1573-756X. [CrossRef]
- Hwang, Heungsun, Hec Montreal´, William R. Dillon, and Yoshio Takane. “An Extension of Multiple Correspondence Analysis for Identifying Heterogeneous Subgroups of Respondents”. In: Psychometrika 71.1 (Mar. 1, 2006), pp. 161–171. ISSN: 1860-0980. [CrossRef]
- Hwang, Heungsun and Yoshio Takane. Generalized Constrained Canonical Correlation Analysis: Multivariate Behavioral Research: Vol 37, No 2. 2010. URL: https://www.tandfonline. com/doi/abs/10.1207/S15327906MBR3702_01 (visited on 12/16/2021).
- Jain, Anil. “Data clustering: 50 years beyond K-means”. In: Pattern Recognition Letters. Award winning papers from the 19th International Conference on Pattern Recognition (ICPR) 31.8 (June 1, 2010), pp. 651–666. ISSN: 0167-8655. [CrossRef]
- Jin, Xin and Jiawei Han. “K-Medoids Clustering”. In: Encyclopedia of Machine Learning. Ed. by Claude Sammut and Geoffrey I. Webb. Boston, MA: Springer US, 2010, pp. 564–565. ISBN: 978-0-387-30164-8. [CrossRef]
- Josse, Julie, Marie Chavent, Benot Liquet, and Franc¸ois Husson. “Handling Missing Values with Regularized Iterative Multiple Correspondence Analysis”. en. In: Journal of Classification 29.1 (Apr. 2012), pp. 91–116. ISSN: 1432-1343. [CrossRef]
- Josse, Julie, Jer´ omeˆ Pages`, and Franc¸ois Husson. “Multiple imputation in principal component analysis”. In: Advances in Data Analysis and Classification 5.3 (Oct. 1, 2011), pp. 231–246. ISSN: 1862-5355. [CrossRef]
- Kamoshida, Ryota and Fuyuki Ishikawa. “Automated Clustering and Knowledge Acquisition Support for Beginners”. In: Procedia Computer Science. Knowledge-Based and Intelligent Information & Engineering Systems: Proceedings of the 24th International Conference KES2020 176 (Jan. 1, 2020), pp. 1596–1605. ISSN: 1877-0509. [CrossRef]
- Kaufman, Leonard and Peter J. Rousseeuw. Finding Groups in Data: An Introduction to Cluster Analysis. en. Google-Books-ID: YeFQHiikNo0C. John Wiley & Sons, Sept. 2009. ISBN: 978-0470-31748-8.
- “Partitioning Around Medoids (Program PAM)”. en. In: Finding Groups in Data. Section: 2 eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/9780470316801.ch2. John Wiley & Sons, Ltd, 1990, pp. 68–125. ISBN: 978-0-470-31680-1. [CrossRef]
- Kaushik, Manju and Bhawana Mathur. “Comparative Study of K-Means and Hierarchical Clustering Techniques”. In: Internatioonal journal of Software and Hardware Research in Engineering 2 (June 1, 2014), pp. 93–98. Keim, Daniel A. and Alexander Hinneburg. An efficient approach to clustering in large multimedia databases with noise — Proceedings of the Fourth International Conference on Knowledge Discovery and Data Mining. 1998. URL: https://dl.acm.org/doi/10.5555/3000292. 3000302 (visited on 12/15/2021).
- Kiselev, Vladimir Yu, Tallulah S. Andrews, and Martin Hemberg. “Challenges in unsupervised clustering of single-cell RNA-seq data”. In: Nature Reviews Genetics 20.5 (May 2019). Number: 5 Publisher: Nature Publishing Group, pp. 273–282. ISSN: 1471-0064. [CrossRef]
- Kleinberg, Jon M. “An Impossibility Theorem for Clustering.” In: Advances in Neural Information Processing Systems (2002).
- Lange, Tilman, Volker Roth, Mikio L. Braun, and Joachim Buhmannand. “Stability-Based Validation of Clustering Solutions”. In: Neural Computation 16.6 (June 1, 2004), pp. 1299–1323. ISSN: 0899-7667. [CrossRef]
- Le Roux, Brigitte and Henry Rouanet. Multiple Correspondence Analysis. Google-Books-ID: GWsHakQGEHsC. SAGE, 2010. 129 pp. ISBN: 978-1-4129-6897-3.
- Lee, Young, Lee, Eun and Byeong, Park. (2012). Principal component analysis in very high-dimensional spaces. Statistica Sinica. 22. 10.5705/ss.2010.149.
- Li, Nan and Longin Jan Latecki. “Affinity Learning for Mixed Data Clustering.” In: IJCAI (2017).
- Lorenzo-Seva, Urbano. “Horn’s Parallel Analysis for Selecting the Number of Dimensions in Correspondence Analysis”. In: Methodology 7.3 (Jan. 2011). Publisher: Hogrefe Publishing, pp. 96–102. ISSN: 1614-1881. [CrossRef]
- Maaten, L.d and G. Hinton. ”Visualizing data using t-SNE”. In: J. Mach. Learn. Res., 9 (2008), pp. 2579-2605.
- MacQueen, J. “Some methods for classification and analysis of multivariate observations”. In: Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics 5.1 (Jan. 1967). Publisher: University of California Press, pp. 281–298. URL: https://projecteuclid.org/ebooks/berkeley-symposium-on-mathematicalstatistics-and-probability/Proceedings-of-the-Fifth-BerkeleySymposium-on-Mathematical-Statistics-and/chapter/Some-methodsfor-classification-and-analysis-of-multivariate-observations/ bsmsp/1200512992 (visited on 12/05/2021).
- McCallum, Andrew, Kamal Nigam, and Lyle H. Ungar. “Efficient clustering of high-dimensional data sets with application to reference matching”. In: KDD ’00: Proceedings of the sixth ACM SIGKDD international conference on Knowledge discovery and data mining. Boston, Massachusetts, United States: ACM Press, 2000, pp. 169–178. ISBN: 1-58113-233-6. [CrossRef]
- McCane, Brendan and Michael Albert. “Distance functions for categorical and mixed variables”. en. In: Pattern Recognition Letters 29.7 (May 2008), pp. 986–993. ISSN: 0167-8655. [CrossRef]
- McInnes, L., J. Healy and J. Melville. “Umap: Uniform manifold approximation and projection for dimension reduction”. In: arXiv, 1802 (2018), p. 03426.
- Meila˘, Marina. “Comparing clusterings—an information based distance”. In: Journal of Multivariate Analysis 98.5 (May 1, 2007), pp. 873–895. ISSN: 0047-259X. [CrossRef]
- Milligan, Glenn W. and Martha C. Cooper. “An examination of procedures for determining the number of clusters in a data set”. In: Psychometrika 50.2 (June 1, 1985), pp. 159–179. ISSN: 1860-0980. [CrossRef]
- Mitsuhiro, Masaki and Hiroshi Yadohisa. “Reduced k-means clustering with MCA in a lowdimensional space”. In: Computational Statistics 30.2 (June 1, 2015), pp. 463–475. ISSN: 1613-9658. [CrossRef]
- Mittal, Mamta, Lalit M Goyal, Duraisamy Jude Hemanth, and Jasleen K Sethi. “Clustering approaches for high-dimensional databases: A review”. In: Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery 9.3 (May 2019), e1300.
- Murtagh, Fionn. Correspondence Analysis and Data Coding with Java and R. en. Google-Books-ID: wIK7p0 NyzoC. CRC Press, May 2005. ISBN: 978-1-4200-3494-3.
- Nagpal, Arpita, Arnan Jatain, and Deepti Gaur. “Review based on data clustering algorithms”. In: 2013 IEEE Conference on Information Communication Technologies. 2013 IEEE Conference on Information Communication Technologies. Apr. 2013, pp. 298–303. [CrossRef]
- Ng, Raymond and Jiawei Han. “CLARANS: a method for clustering objects for spatial data mining”. In: IEEE Transactions on Knowledge and Data Engineering 14.5 (Sept. 2002). Conference Name: IEEE Transactions on Knowledge and Data Engineering, pp. 1003–1016. ISSN: 1558-2191. [CrossRef]
- “Efficient and Effective Clustering Methods for Spatial Data Mining”. In: VLDB. 1994.
- Nietto, Paulo Rogerio and Maria do Carmo Nicoletti. “Estimating the Number of Clusters as a Preprocessing Step to Unsupervised Learning”. In: Intelligent Systems Design and Applications. Ed. by Ana Maria Madureira, Ajith Abraham, Dorabela Gamboa, and Paulo Novais. Advances in Intelligent Systems and Computing. Cham: Springer International Publishing, 2017, pp. 25–34. ISBN: 978-3-319-53480-0. [CrossRef]
- Nishisato, Shizuhiko. Analysis of Categorical Data: Dual Scaling and its Applications. en. Publication Title: Analysis of Categorical Data. University of Toronto Press, Nov. 2019. [CrossRef]
- Obembe, Olawole and Jelili Oyelade. Data Clustering: Algorithms and Its Applications. IEEE Xplore. 2019. URL: https://ieeexplore.ieee.org/document/8853526 (visited on 03/16/2021).
- Ortega, Francisco B., Carl J. Lavie, and Steven N. Blair. “Obesity and Cardiovascular Disease”. In: Circulation Research 118.11 (May 27, 2016). Publisher: American Heart Association, pp. 1752–1770. [CrossRef]
- Oyelade, J et al. "Data Clustering: Algorithms and Its Applications," 2019 19th International Conference on Computational Science and Its Applications (ICCSA), 2019, pp. 71-81. [CrossRef]
- Pagès, Jerome. Analyse factorielle de donnees mixtes´. 2004. URL: http://www.numdam.org/ article/RSA_2004__52_4_93_0.pdf (visited on 03/19/2021).
- Pagès, Jerome and Francois Husson. Exploratory Multivariate Analysis by Example Using R 2nd Edition - F. 2017. URL: https://www.routledge.com/ExploratoryMultivariate-Analysis-by-Example-Using-R/Husson-Le-Pages/p/ book/9780367658021 (visited on 12/16/2021).
- Pasi Franti¨, Mohammad Rezaei. Can the Number of Clusters Be Determined by External Indices? 2020. URL: https://ieeexplore.ieee.org/document/9090190 (visited on 05/12/2022).
- Rezaei, Mohammad and Pasi Franti¨. “Set Matching Measures for External Cluster Validity”. In: IEEE Transactions on Knowledge and Data Engineering 28.8 (Aug. 2016). Conference Name: IEEE Transactions on Knowledge and Data Engineering, pp. 2173–2186. ISSN: 1558-2191. [CrossRef]
- Rousseeuw, Peter J. “Silhouettes: A graphical aid to the interpretation and validation of cluster analysis”. en. In: Journal of Computational and Applied Mathematics 20 (Nov. 1987), pp. 53–65. ISSN: 0377-0427. [CrossRef]
- Saint Pierre, Aude, Joanna Giemza, Isabel Alves, Matilde Karakachoff, Marinna Gaudin, Philippe Amouyel, Jean-Franc¸ois Dartigues, Christophe Tzourio, Martial Monteil, Pilar Galan, Serge Hercberg, Iain Mathieson, Richard Redon, Emmanuelle Genin´ , and Christian Dina. “The genetic history of France”. In: European Journal of Human Genetics 28.7 (July 2020). Bandiera abtest: a Cg type: Nature Research Journals Number: 7 Primary atype: Research Publisher: Nature Publishing Group Subject term: Genetics;Population genetics Subject term id: genetics;populationgenetics, pp. 853–865. ISSN: 1476-5438. [CrossRef]
- Saisubramanian, Sandhya, Sainyam Galhotra, and Shlomo Zilberstein. “Balancing the Tradeoff Between Clustering Value and Interpretability”. In arXiv:1912.07820 [cs, stat] (Jan. 30, 2020). arXiv: 1912.07820. URL: http://arxiv.org/abs/1912.07820 (visited on 03/17/2021).
- Sculley, D. “Web-scale k-means clustering”. In: Proceedings of the 19th international conference on World wide web. WWW ’10. New York, NY, USA: Association for Computing Machinery, Apr. 2010, pp. 1177–1178. ISBN: 978-1-60558-799-8. [CrossRef]
- Sheikholeslami, Gholamhosein, Surojit Chatterjee, and Aidong Zhang. WaveCluster: A MultiResolution Clustering Approach for Very Large Spatial Databases — Semantic Scholar. 1998. URL: https://www.semanticscholar.org/paper/WaveCluster%5C%3A-A-Multi-Resolution-Clustering-Approach-Sheikholeslami-Chatterjee/ f0015f0e834a84699a9b83c6c9af33acdac05069 (visited on 12/16/2021).
- Sieranoja, Sami and Pasi Franti¨. “Adapting k-means for graph clustering”. en. In: Knowledge and Information Systems 64.1 (Jan. 2022), pp. 115–142. ISSN: 0219-3116. [CrossRef]
- “Fast and general density peaks clustering”. en. In: Pattern Recognition Letters 128 (Dec. 2019), pp. “Fast and general density peaks clustering”. en. In: Pattern Recognition Letters 128 (Dec. 2019), pp. 551–558. ISSN: 0167-8655. [CrossRef]
- Testa, Damien and Laurent Chiche. Unsupervised clustering analysis of data from an online community to identify lupus patient profiles with regards to treatment preferences - Damien Testa, Noemie´ Jourde-Chiche, Julien Mancini, Pasquale Varriale, Lise Radoszycki, Laurent Chiche, 2021. URL: https://journals.sagepub.com/doi/10.1177/09612033211033977 (visited on 11/15/2021).
- Tibshirani, Robert, Guenther Walther, and Trevor Hastie. “Estimating the number of clusters in a data set via the gap statistic”. en. In: Journal of the Royal Statistical Society: Series B (Statistical Methodology) 63.2 (2001). eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1111/14679868.00293, pp. 411–423. ISSN: 1467-9868. [CrossRef]
- Torgerson, W.S. “Multidimensional scaling: I. Theory and method Psychometrika”, 17 (1952), pp.
- Van de Velden, Michel, A. Iodice D’ Enza, and F. Palumbo. “Cluster Correspondence Analysis”. In: (Oct. 1, 2014). Number: EI 2014-24. URL: https://repub.eur.nl/pub/77010 (visited on 09/09/2021).
- Vellido, Alfredo. “The importance of interpretability and visualization in machine learning for applications in medicine and health care”. In: Neural Computing and Applications 32.24 (Dec. 1, 2020), pp. 18069–18083. ISSN: 1433-3058. [CrossRef]
- Wang, Wei, Jiong Yang, and Richard Muntz. STING : A Statistical Information Grid Approach to Spatial Data Mining. 1997.
- Warwick, K. M. and L. Lebart. “Multivariate descriptive statistical analysis (correspondence analysis and related techniques for large matrices)”. en. In: Applied Stochastic Models and Data Analysis 5.2 (1989). eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/asm.3150050207, pp. 175–175. ISSN: 1099-0747. [CrossRef]
- Windgassen, Sula, Rona Moss-Morris, Kimberley Goldsmith, and Trudie Chalder. “The importance of cluster analysis for enhancing clinical practice: an example from irritable bowel syndrome”. In: Journal of Mental Health 27.2 (Mar. 4, 2018). Publisher: Routledge eprint. pp. 94-96. ISSN: 0963-8237. Publisher: Routledge eprint. [CrossRef]
- Wiwie, Christian, Jan Baumbach, and Richard Rottger¨. “Comparing the performance of biomedical clustering methods”. In: Nature Methods 12.11 (Nov. 2015). Bandiera abtest: a Cg type: Nature Research Journals Number: 11 Primary atype: Research Publisher: Nature Publishing Group Subject term: Computational biology and bioinformatics;Databases Subject term id: computationalbiology-and-bioinformatics;databases, pp. 1033–1038. ISSN: 1548-7105. [CrossRef]
- Xu, Rui and Donald C. Wunsch. “Clustering Algorithms in Biomedical Research: A Review”. In: IEEE Reviews in Biomedical Engineering 3 (2010). Conference Name: IEEE Reviews in Biomedical Engineering, pp. 120–154. ISSN: 1941-1189. [CrossRef]
- Yang, Jiawei, Susanto Rahardja, and Pasi Franti¨ . “Mean-shift outlier detection and filtering”. en. In: Pattern Recognition 115 (July 2021), p. 107874. ISSN: 0031-3203. [CrossRef]
- Zhang, Tian, Raghu Ramakrishnan, and Miron Livny. “BIRCH: an efficient data clustering method for very large databases”. In: ACM SIGMOD Record 25.2 (June 1996), pp. 103–114. ISSN: 01635808. [CrossRef]
- Zhao, Ying and George Karypis. Comparison of Agglomerative and Partitional Document Clustering Algorithms. Section: Technical Reports. MINNESOTA UNIV MINNEAPOLIS DEPT OF COMPUTER SCIENCE, Apr. 17, 2002. URL: https://apps.dtic.mil/sti/ citations/ADA439503 (visited on 09/09/2021).
- Zhou, Fang L., Hirotaka Watada, Yuki Tajima, Mathilde Berthelot, Dian Kang, Cyril Esnault, Yujin Shuto, Hiroshi Maegawa, and Daisuke Koya. “Identification of subgroups of patients with type 2 diabetes with differences in renal function preservation, comparing patients receiving sodium-glucose co-transporter-2 inhibitors with those receiving dipeptidyl peptidase-4 inhibitors, using a supervised machine-learning algorithm (PROFILE study): A retrospective analysis of a Japanese commercial medical database”. en. In: Diabetes, Obesity and Metabolism 21.8 (2019). eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1111/dom.13753, pp. 1925–1934. ISSN: 1463-1326. [CrossRef]
- Zwick, William R. and Wayne F. Velicer. “Comparison of five rules for determining the number of components to retain”. In: Psychological Bulletin 99.3 (1986). Place: US Publisher: American Psychological Association, pp. 432–442. ISSN: 1939-1455. [CrossRef]


 |
| Name | Description | Modalities | Accepted value range |
|---|---|---|---|
| age | Age | age ≤55; age > 55 | [18, 120[ |
| BMI | BMI | Underweight: < 18.5 kg/m2; Normal: between 18.5 and 24.9 kg/m2; Overweight: between 25.0 and 29.9 kg/m2; Obese: ≥30.0 kg/m2 |
[10, 100[ |
| high sbp | High systolic blood pressure | 1: SBP > 130 mmHg; 0: SBP ≤130 mmHg | ≤400 |
| high dbp | High diastolic blood pressure | 1: DBP > 80 mmHg; 0: DBP ≤80 mmHg | ≤200 |
| Prevalence (% of patients) | Lift values | ||||||
|---|---|---|---|---|---|---|---|
| Modality | C1 | C2 | C3 | Cohort | |||
| n = 12272 | n = 15477 | n = 6385 | n = 34134 | C1 | C2 | C3 | |
| (36.0%) | (45.3%) | (18.7%) | (100%) | ||||
| Female | 62.1 | 64.2 | 71.4 | 64.8 | 0.96 | 0.99 | 1.10 |
| Male | 37.9 | 35.8 | 28.6 | 35.2 | 1.08 | 1.02 | 0.81 |
| Cholesterol normal | 60.6 | 90.7 | 16.5 | 66.0 | 0.92 | 1.37 | 0.25 |
| Cholesterol above normal | 20.8 | 8.5 | 26.6 | 16.3 | 1.28 | 0.52 | 1.63 |
| Cholesterol well above normal | 18.5 | 0.7 | 56.9 | 17.7 | 1.05 | 0.04 | 3.22 |
| Glucose normal | 80.0 | 99.2 | 42.7 | 81.7 | 0.98 | 1.21 | 0.52 |
| Glucose above normal | 20.0 | 0.8 | 57.3 | 18.3 | 1.10 | 0.04 | 3.13 |
| Physical activity | 81.0 | 76.8 | 79.6 | 78.8 | 1.03 | 0.97 | 1.01 |
| Age ≤55 | 45.3 | 48.2 | 34.1 | 44.5 | 1.02 | 1.08 | 0.77 |
| Age > 55 | 54.7 | 51.8 | 65.9 | 55.5 | 0.99 | 0.93 | 1.19 |
| BMI obese | 41.4 | 21.8 | 44.5 | 33.1 | 1.25 | 0.66 | 1.34 |
| BMI overweight | 36.5 | 37.3 | 36.1 | 36.8 | 0.99 | 1.01 | 0.98 |
| BMI normal or underweight | 22.1 | 40.9 | 19.5 | 30.1 | 0.73 | 1.36 | 0.65 |
| High Systolic blood pressure | 100.0 | 14.3 | 18.5 | 45.9 | 2.18 | 0.31 | 0.40 |
| High Diastolic blood pressure | 100.0 | 12.9 | 16.8 | 44.9 | 2.23 | 0.29 | 0.37 |
| Hypertension | 100.0 | 0 | 0 | 36.0 | 2.78 | 0 | 0 |
| Smoke | 10.1 | 7.2 | 7.6 | 8.3 | 1.21 | 0.87 | 0.91 |
| Alcohol | 6.6 | 3.8 | 5.7 | 5.2 | 1.28 | 0.74 | 1.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





