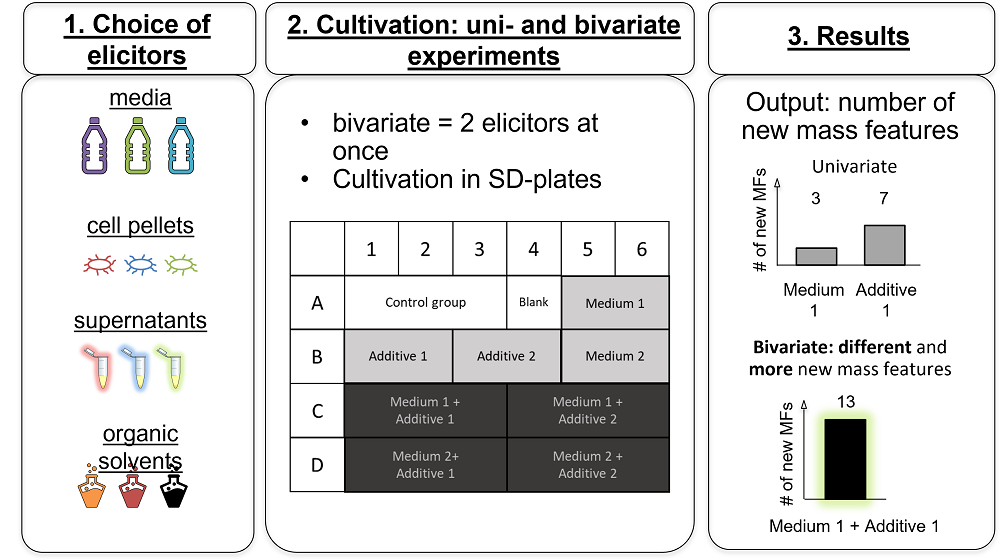
The scarcely investigated myxobacterium Corallococcus coralloides holds a large genome containing many uncharacterized biosynthetic gene clusters (BGCs) that potentially encode the synthesis of entirely new natural products. Despite its promising genomic potential, suitable cultivation conditions have not yet been found to activate the synthesis of new secondary metabolites (SMs). Finding the right cultivation conditions to activate BGCs in the genome remained a major bottleneck and its full biosynthetic potential was so far not retrieved. Here, we therefore applied a bivariate OSMAC approach, using a combination of two elicitor changes at once for activation of BGCs and concomitant SM production by C. coralloides. The bivariate OSMAC screening was carried out in 24-well System Duetz-plates, applying univariate and bivariate OSMAC conditions. We combined biotic additives and organic solvents with minimal media and complex growth medium. The success of the method was evaluated by the number of new mass features detected in the respective extracts. We found synergistic effects in bivariate OSMAC designs. The number of new mass features detected in bivariate OSMAC exceeded the sum of new mass features found in the respective univariate OSMAC with only one elicitor. Overall, the bivariate OSMAC screening led to 26 new mass features, which were not detected in the univariate OSMAC design. Hence, the presence of multiple elicitors in the bivariate OSMAC designs successfully activated the biosynthetic potential in C. coralloides. We propose the bivariate OSMAC designs with a complex combination of elicitors as a straightforward strategy to robustly expand the SM space of microorganisms with large genomes.