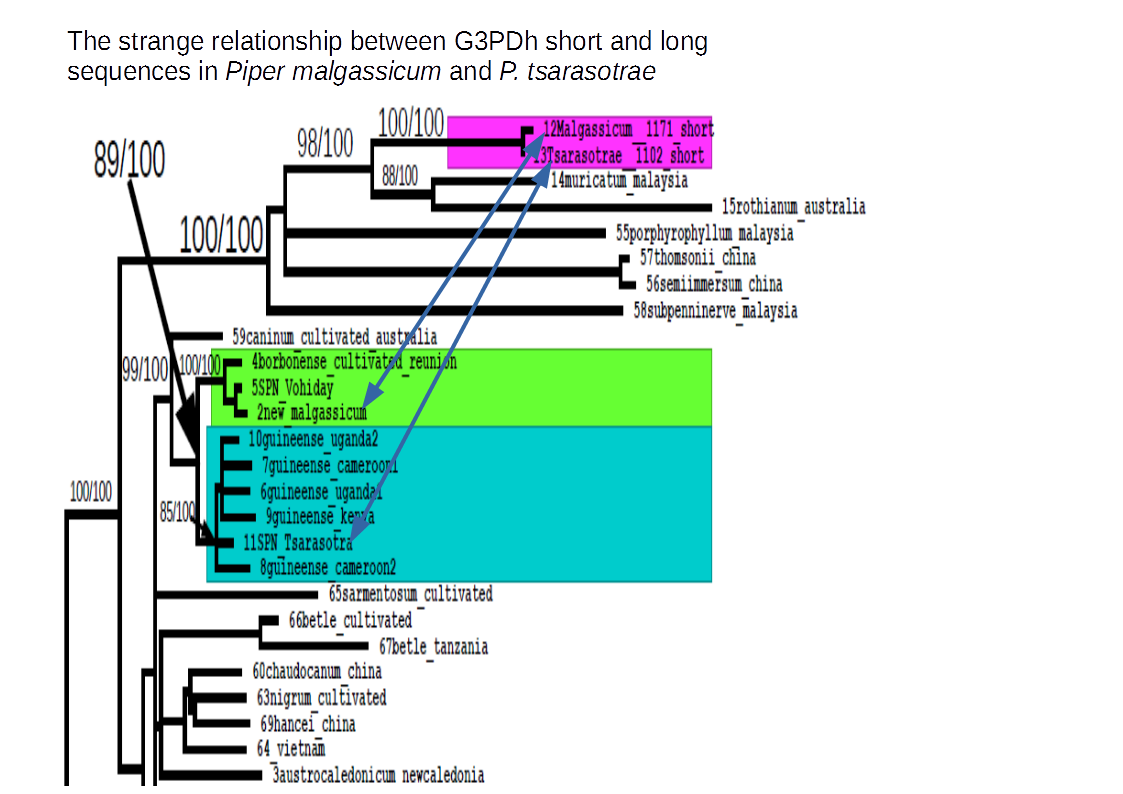
two new species of genus Piper L. from Madagascar: Piper malgassicum and Piper tsarasotrae, were analyzed to investigate their phylogenetic position and evolutionary history. Both plastidial and nuclear markers were used for sequencing. The plastidial markers (ndhF and the trnL intron) showed a close relationship between the two species with respect to the other species of Piper. Both species appeared phylogenetically related to the African P. guineense and the Malagasian/Mascarenhas endemic P. borbonense. The nuclear marker (G3PDH) amplification produced two separate sets of sequences: “long” sequences, that could be easily translated into an amino acid chain, and “short” sequences, characterized by deletions that did not allowed to translate them correctly to an amino acid sequence. Analyzing together the nuclear sequences, we observed that the “long” sequence of P. tsarasotrae had a stricter relationship to the African accessions of P. guineense, while the accession of P. malgassicum was more strictly related to P. borbonense. On the contrary both “short” sequences of Piper malgassicum and Piper tsaratsotrae resulted phylogenetically related to Asian accessions and more distantly related to the formerly cited species. This unexpected result was tentatively explained with a more ancient hybridization event between an ancestor of P. malgassicum and P. tsarasotrae (and possibly P. borbonense) and an Asian species of Piper. The Asian contribution would have produced the ancestors of the “short” sequences that would eventually have lost functionality by deletions, becoming paralogs. A more recent hybridization event would have led to the separation of Piper malgassicum from Piper tsarasotrae with an African pollen-derived genome contribution from P. guineense or, more probably, an ancestor thereof, to an ancestor of P. tsarasotrae. The chromosome numbers of P. tsarasotrae (2n = about 38) and P. malgassicum (2n = about 46), were more like the Asian species than to the American species. Unfortunately, no chromosome number of the African species P. guineense is currently available, to analyze eventual chromosomal connections.