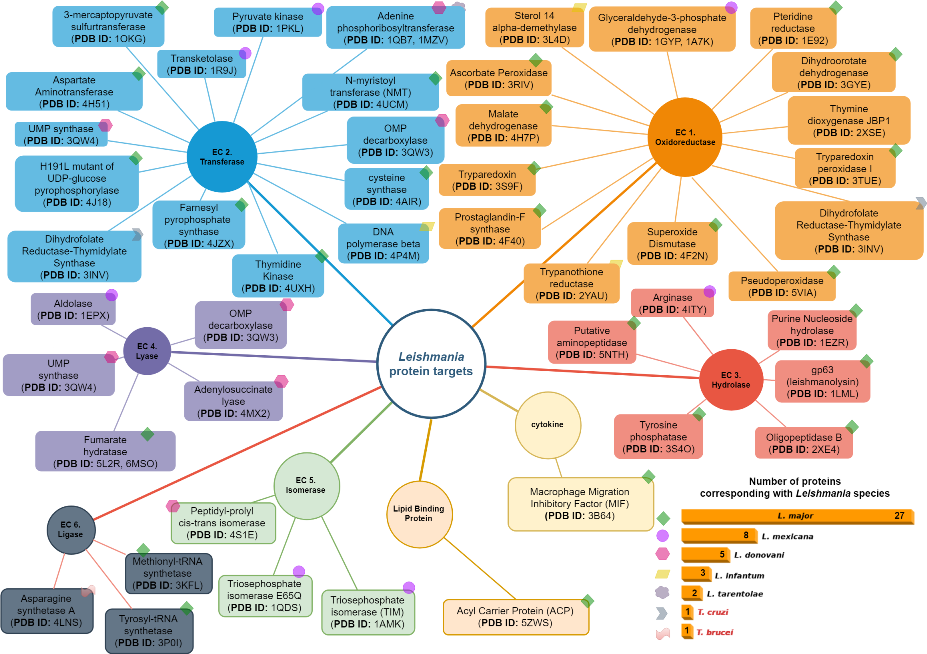
We report on the state of the art of proteins recognized as potential targets for the development of leishmania treatments through the search of biologically active chemical species, either from experimental in vitro, in vivo, or in silico sources. We classify the gathered information, in several ways: vector taxonomy and geographical distribution, leishmania parasite taxonomic and geographical distribution and enzymatic function (oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases and cytokines). Our aim is to provide a much needed reference layout for research efforts aimed to understand the background of ligand-protein activation/inactivation processes, in this specific case, related with enzymes known to be part of biochemical cascade reactions initiated following a leishmania infectious episode.