1. Introduction
BCa is one of the common malignant tumors worldwide. According to the latest statistics from GLOBOCAN, the number of new cases worldwide in 2022 reached 614298, accounting for 3.1% of all cancers and caused 220596 deaths, accounting for 2.3% of all cancer deaths [
1]. Globally, BCa exhibits a substantially higher incidence and mortality burden among males, with the incidence rate approximately four times greater than that observed in females [
1]. It is the sixth most common cancer in males, and the incidence rate increases with age, especially in elderly people over 65 years old [
2]. BCa is associated with a multitude of lifestyle and environmental factors, among which smoking is a major known risk factor. Specially, smokers exhibit a 2-4 times increased risk of BCa relative to non-smokers, accounting for 65% of incident BCa cases [
3]. Occupational exposure to aromatic amines and other chemical carcinogens is also a crucial risk factor. Furthermore, chronic infections, such as schistosomiasis-induced chronic cystitis, are also related to the occurrence of BCa. According to tumor invasion depth, BCa is divided into non-muscular invasive BCa (NMIBC) which accounts for more than 75% and muscular invasive BCa (MIBC) [
3]. The main treatment method of NMIBC is transurethral bladder tumor electrolysis combined with chemotherapy and immunotherapy, while MIBC is mainly treated by radical cystectomy [
4]. As BCa advances through its clinical stages, the risk of cystectomy increases and the quality of life for BCa patients will deteriorate physically, psychologically and socially [
5]. Despite improvements in surgical techniques and improved 5-year survival rates in recent years, the risk of recurrence remains high (approximately 50-70%) [
6,
7]. In addition, 10% to 15% of NMIBC patients experience tumor progression or metastasis within 5 years and lead to poor prognosis [
6,
8]. 50% of MIBC patients experience metastasis within two years and die within five years [
9]. Screening of high-risk individuals can help diagnose BCa at an early stage (Ta/T1 stage), so that it can be treated more effectively [
10]. Therefore, effective early diagnosis methods are crucial to reduce mortality, but there is still a lack of effective non-invasive early screening methods for BCa.
The traditional gold standard for the diagnosis and surveillance of BCa is cystoscopy combined with tissue biopsy [
11]. However, this approach is invasive and is frequently associated with risks such as urethral injury and infection [
11]. The procedural risks and costs are particularly elevated for patients with NMIBC who require frequent monitoring for disease recurrence [
11,
12]. Furthermore, variability in operator technique can lead to inconsistencies in the sensitivity and specificity of detection [
13]. Since early-stage BCa is often asymptomatic, cystoscopy is not suitable for population-wide early screening [
14]. Urine cytology represents the most convenient non-invasive diagnostic tool, with a specificity exceeding 90% [
15,
16], and thus serves as an important adjunct to cystoscopy [
11]. Its use has been shown to improve patient survival and is cost-effective [
17]. However, for early-stage, particularly low-grade (G1/G2) BCa, its sensitivity is significantly limited (approximately 42%) due to factors including the lack of definitive morphological atypia in malignant cells, interference from excessive inflammatory or blood cells, low cellular yield, and variability in sample preservation and processing [
18,
19,
20,
21,
22,
23]. Consequently, more than half of BCa cases may be missed, delaying early intervention [
18]. Additionally, the diagnostic criteria and procedural protocols for conventional urine cytology are not standardized across institutions, and inter-pathologist subjectivity in assessing cellular atypia further compromises the consistency and accuracy of reports [
21]. Therefore, despite its widespread clinical use, the inherent limitations of traditional urine cytology fail to meet the demands of early cancer detection.
To address these shortcomings, numerous urine-based BCa detection methods have been developed. Several have received FDA approval and been integrated into clinical practice, including assays for urinary nuclear matrix proteins [
24,
25,
26,
27,
28], human bladder tumor antigen (BTA) [
24,
27,
28,
29], fluorescent immunohistochemistry (uCyt+/ImmunoCyt) [
24,
27,
28], fluorescence in situ hybridization (FISH) UroVysion [
24,
27,
30]. Although these biomarker-based tests are approved for BCa monitoring, their sensitivity and specificity for early-stage detection remain suboptimal. Moreover, conditions such as cystitis and hematuria can lead to false-positive results [
24,
27,
30,
31,
32]. Thus, there is a pressing clinical need to enhance the technical capabilities of urine-based diagnostics, either by refining cytological methodologies or by developing novel urinary biomarkers with high sensitivity and specificity, to effectively meet the requirements for the early diagnosis of BCa.
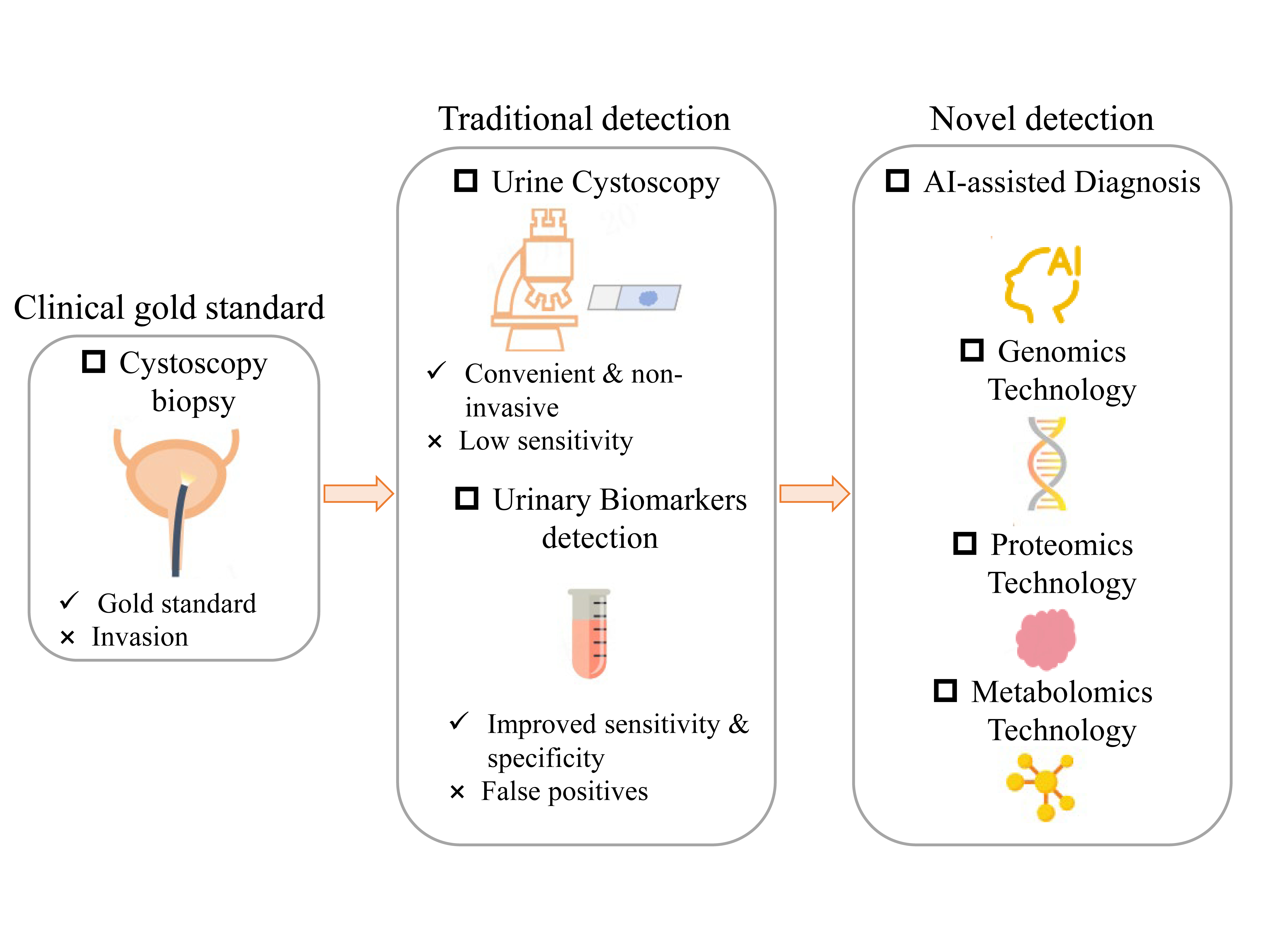
This review will elaborate on the evolutionary trajectory of BCa detection technologies, ranging from cytology which is clinical gold standard but invasive, to traditional non-invasive urine detection including urine cytology and DNA/RNA biomarker assays, to novel detection such as AI-assisted detection and multi-omics analyses, as shown in
Figure 1.
2. AI Empowered Urine Cytological Detection
In order to address the limitations of traditional urine cytology—such as low sensitivity, strong subjectivity, poor reproducibility, and time consumption, AI technology has been introduced into urine cytological detection. It has brought new breakthroughs to urine cytological detection in recent years [
33]. The AI system reduces artificial errors by efficiently analyzing urine cytological images [
34,
35,
36,
37], automating cell-level classification [
38,
39] and predicting patient outcomes [
40,
41,
42]. These improvements enhance diagnostic accuracy, consistency, and efficiency while optimizing diagnostic workflow [
43]. Currently, the application of AI in urinary cytological detection mainly focused on cell-level classification and patient-level diagnosis.
2.1. Single-Cell Level Classification
In 2015, the DL model based on artificial neural networks (ANN) was first applied to urine cytological detection [
37]. ANN is a computational model inspired by biological neurons, consisting of multi-layer interconnected artificial neurons that achieve nonlinear data mapping by adjusting synaptic weights and activation functions, thus completing pattern recognition [
44,
45]. Its training process is based on a backpropagation algorithm, which uses gradient descent to optimize the loss function to minimize prediction errors, and extracts hierarchical features of data through hidden layers [
46]. However, the ANN model with a fully connected structure cannot take the image directly as input, which is prone to parameter explosion and overfitting problems [
45]. Multi-layer convolutional neural network (CNN) optimizes the network structure and extraction method of input features based on ANN [
47], as shown in
Figure 2. It efficiently captures spatial local features (such as textures, edges) through the local receptive field, parameter sharing and pooling operations of the convolution layer. Thus, it significantly reduces the number of parameters and retaining the topological structure of the input data [
45]. A multilayer CNN model using 2405 urine cytological full-slide images as input achieved an area under the curve (AUC) of 0.88 in the diagnosis of high-grade BCa, with a sensitivity of 79.5% and a specificity of 84.5% [
36]. The issues of parameter explosion and overfitting problems caused by directly adaption of images as input in the ANN model are solved.
To improve the interpretability of the model, it is necessary to identify urinary cell types. A study enhanced the accuracy of urinary cell classification by a fine-grained CNN model, as shown in
Figure 2. They reached an AUC of 0.989 in detection of high-grade urothelial cancer cells, with an accuracy rate of 95.36% [
38]. Wu et al. used a multi-scale feature fusion strategy to eliminate background noise interference, thereby improving the accuracy of urinary cell segmentation [
48]. In addition, by combining different types of cell morphological characteristics with CNN, such as nucleoplasm ratio, nuclear area and chromatin atypicality, the sensitivity to recognition of malignant tumor cells reached 0.896-1.000. With the same CNN model, at least 7 abnormal cells were identified in each urine cytological full-slide image of patient with BCa [
39]. In order to improve cell segmentation accuracy, Kaneko et al. reduced boundary artifacts based on the superimposed segmentation tool named “CutMix” and adjusted the weight of the label [
35]. A cell segmentation tool named “CircleCut” that handles irregular cell boundaries was developed by using 4637 cell annotation images. And combining with refined feature data augmentation technology and CNN, cell classification model training was completed. The AUC of urinary cell classification reached 0.99, with an accuracy rate of 95%, a sensitivity of 96.7%, and a specificity of 95%. AI classification based on urine cytology improves the accuracy and efficiency of diagnosis, providing a solid foundation for the development of AI-based urine cytological diagnostic platform.
2.2. Patient Level Diagnostics
The deep learning system based on CNN can effectively predict the existence of malignant tissue in urine cytopathological images, and its prediction is interpretable based on the abnormal cells selected by the model [
49]. The fully automatic AI pathological prediction system was achieved by analyzing digitized urinary cytology images and combining semi-supervised learning methods. In the test of 315 digitized urine cytology slides, with a comparable specificity and a consistency of 86%, this AI system achieved significantly higher sensitivity (63%) than cytopathologist (46%) [
50]. The latest research also shows that the application of AI in pathological prediction is not limited to the field of cytology. The Virchow model is by far the largest fundamental model of computational pathology, which is able to achieve an AUC of 0.95 at the individual level in nine common cancers and seven rare cancers [
51]. This demonstrates its outstanding performance in pan-cancer detection. And it shows that AI technology has broad application and great potential in the field of pathological prediction.
Based on AI segmentation and classification of urinary cells, a fully automated BCa detection platform based on urine cytology named VisioCyt has been developed. This approach enabled the differentiation of urothelial cells, squamous cells, and inflammatory cells. In a multicenter prospective study, the assay achieved a sensitivity of 84.9%; notably, the sensitivity for low-grade tumors reached 77%, which was substantially higher than the 26.3% observed with conventional urine cytology in this study [
52]. Recently, a commercial BCa AI detection platform based on urine cytology named AIxURO has been developed, which is used to identify different types of cells in urinary cytology images and provide quantitative analysis. The AIxURO platform integrates the morphological diagnostic criteria from “Paris System Report urine cytology” (TPS 2.0) [
53], including nucleoplasm ratio, nuclear area and chromatin abnormal characteristics, thereby enabling high-throughput digital analysis of urothelial cells. Based on the contrast learning framework, its multi-scale feature fusion mechanism can eliminate background noise interference and optimize diagnostic specificity. The AIxURO platform-assisted diagnosis achieved a sensitivity of 92.3% and a specificity of 100%, with a high concordance with expert consensus. Specifically, an average of 686 abnormal cells were identified in positive cases, whereas only 8 abnormal cells were detected in negative cases [
54]. Combined with anchor quality sampling strategy and embedding model, the AIxURO platform’s recognition sensitivity to a typical urothelial cell was increased from 25.0% to 63.9%, while significantly shortening the screening time [
42]. For postoperative follow-up of upper urinary tract urothelial carcinoma (UTUC), the AIxURO platform increased the detection rate of early recurrence from 29.6% (8/27) to 37.0% (10/27), enhancing the credibility of urinary cytology reports and helping to detect intravesical recurrence in the early stage [
41].
Conventional urine cytology exhibits insufficient diagnostic sensitivity for early-stage and low-grade BCa. AI technologies, particularly those based on CNNs, significantly enhance diagnostic performance by enabling automated cell segmentation, classification, and quantitative morphological analysis. The key lies in their ability to accurately identify subtle cytological features such as increased nuclear-to-cytoplasmic ratio, enlarged nuclear area, and chromatin abnormalities, thereby achieving highly sensitive detection of early-stage tumors. For example, the VisioCyt platform have improved the sensitivity for low-grade tumors from 26.3% with conventional cytology to 77%. In conclusion, by enhancing the capture and interpretation of early biological signals, AI is poised to redefine urine cytology as a highly sensitive, non-invasive tool for screening and monitoring, offering substantial promise for the early diagnosis and postoperative surveillance of BCa.
3. Detection Technologies of Urine Genome Technology
Urine samples of BCa patients contain a large amount of tumor-derived DNA/RNA that can be used as liquid biomarkers for BC diagnosis and monitoring [
55,
56,
57]. The progress of genetic detection technology for early diagnosis of BCa revolves around “improving the detection rate of small tumors” and “breaking through early missed bottlenecks”. Different technologies form complementary systems through targeted optimization and multi-dimensional integration.
3.1. Detection Technologies of Urine DNA
3.1.1. Polymerase Chain Reaction (PCR) Detection Technology
The PCR technique provides a foundation for precise detection based on gene expression. Building upon this, the two-gene ratio model (with an upregulated gene as the numerator and a downregulated gene as the denominator) represents a significant advancement in the exploration of early diagnostic strategies. A study established a DNA discriminant score based on the urinary ratios of IQGAP3/BMP4 and IQGAP3/FAM107A [
58]. This model achieved an AUC of 0.862 for the diagnosis of early-stage, particularly low-grade BCa, with a sensitivity of 71.0% and specificity of 88.6%. This study provides the first evidence that gene ratio analysis can effectively differentiate between early-stage cancer and hematuria, laying the foundation for the subsequent application of PCR technology in NMIBC. Moreover, the IQGAP3/BMP4 ratio is strongly correlated with the recurrence-free survival in patients with NMIBC, suggesting that this index can not only be used for early diagnosis, but also predict the risk of recurrence 1-2 years post-surgery [
59].
Based on previous studies on BCa DNA mutations, telomerase reverse transcriptase (TERT) gene promoter mutations [
60,
61], and fibroblast growth factor receptor 3 (FGFR3) mutation [
62,
63] have been the most common somatic mutation. By integrating the two-gene model and orthodenticle homeobox 1 (OTX1) gene mutation detection, a three-genes urine assay for early BCa detection was developed [
64,
65]. In a prospective multicenter trial, this assay achieved a sensitivity of 57% for recurrence monitoring of early BCa, covering 80% of early recurrence cases [
66]. And it further proved that “multigene collaboration” is better than single-gene detection, thereby providing ideas for missed diagnosis and prevention of early stage. This idea directly inspired the development of Uromonitor® detection [
67]. To further enhance diagnostic sensitivity, OTX1 gene mutation detection was replaced with the KRAS gene mutation-based Uromonitor® detection [
67,
68], which increased the overall sensitivity for BCa recurrence to 100% (although the sample size needs to be considered). Especially for low-grade tumors, it has a significant higher sensitivity (65%) than urinary cytology (0%) in this study [
67]. For the detection of early recurrence with 6-12 months postoperatively, this assay achieved a sensitivity of 93.1% and a specificity of 85.4%, while enabling the detection of small tumors (diameter <5mm) [
69]. While, the sensitivity of urine cytology for early postoperative recurrence in this study was only 26.3%. This solved the problem of insufficient sensitivity of OTX1 gene mutation in early multigene combination detection. In addition, the negative predictive value (NPV) of Uromonitor® test is as high as 98.8% [
70]. This robust NPV supports the safe reduction of cystoscopy frequency by approximately 30%, especially suitable for non-invasive long-term follow-up of patients with early BCa.
PCR technology has become the first choice for early screening due to its fast and low-cost characteristics. In particular, Uromonitor® with high NPV (>95%) can effectively eliminate early recurrence and reduce invasive examinations; its limitation is that it is insufficient sensitivity to small tumors (about 60%), and it depends on sequencing technology to supplement it.
3.1.2. Next-Generation Sequencing (NGS) Technology
NGS technology breaks through the bottleneck of small-tumors detection through multi-dimensional molecular characteristics. It can perform rapid and high-throughput sequencing of large amounts of DNA or RNA at the same time [
71], with workflow involving library construction, template preparation, sequencing reactions, and bioinformatic data analysis [
72]. And it is divided into whole genome sequencing (WGS) technology [
73] and deep sequencing technology [
74].
WGS is an important application of NGS, which performs comprehensive and non-selective sequencing of the entire genome of an organism [
73]. Based on WGS technology, it was found that mutations of 11 genes in urine and copy number variations (CNVs) of 39 chromosomal arms were related to BCa. And the UroSEEK detection scheme was developed with a sensitivity of 67% for low-grade BCa detection-far exceeding urinary cytology (0%) in this study. It can detect early tumors containing only 1-2 gene mutations, and effectively identify small tumors missed by traditional methods [
75], solving the problem of “high missed detection rate” by PCR technology in low-grade tumors. Moreover, it is confirmed that the frequency of TERT promoter mutations in low-level non-invasive BCa is as high as 77% [
76], becoming the core detection target of this technology. However, WGS technology is expensive, limiting its widespread use. To address this issue, shallow whole-genome sequencing (sWGS) was introduced, reducing per-base coverage while maintaining genome-wide information through imputation algorithms and population genetics modeling [
77,
78,
79]. This approach cuts sequencing costs to ~20% of WGS while preserving high-resolution variant detection, especially in large cohorts (>2000 samples) [
78]. Based on sWGS technology, a diagnostic model named UCdetector was developed [
80]. The UCdetector model was sensitive to early stage with a specificity of 94.7%, and especially for small-tumors detection rate reached 78%, solving the limitation of PCR technology relying on “high abundance mutations”. With the joint development of sequencing technology and AI technology, the UCseek technology was developed and it had good performance at aWGS sequencing depths of 0.3X-0.5X [
81], The detection accuracy of low-grade and early-stage tumor is 91.8% and 94.3%, respectively. And the sensitivity of monitoring recurrence is 90.91%, which was significantly better than cystoscopic imaging (59.09%), proving that gene mutation analysis can improve the detection rate of small tumors and become a potential tool for early screening in grassroots hospitals. When the sequencing depth is further reduced to 0.1X, low coverage WGS (LC-WGS) was developed, and the sequencing cost was further reduced. The sensitivity to early-stage BCa detection was 82.5%, specificity was 96.9%, and the AUC in T1 stage of BCa reached 0.91 [
82], which was suitable for large-scale early screening.
Another development direction of NGS technology is to obtain more accurate genomic information, so deep sequencing technology has been developed [
83]. By performing multiple independent sequencing reactions on DNA or RNA fragments in the same region, false positive results due to random errors are reduced, and detection sensitivity and accuracy are improved [
84]. During data analysis, specific algorithms and statistical models [
83] are employed to integrate and interpret the repeated sequencing data, thereby enabling the more reliably identification of low-frequency mutations, rare variants, and complex gene expression patterns. It is particularly valuable in studying tumor heterogeneity [
85], pathogen mutation [
86], and evolution [
87,
88]. For patients with hematuria, an ultra-deep sequencing (23 genes, 443 mutations) yields a sensitivity of 87.3% and specificity of 84.8% for the detection of early-stage disease. Additionally, this assay achieved a sensitivity of 86.2% for the NMIBC recurrence, with the false-positive rate was controlled at 15.2% [
89]. uCAPP-Seq technology also has good effect in predicting the applicability of immunotherapy [
90], extending deep sequencing from “early diagnosis” to “prognostic stratification” for the first time [
91]. To detect residual disease with high sensitivity, uCAPP-Seq technology was employed to integrate ultra-low throughput WGS (ULP-WGS) with personalized profiling of BCa. Using leave-one-out cross-validation, a random forest model incorporating urine cell-free DNA (cfDNA)-derived factors yielded 87% sensitivity in predicting residual disease. This proved the superiority of urinary multitopic analysis in tumor residue detection, and provides a molecular basis for personalized treatment decisions.
PCR techniques have become important tools for initial screening, leveraging their advantages of speed and cost-effectiveness. NGS technologies have further enhanced detection sensitivity for low-grade and small tumor lesions (exceeding 90% in some assays) by integrating multidimensional features such as mutations and CNVs. The technological evolution has progressed along two main trajectories: first, reducing costs through shallow or low-coverage sequencing to facilitate application in primary care settings; second, utilizing ultra-deep sequencing and multi-omics integration to achieve high-sensitivity detection and prognostic stratification. Collectively, these advancements are transforming urine-based testing from an auxiliary method into a highly sensitive, standardized tool for early screening and recurrence monitoring, providing crucial support for precision diagnosis and comprehensive management of BCa.
3.2. Urine DNA Methylation Detection Technologies
3.2.1. Methylation-Specific PCR (MSP) Technology
With its high sensitivity and specificity [
92], MSP technology based on PCR technology has become the basic platform for urine methylation detection [
93,
94]. By targeting the methylation status of key genes, it has built a two-marker or multi-marker models have been developed to overcome the limitations of traditional methods in early BCa especially low-grade, NMIBC detection. Through MSP technology, the methylation level of DMRTA2 showed higher sensitivity in the early stage of detection, with T1 at 94.1%, T2 at 96.4%, T3 at 77.8%, and T4 at 71.4% [
95]. MSP technology detects six methylation markers (ASTN1, DLX1, etc.) to build a GynTect test solution, with 60% sensitivity and 96.7% specificity in NMIBC detection. The sensitivity can be increased to more than 90% through algorithm optimization [
94], confirm the potential of MSP technology in low-grade tumor screening. Reduce six methylation markers to four (HOXA9, PCDH17, POU4F2, ONECUT2), combined with high-resolution melting curve, detection sensitivity reaches 90.5% and PPV reaches 100% [
96]. Through bioinformatics-driven optimization, the panel was reduced to two markers, improving sensitivity to 91.2%. For early-stage BCa, MSP demonstrated superior sensitivity compared with urine cytology (88.1% vs. 55.6%) and FISH (89.7% vs. 72.2%). Importantly, its application could reduce unnecessary cystoscopies by approximately 30% [
97], verifying the efficient detection ability of MSP technology for early BCa.
However, single-marker assays show limited sensitivity across different BCa stages, indicating the need for the combined use of different markers. Through methylation detection and gene mutation detection, the AUC of different stages was 0.96, sensitivity was 93%, specificity was 86%, and NPV was as high as 99%, indicating that 77% reduction in unnecessary cystoscopy [
98]. Later, NGS technology was introduced to combine with MSP, and it was found that the three markers combination of NRN1 methylation combined with TERT and FGFR3 mutations showed good performance in differentiating BCa from controls (
Figure 3a). In external verification comparing with cytology and FISH, as shown in
Figure 3b, the model was achieved with a superior sensitivity of 86.4% and a comparable specificity of 89.5% [
99]. MSP technology can accurately capture early tumors by targeting methylation sites of known cancer-related genes (such as TWIST1, RUNX3, HOXA9). Especially in NMIBC monitoring, EpiCheck series of studies have shown that its NPV is as high as 95.1%~99.3%, which can reduce unnecessary microscopy by more than 80% [
100,
101,
102,
103,
104,
105,
106]. Its core advantage is its simplicity of operation, high throughput, and is suitable for routine clinical screening. Multi-marker combinations (such as the five-marker stratification model) further enhance the recognition ability of high-risk NMIBC and MIBC (sensitivity 90.5%, specificity 86.8%), providing candidate markers for subsequent in-depth analysis at the genomic level.
3.2.2. DNA Methylation Detection Technology
By high-throughput analyzing multiple gene methylation patterns, combined with mutation status and clinical parameters, it constructs more complex prediction models, demonstrating high application value in early diagnosis, recurrence monitoring, and prognosis evaluation.
A study screened three biomarkers (a combination of hypermethylation and hypomethylation) using pyrosequencing. The AUC for predicting NMIBC recurrence reached 0.95, with a sensitivity of 89% and specificity of 97%, significantly outperforming cytology (35%) and cystoscopy (15%) [
107]. This study first confirmed the predictive efficacy of dynamic methylation changes for recurrence risk. Subsequently, another study found that hypermethylation of the HIST1H4F gene was significantly increased in BCa [
108]. Although not directly associated with clinicopathological parameters, its stable methylation status provides a new candidate biomarker for early screening. Meanwhile, a separate study combined SNaPshot methylation analysis with gene mutation detection to construct a prediction model incorporating age factors [
64]. The NPV reached 99.6%~99.9% in hematuria patients, indicating an extremely low false-negative rate. Later, a study reported that the combination of EpiCheck detection and urine cytology achieved a sensitivity of 83.3% and specificity of 86.3% in monitoring high-grade BCa, confirming that methylation biomarkers can compensate for the missed diagnosis of low-grade tumors by cytology [
103]. More recently, a study developed an eight-biomarker model (FGFR3 and TERT mutations + OTX1 and ONECUT2 methylation) based on a cohort of high-risk hematuria patients following AUA guidelines [
109]. The AUC ranged from 0.929 to 0.971, and the probability of BCa in negative cases of the low-risk group was only 0.3%~2%, enabling safe avoidance of cystoscopy, while positive cases in the medium-high risk group required accelerated evaluation to achieve precise stratification. The success of such models relies on the analysis of methylation regulation of key signaling pathways (e.g., Wnt and Notch pathway-related genes) in BCa initiation and progression, providing a basis for personalized diagnosis and treatment.
3.2.3. DNA NGS Technology
NGS technology enables unbiased scanning of genome-wide methylation patterns, identifying differentially methylated regions (DMRs) and tissue-specific biomarkers, and has achieved breakthroughs in tumor grading, localization, and ultra-sensitive detection, serving as a bridge connecting basic research and clinical applications.
A study identified methylation signatures of 32 genes (e.g., EOMES, GP5, ZSCAN12) using microarray-based whole-genome bisulfite sequencing (WGBS), which could completely distinguish low-grade from high-grade BCa [
110]. Among these, GP5 and ZSCAN12 were newly discovered high-grade specific hypermethylated genes, providing molecular biomarkers for pathological grading. Another study developed the GUseek system, which integrated methylation and CNVs through shallow WGBS [
111], its accuracy exceeded 95% in detecting genitourinary system cancers, with a sensitivity of over 72% for distinguishing BCa, prostate cancer, and renal cell carcinoma, realizing “one-stop” non-invasive localization. Additionally, UroMark test, based on NGS analysis of 150 CpG sites, achieved an AUC of 97%, sensitivity of 98%, specificity of 97%, and NPV of 97% in the validation cohort, becoming the first genome-wide methylation detection protocol approaching clinical application standards [
112]. Moreover, the UCseek technology achieved an accuracy of 94.3% for early UC at an ultra-low sequencing depth (0.3X-0.5X), with a recurrence monitoring sensitivity of 90.91%, far exceeding cystoscopy (59.09%) [
81]. This confirmed that genome-wide methylation signatures can be stably detected in trace amounts of DNA, enabling rapid outpatient testing.
In addition, a study identified FRAGMA technology, which inferred methylation status through cfDNA fragmentation patterns without bisulfite treatment, achieving an AUC of 0.98 in hepatocellular carcinoma and nasopharyngeal carcinoma [
113]. This provides a new methodological approach for BCa cfDNA methylation detection, especially suitable for analyzing trace cell-free DNA released by small tumors, solving the problem of traditional detection’s dependence on sample volume.
From PCR to whole-genome sequencing, BCa urine DNA methylation detection technology has gradually achieved a leap from “targeted validation” to “panoramic analysis.” PCR technology remains the mainstay of clinical translation due to its high sensitivity and operability; genomic sequencing technology improves the detection rate of small tumors through multi-omics integration; whole-genome sequencing technology lays the foundation for precision medicine. In the future, it is necessary to further optimize biomarker combinations, reduce sequencing costs, and verify their clinical utility through large-scale multi-center prospective studies, ultimately realizing a comprehensive revolution in early BCa diagnosis.
3.3. Urine RNA Detection Technologies
3.3.1. mRNA Detection Technology
Establishing standardized detection workflows has laid the foundation for reliable urine mRNA biomarker analysis. However, early studies were hindered by sample heterogeneity and donor variability. In 2007, a study first introduced a standardized workflow with external RNA controls (RNAₗᵤC), analyzing RNA from whole urine, cell pellets, and cell-free fractions via RT-qPCR to address the interference of urine components [
114]. The ETS2/uPA mRNA ratio screened in this study achieved a specificity of 100% and AUC of 0.929, outperforming the traditional tissue marker hTERT. However, its sensitivity for low-grade tumors was only 53.9%, indicating that single biomarkers have limited ability to detect early small tumors, which requires technical innovation and combination optimization. This standardized technology provided a methodological template for subsequent studies, promoting the transition of urine mRNA detection from experimental exploration to clinical translation.
With the development of high-throughput technologies, researchers have shifted from single-gene analysis to multi-gene combination modeling. A study quantified 45 genes using TaqMan Arrays and verified the superiority of the GS_D2 model (IGF2 and MAGEA3), which achieved an AUC of 0.918 and a sensitivity of 94.74% for high-risk tumors with strong cross-center stability [
115]. However, its sensitivity for low-risk tumors was 67.86%, which still needs improvement. In parallel, another study focused on carbonic anhydrase IX (CAIX) splice variants and found that the proportion of full-length isoforms (FL%) was significantly increased in cancer patients (median 70.8% vs. 2.6% in controls) with an AUC of 0.837 and sensitivity of 90%, providing a new dimension of “splice variant ratio” for biomarker screening [
116]. Furthermore, a study discovered that NDRG2 mRNA was significantly downregulated in the urine of BCa patients [
117]. Combined with protein level verification, the AUC reached 0.888 with a sensitivity of 85.5%, and it was associated with tumor grade and stage, demonstrating the clinical value of in-depth single-gene mining.
In the exploration of clinical utility, a study demonstrated through Cxbladder Monitor in 2017 that gene expression analysis combined with clinical data can construct an efficient “rule-out” detection model, achieving a sensitivity of 91% and NPV of 96%, with a sensitivity of 84% for low-grade tumors, significantly reducing unnecessary cystoscopies [
118]. Building on this, a study developed an 8-gene classifier (including ANXA10 and IGF2) optimized for follow-up scenarios, achieving a sensitivity of 94% and NPV of 98% for low-risk NMIBC, which could reduce invasive procedures by 17% [
119]. Later, a machine learning (ML)-based UriBLAD model (32 genes) further broke through performance bottlenecks, with an AUC of 0.93 in the validation cohort and a sensitivity exceeding 80% for both non-muscle-invasive and low-grade cancers, demonstrating the strong generalization ability of multi-omics data and algorithm integration [
120].
3.3.2. miRNA Detection Technology
As key molecules regulating gene expression, miRNAs are stably present in urine, and early studies screened differential molecules through microarray chips combined with qPCR verification. A study identified 15 differential miRNAs in BCa tissues, among which 4 (e.g., miR-141 and miR-205) could distinguish NMIBC from MIBC, with a combined detection accuracy of >75%, first proving the two diagnostic and prognostic value of miRNA profiles [
121]. In the same vein, another study found that urinary miR-1255b-5p had a sensitivity of 85% for invasive cancer, initiating the exploration of miRNA detection in non-invasive samples and establishing the classic “chip screening - qPCR verification” workflow [
122].
NGS has promoted miRNA detection into the genome-wide era. A study screened 3 core miRNAs (e.g., let-7c-5p and miR-30a-5p) through NGS, and the combined model achieved 0.70 of AUC in distinguishing BCa from controls while being able to identify tumor grade and invasiveness [
123]. To address the low miRNA content in early small tumors, a study combined telomerase extension reaction with graphene oxide (GO) signal amplification in 2017 to construct a combined detection method for proteins and miRNAs [
124]. The 5 selected miRNAs (e.g., let-7b-5p) significantly improved the discrimination efficiency between NMIBC and MIBC. Innovatively, a study innovatively integrated miRNAs (e.g., miR-34a-5p) with surface-enhanced Raman scattering (SERS) and constructed a combined model using ML, achieving an AUC of 0.92 in distinguishing BCa from controls and 0.95 in distinguishing molecular subtypes, providing a new strategy for point-of-care rapid diagnosis [
125].
In addition, miRNAs in urinary extracellular vesicles (EVs) are more stable due to vesicle protection. A study identified miR-21-5p from urinary EVs, which achieved a diagnostic sensitivity of 75.0% and specificity of 95.8%, with a sensitivity of 75.0% even in urine cytology-negative patients [
126]. Its expression significantly decreased after surgery, demonstrating the advantage of EVs as biomarker carriers. Another study established a 6-miRNA signature (including let-7c and miR-135a) through TaqMan microarrays, achieving an overall diagnostic AUC of 88.3% and an AUC of 92.9% for high-grade NMIBC, which was not interfered by clinical factors [
127]. A meta-analysis showed that the combined AUC of urinary miRNAs was 0.88, and multi-miRNA combinations were superior to single biomarkers, among which miR-143 performed prominently (AUC 0.88), verifying the universal advantage of combined models [
128].
3.3.3. lncRNA Detection Technology
Long non-coding RNAs (lncRNAs) have become emerging targets for non-invasive detection due to their long sequences and high stability. A study developed a gold nanoparticle (Au-NP) hybridization method for detecting lncRNA-UCA1 without PCR, which could complete detection within 30 minutes through color change, achieving a sensitivity of 92.1% and specificity of 93.3%, with a consistency of 98% with qRT-PCR [
129]. It is particularly suitable for early screening in schistosomiasis-endemic areas, addressing the demand for convenient detection in primary medical care.
Based on qRT-PCR quantitative analysis and microarray screening, a study screened a two-lncRNA combination (uc004cox.4 upregulation and GAS5 downregulation). The validation cohort achieved an AUC of 0.885, sensitivity of 84.5%, and specificity of 78.2%, significantly outperforming urine cytology [
130]. High expression of uc004cox.4 was associated with NMIBC recurrence. Another meta-analysis showed that the overall combined AUC of urinary lncRNAs was 0.86, and intracellular lncRNAs (e.g., UCA1) had better performance than free forms due to stability advantages. Multi-lncRNA combinations (AUC 0.87) were slightly superior to single indicators (0.85), confirming core biomarkers such as UCA1 and H19 [
131].
Based on bioinformatics analysis of genomic instability, a study constructed a 5-lncRNA signature (GllncSig), which was associated with overall survival and chemotherapy sensitivity of BCa. The low-risk group was more sensitive to drugs such as cisplatin (lower IC50) [
132]. Although this study was based on tissue samples, the screened molecules (e.g., CFAP58-DT and MIR100HG) provide potential targets for urinary biomarker combinations, promoting the extension of lncRNA detection from simple diagnosis to “diagnosis-prosmallgnosis-treatment” whole-process management.
Urine RNA detection technology has achieved significant progress in early BCa diagnosis, especially in the detection of tumors and NMIBC, through mRNA standardization and multi-gene modeling, miRNA high-throughput screening and signal amplification, and lncRNA nano-visualization and combination optimization. Each of the three types of RNA technologies has unique advantages: mRNA biomarkers (e.g., ETS2/uPA, IGF2/MAGEA3) have shown high specificity in clinical validation, suitable for rule-out detection; miRNAs (e.g., miR-21-5p, 6-miRNA signature) rely on their stability and multi-modal combination to improve the capture ability of trace tumors; lncRNAs (e.g., UCA1, uc004cox.4 + GAS5) have expanded convenient detection and prognostic prediction scenarios through nanotechnology and bioinformatics. However, current technologies still face common challenges: insufficient standardization, as differences in sample processing, internal reference selection, and detection platforms among different studies lead to result heterogeneity, requiring the establishment of unified technical specifications; early sensitivity bottlenecks, as the molecular enrichment efficiency for extremely early micro-tumors such as carcinoma in situ still needs improvement, requiring the analysis of tumor heterogeneity through single-cell sequencing and spatial transcriptomics; and the need for breakthroughs in multi-omics integration. Currently, the three types of RNA technologies develop independently. In the future, it is necessary to construct mRNA-miRNA-lncRNA combined models, combined with DNA methylation, protein biomarkers, and clinical parameters, to form a multi-dimensional diagnostic system.
4. Protein/Peptide Biomarker Detection Technology
Protein/peptide biomarkers have become core targets for non-invasive BCa detection due to their strong functional correlation with tumor initiation and invasion (e.g., NMP22, BTA, survivin). Core breakthroughs in detection technologies have focused on three directions: low-abundance protein capture, signal amplification, and multi-biomarker integration. Over the past 5 years, they have shown a convergent innovation trend of “nanotechnology + AI.”
4.1. Optimization and Clinical Application of Traditional Detection Technologies
Enzyme-Linked Immunosorbent Assay (ELISA) is a widely used non-invasive technique for urinary biomarker detection in BCa, facilitating early screening, recurrence monitoring, and prognostic assessment via quantifying biomarkers such as nuclear matrix protein 22 (NMP22) [
133], survivin [
134], and vascular endothelial growth factor (VEGF) [
135]. Clinically, its cost-effectiveness and compatibility with standard laboratory equipment enable broad adoption across healthcare settings, while its pg/mL to ng/mL detection limits accommodate low-abundance biomarkers in early-stage urine samples [
134]. Commercial ELISA kits like Alere NMP22® have demonstrated utility in BCa surveillance, achieving 58.3% sensitivity for low-grade BCa and 74.6% for high-grade (G3) BCa—outperforming traditional urine cytology [
28]. Serial ELISA measurements of urinary NMP22 can also identify recurrence 2–3 months earlier than cystoscopy, reducing invasive procedures [
136]. However, ELISA has inherent limitations: urinary matrix factors (e.g., high salt, proteases) cause poor reproducibility with coefficients of variation (CV) often exceeding 20% [
137,
138], while cross-reactivity with non-target proteins and biomarker isoform heterogeneity lead to false positives, especially in patients with urinary tract infections [
28]. Variations in the critical value of NMP22 within a certain range lead to decreased sensitivity and increased specificity, thereby enhancing the diagnostic accuracy for BC patients, however, this adjustment also reduces diagnostic efficacy and lowers the negative predictive value for BCa patients [
139]. In summary, ELISA serves as a valuable first-line tool for BCa urinary detection, but its clinical utility is constrained by analytical challenges, highlighting the need for assay optimization and standardization to enhance accuracy in precision BCa management.
4.2. Cutting-Edge Breakthroughs in Highly Sensitive Detection Technologies
4.2.1. Proteomic Technologies
A series of studies have continuously advanced its clinical utility, with progressive breakthroughs in diagnostic accuracy and standardization. Notably, a classic multicenter external validation study laid a foundational basis for the clinical application of multiplex urinary protein panels [
140]. This study validated the panel in a large cohort across multiple institutions, demonstrating robust diagnostic performance with an AUC of 0.87 for BCa detection, particularly excellent sensitivity (83%) for early-stage tumors, and good reproducibility across different clinical laboratories, which is crucial for promoting the standardized application of proteomic assays in clinical practice [
140]. Further clinical validation of multiplex protein assays was subsequently conducted. A 10-plex urinary protein assay (Oncuria assay) targeting 10 key analytes (including angiogenin, IL-8, and VEGF-A) was clinically validated, achieving high diagnostic accuracy with consistent performance across different Luminex instrumentation platforms, significantly outperforming traditional single-biomarker detection and providing a more reliable tool for non-invasive BCa detection and progression tracking [
141]. Most recently, core breakthroughs have focused on in-depth capture of low-abundance proteins and integration of multi-source data. A study constructed a urinary nanoparticle protein corona using Fe3O4@SiO2 nanoparticles and integrated serum and urine proteomic data by combining the transfer learning algorithm (ProteoTransNet) [
142]. The constructed diagnostic model achieved an AUC of 0.996 in 1056 multi-center samples, with AUC values of 0.902, 0.926, and 0.941 for staging classification of Ta, T1, and MIBC, respectively. Moreover, stable performance with AUC ≥ 0.98 was maintained in the subgroup of patients with comorbid underlying diseases, representing the latest advancement in proteomic-based BCa detection.
4.2.2. Surface-Enhanced Raman Scattering (SERS) Sensors
SERS is a sensitive, label-free, and molecularly specific noninvasive detection technique, making it a desirable method for BCa diagnosis by enabling single-molecule-level detection of biomolecules in urine.
A serum-based SERS platform has been developed to address the clinical need for non-invasive BCa staging, focusing on distinguishing NMIBC from MIBC. Using gold-silver core-shell nanoparticles as SERS substrates and partial least squares-linear discriminant analysis (PLS-LDA) for spectral data mining, the system achieved 93.3% overall diagnostic accuracy in 90 participants (30 healthy volunteers, 28 NMIBC patients, 32 MIBC patients) [
143]. For NMIBC vs. MIBC discrimination, the AUC reached 0.983 with 93.2% accuracy, 90.6% sensitivity, and 96.3% specificity. It identified staging-specific spectral markers: MIBC patients exhibited higher intensities at 494 cm⁻¹ (L-arginine), 589 cm⁻¹ (amide-VI), 639 cm⁻¹ (L-tyrosine), and 1654 cm⁻¹ (amide-I/α-Helix) compared to NMIBC, reflecting amino acid metabolism disorder and protein dysregulation in invasive tumors. The platform maintained >88% accuracy in patients with comorbidities, outperforming traditional serum biomarkers such as Carcinoembryonic Antigen (CEA, AUC 0.71).
Preclinical validation of SERS for early-stage BCa detection has been conducted using a chemically induced rat model, providing critical evidence for translational potential. Combining gold nanoparticle SERS substrates with principal component analysis (PCA) and PLS-LDA, the platform analyzed urine samples to achieve 99.6% accuracy for diagnosing early-stage and polyp-form BCa, with an AUC >0.996 [
144]. It captured subtle nucleoside changes (732 cm⁻¹, C-N stretching of adenosine) before visible tumor formation, detecting BCa earlier than cystoscopy, and its spectral signature correlated strongly with human early-stage BCa (r=0.87)—supporting its relevance for clinical early screening.
To address the underdiagnosis of low-grade BCa, a landmark advancement introduced “SERSomes”—spectral sets of multiple individual spectra per sample—to comprehensively profile urine metabolites. Using citrate-reduced Ag NPs as SERS substrates, the platform mixed pretreated urine with NPs and acquired spectra for analysis. In a clinical cohort of 116 participants (31 low-grade BCa T1 patients, 38 low-grade BCa T2 patients, 47 healthy controls), the SERSomes combined with random forest (RF) ML achieved 89.47% accuracy for low-grade BCa diagnosis (AUC 0.88) and 90% accuracy for T1/T2 stratification (AUC 0.83) [
145]. Key contributing spectral bands included 749–767 cm⁻¹ (tryptophan/ethanolamine) for diagnosis and 1440–1461 cm⁻¹ (deoxyribose/lipid CH₂ deformation) for stratification. The entire workflow took <30 minute, enabling point-of-care application (POC), and avoided the need for deproteinization pretreatment.
Complementing these targeted advancements, a urine-based SERS sensor leveraging MXene@gold nanoparticle composite substrates further expanded clinical utility by focusing on protein biomarker detection. Prepared via hydrothermal synthesis, the substrate was loaded with 15 nm gold nanoparticles and modified with anti-BTA monoclonal antibodies, achieving a detection limit of 0.1 pg/mL for BTA—1000-fold lower than traditional ELISA [
146]. In 210 clinical samples, it distinguished BCa patients from healthy controls with 91.7% sensitivity and 93.3% specificity, and differentiated BCa from urinary tract infections with 89.2% sensitivity and 90.0% specificity—effectively mitigating interference from inflammatory factors in the urine matrix.
Across these advancements, SERS sensors have evolved from staging and early detection to targeted solutions for low-grade BCa screening, with inherent advantages of high sensitivity, label-free detection, molecular specificity, and minimal invasiveness. These features make them promising for POC and large-scale screening applications. Future directions will focus on standardizing substrate fabrication and data analysis protocols, integrating SERS with multi-omics techniques, and conducting large-scale prospective trials to accelerate clinical adoption—overcoming current limitations such as matrix interference and inter-study variability.
The phase-separation and enzyme-responsive POC testing technology, which requires no sample pretreatment, establishes a new paradigm for the early diagnosis of BCa. The recently developed BLOOM system utilizes the BCa-specific biomarker hyaluronidase to degrade a biphasic gel film, releasing encapsulated low-density organogel signal particles. Driven by buoyancy, these particles autonomically migrate into the oil phase, where they generate solvatochromic fluorescence [
147]. This process achieves spatial separation and reading of the signal, thereby completely circumventing interference from complex matrices such as hematuria. The system can directly analyze raw urine samples. Coupled with a smartphone-based fluorescence reader at a cost of approximately US
$2.5 per test, it is suitable for home-based self-testing. In a double-blind validation study involving 105 samples, it achieved AUC of 0.93 for detecting early-stage BCa, with both sensitivity and specificity around 88%. By converting enzymatic activity into a physical phase-transition signal, this technology enables robust, low-cost POC early screening, representing a significant frontier breakthrough alongside proteomics and SERS.
The landscape of early non-invasive diagnostic technologies for BCa has evolved from the optimization of traditional ELISA to breakthroughs in high-sensitivity detection. Proteomics has enhanced assay stability and interference resistance through multi-marker integration, while surface-enhanced Raman scattering (SERS) technology has achieved detection limits at the pg/mL level, addressing the challenge of identifying low-abundance proteins. Notably, novel POC systems based on phase separation and enzyme response, such as the BLOOM system, enable pretreatment-free, high-fidelity detection of raw urine by converting hyaluronidase activity into a spatially segregated physical signal, unaffected by hematuria. This provides a novel pathway for low-cost home-based screening. Collectively, these advances have elevated the detection sensitivity for low-grade tumors from approximately 40% with conventional methods to over 80%, laying a solid foundation for the early diagnosis and precise monitoring of BCa.
5. Metabolomic Diagnostic Technologies
Metabolomic-related technologies detect changes in small-molecule metabolites to identify potential diagnostic biomarkers [
148], providing new insights for early disease screening and diagnosis. In recent years, various metabolomic detection technologies, such as high-performance liquid chromatography-mass spectrometry (HPLC-MS) and gas chromatography-mass spectrometry (GC-MS), have continuously advanced and been applied in BCa diagnostic research.
5.1. High-Performance Liquid Chromatography-Mass Spectrometry (HPLC-MS) Technology
In early studies, researchers employed a combination of a high-performance liquid chromatography system and a hybrid triple quadrupole time-of-flight mass spectrometer for metabolomic analysis of urine samples to distinguish between healthy individuals and BCa patients. The study collected urine samples from 48 healthy individuals and 41 BCa patients, using two statistical methods: PCA and orthogonal partial least squares discriminant analysis (OPLS-DA). Results showed that in positive ionization mass spectrometry mode, OPLS-DA accurately predicted all 48 healthy samples and 41 BCa urine samples [
149]. As an unsupervised analysis method, PCA also correctly predicted 46 healthy samples and 40 BCa samples, initially demonstrating the potential of HPLC-MS metabolomic technology for non-invasive early detection of BCa.
With technological advancements, ultra-performance liquid chromatography (UPLC) combined with mass spectrometry has been increasingly applied in urinary metabolomic research on BCa. A study conducted untargeted metabolomic analysis of urine from BCa patients and healthy controls using UPLC-MS to screen for differential metabolites. The study included 57 BCa patients and 38 cancer-free controls, identifying 27 statistically significant differential metabolites (VIP > 1.5 and FDR < 0.05) via PCA and OPLS-DA [
150]. This panel of metabolites achieved an AUC of 0.951, sensitivity of 91.2%, specificity of 86.8%, and accuracy of 89.5% in distinguishing BCa patients from cancer-free controls. For differentiating NMIBC from cancer-free controls, the AUC was 0.943, sensitivity 81.6%, specificity 94.6%, and accuracy 88%. Additionally, Pearson correlation coefficient analysis revealed that age, gender, and BMI had weak correlations with these 27 differential metabolites, indicating minimal impact of these factors on the test results. Another study utilized UPLC-MS to analyzed urine from 198 BCa patients and 98 healthy volunteers [
151]. Results confirmed that p-cresol glucuronide could serve as a diagnostic biomarker for BCa (AUC = 0.79) and had staging value for NMIBC (AUC = 0.803).
Furthermore, a study used UPLC-MS for metabolomic analysis of urine from 29 BCa patients and 15 healthy controls, identifying 208 metabolites in total [
152], the workflow is shown in
Figure 4. OPLS-DA analysis showed significant discriminative potential between the metabolic profiles of BCa patients and healthy controls. And permutational multivariate analysis of variance validated that the probability of the model occurring randomly was < 0.001. Nineteen differential metabolites were screened, mainly involving pathways such as phenylacetate metabolism, propanoate metabolism, and fatty acid metabolism. Eleven potential biomarkers were further selected through random forest (RF), support vector machine (SVM), and Boruta analysis, and a logistic regression model was constructed. The ROC curve of this model showed an AUC of 0.983, sensitivity of 95.3%, and specificity of 100%, demonstrating excellent diagnostic performance for BCa.
This technology offers advantages such as high separation efficiency, strong detection sensitivity, and the ability to identify multiple metabolites simultaneously. However, it also has limitations including high instrument costs, complex sample pretreatment, and long detection cycles, which restrict its widespread application in primary medical institutions.
5.2. Gas Chromatography-Mass Spectrometry (GC-MS) Technology
Gas chromatography-time-of-flight mass spectrometry (GC×GC TOF MS) has been used to detect urinary volatile organic compounds (VOCs) for identifying BCa diagnostic biomarkers due to its high-resolution advantage. A study employed headspace solid-phase microextraction (HS-SPME) combined with comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GC×GC TOF MS) to analyze 218 VOCs in urine from 75 BCa patients (48 NMIBC, 27 MIBC) and 50 healthy controls. Orthogonal partial least squares discriminant analysis (OPLS-DA) screened 12 significantly differential VOCs, among which 6 metabolites (2-methoxyphenol, phenylacetaldehyde, nonanal, decanal, isovaleric acid, and phenylpropionic acid) were only detected in the urine of BCa patients. A random forest diagnostic model built on these 12 VOCs achieved a sensitivity of 89% and specificity of 94% in the training set (80% of samples) and a sensitivity of 85% and specificity of 91% in the validation set (20% of samples). Additionally, the model’s sensitivity for MIBC (92.6%) was slightly higher than that for NMIBC (82.3%) [
153].
A subsequent multicenter cohort study validated urinary VOCs in 230 participants (110 BCa patients, 120 healthy controls), confirming 8 previously identified differential VOCs (including 2-methoxyphenol and nonanal) with consistent expression patterns. A logistic regression model based on these VOCs achieved an AUC of 0.77, sensitivity of 71%, and specificity of 72% in external validation. Meanwhile, it was found that the VOC profile remained stable within 48 hours after urine collection, providing a basis for standardized sample processing [
153]. Another detection protocol constructed with 6 VOCs of statistically significant abundance achieved an AUC of 0.80, sensitivity of 71%, and specificity of 80%, offering a new non-invasive tool for BCa diagnosis and surveillance [
153]. This technology excels in detecting volatile metabolites with high separation efficiency. However, it has drawbacks such as difficulty in detecting non-volatile metabolites, cumbersome sample pretreatment, long detection times, and poor result reproducibility easily affected by experimental conditions.
5.3. Gas Chromatography-Ion Mobility Spectrometry (GC-IMS) Technology
GC-IMS has become an important technology for rapid urinary screening of BCa due to its combined advantages of chromatographic separation capability and high ion mobility spectrometry sensitivity, with a typical detection time of < 30 minutes. A multicenter study enrolled 312 high-risk individuals (smoking history > 10 years, occupational exposure to aromatic amines, or chronic cystitis history) and used a portable GC-IMS device that requires no sample pretreatment, with a detection time of only 18 minutes. By collecting fingerprint spectra of urinary VOCs and combining PCA and linear discriminant analysis models, 47 BCa patients (including 32 early Ta/T1 stage cases) were screened from the high-risk population. The diagnostic model achieved an AUC of 0.86 (95% CI: 0.81–0.91), sensitivity of 80.9%, and specificity of 83.2%. Compared with traditional B-ultrasound screening, this device increased the detection rate of early BCa by 42% and reduced the false-positive rate from 15.3% (B-ultrasound) to 8.7%, making it particularly suitable for large-scale screening of high-risk populations in community hospitals and primary medical institutions [
154].
Furthermore, a prospective trial was conducted in 189 hematuria patients (89 BCa cases, 100 non-cancer cases) to evaluate the device’s performance, identifying 5 BCa-specific VOC peaks. A diagnostic model built on these peaks achieved an AUC of 0.81 (95% CI: 0.75–0.87), sensitivity of 76.4%, and specificity of 78.0%, demonstrating stable performance in distinguishing BCa from benign urinary system diseases such as urinary tract infections and kidney stones [
154]. This technology boasts advantages including fast detection speed, portable equipment, and simple operation. However, it has limitations such as relatively low resolution, insufficient sensitivity for detecting certain low-concentration metabolites, and the need for improved biomarker specificity.
Metabolomic diagnostic technologies for early BCa based on urine have made significant progress. From early HPLC-MS and GC-MS technologies to subsequent GC-IMS technology, each has unique features and advantages. By screening differential metabolites and constructing diagnostic models, these technologies have demonstrated good potential in early BCa diagnosis. Some technologies have been validated in multicenter studies, significantly improving the detection rate of early BCa. Nevertheless, existing technologies still face several challenges, such as high instrument costs, complex sample pretreatment, insufficient biomarker specificity, and unstable detection results. Future efforts should focus on further optimizing technical parameters, reducing costs, improving detection specificity and sensitivity, conducting larger-scale clinical validation, and promoting the development of these technologies toward standardization and portability to realize their widespread application in clinical practice for early BCa diagnosis.
Metabolomic technologies (including HPLC-MS, GC-MS, and GC-IMS) enable non-invasive early detection of BCa by profiling characteristic alterations in urinary small-molecule metabolites, achieving high diagnostic specificity (AUC 0.77–0.98). Among these, GC-IMS offers rapid (<30 minutes) and portable analysis, making it particularly suitable for large-scale screening in high-risk populations within primary care settings. Furthermore, Raman spectroscopy [
155] allows cellular-level discrimination by directly analyzing the molecular vibrational fingerprints of exfoliated urinary cells [
156]. Together, these approaches form a multi-dimensional detection framework spanning metabolites to cells, significantly enhancing the ability to identify early-stage lesions.
Conclusion and Outlook
Noninvasive urine-based diagnostic technologies for early BCa have evolved from traditional cytology, which relies on morphological observation, to the era of molecular diagnosis based on multi-omics biomarkers. Artificial intelligence has significantly enhanced the automation and accuracy of urine cytology image analysis through deep learning. Genomic technologies (PCR, NGS) enable high-throughput and highly sensitive detection of DNA mutations, copy number variations, and methylation patterns. Transcriptomics (mRNA, miRNA, lncRNA) and proteomics/metabolomics further reveal the characteristic molecular profiles of tumors from a functional perspective. Collectively, these advances have elevated the detection sensitivity for low-grade, early-stage tumors from less than 50% with conventional methods to over 80%, with AUC values exceeding 0.95 for some cutting-edge technologies, laying a robust foundation for achieving molecular early-warning of BCa.
However, key challenges remain in this field: First, insufficient technical standardization and clinical validation. Variability across different platforms and procedures leads to significant result heterogeneity, necessitating large-scale, prospective multicenter studies to establish unified protocols. Second, persistent limitations in detecting early, small-volume tumors. The abundance and specificity of existing biomarkers for lesions with low tumor burden or carcinoma in situ still requires improvement. Third, the conflict between cost and accessibility. The high expense of high-sensitivity technologies (e.g., WGS, deep proteomics) hinders their application in primary care and population-wide screening.
Future development is expected to focus on the following directions: Multi-omics integration and AI-driven analysis: Combining genomic, epigenomic, proteomic, and metabolomic data to construct multidimensional diagnostic models, and leveraging AI algorithms to uncover deep-level correlative features for more precise risk stratification and early detection. Innovation in point-of-care (POC) and home-based testing technologies: Continuing to develop portable devices based on phase-separation sensing (e.g., the BLOOM system), microfluidic chips, and smartphone-compatible readers to advance testing toward real-time, convenient, and low-cost solutions, empowering home health management. Liquid biopsy and dynamic monitoring: Deepening the application of urine-based liquid biopsies (e.g., cfDNA, exosomes) to monitor molecular residual disease and clonal evolution during treatment, providing dynamic molecular evidence for treatment response assessment and recurrence warning. Standardization system establishment and large-scale validation: Developing standardized operating procedures encompassing the entire workflow from sample collection and preservation to data analysis, and validating their clinical utility and health-economic value through real-world studies to accelerate the translation of technologies into clinical guidelines.
In summary, through interdisciplinary convergence and technological integration, noninvasive urine-based diagnostics are progressively becoming an indispensable component of the early screening, precise diagnosis, and comprehensive management framework for BCa. This holds the potential to fundamentally improve patient prognosis and alleviate the societal healthcare burden.
Author Contributions
Z.H.: Conceptualization, Data curation, Investigation, Writing – original draft, Visualization. S.Y.: Conceptualization, Writing –review & editing, Funding acquisition, Supervision. Y.G.: Writing – review & editing. J.Y.: Conceptualization, Writing – review & editing, Supervision. L.Y.: Resources, Writing – review & editing, Funding acquisition, Supervision. L.Z.: Resources, Writing – review & editing, Funding acquisition, Supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Fundamental Research Funds for the Central Universities (JKF-2025086684620). This research was supported by the Noncommunicable Chronic Diseases-National Science and Technology Major Project (2024ZD0525700).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
During the preparation of this manuscript, the authors used ChatGPT (OpenAI
https://openai.com/) and Gemini (Google,
https://gemini.google.com/app) for language editing and improving the clarity of the text. The authors reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI |
Artificial Intelligence |
| ML |
Machine Learning |
| DL |
Deep Learning |
| ANN |
Artificial Neural Network |
| CNN |
Convolutional Neural Network |
| BCa |
Bladder Cancer |
| NMIBC |
Non-Muscle Invasive Bladder Cancer |
| MIBC |
Muscle Invasive Bladder Cancer |
| UTUC |
Upper Tract Urothelial Carcinoma |
| PCR |
Polymerase Chain Reaction |
| qPCR |
Quantitative Polymerase Chain Reaction |
| NGS |
Next-Generation Sequencing |
| WGS |
Whole Genome Sequencing |
| sWGS |
Shallow Whole Genome Sequencing |
| LC-WGS |
Low-Coverage Whole Genome Sequencing |
| CNV |
Copy Number Variation |
| MSP |
Methylation-Specific PCR |
| ELISA |
Enzyme-Linked Immunosorbent Assigmnt |
| SERS |
Surface-Enhanced Raman Scattering |
| HPLC-MS |
High-Performance Liquid Chromatography-Mass Spectrometry |
| GC-MS |
Gas Chromatography-Mass Spectrometry |
| GC-IMS |
Gas Chromatography-Ion Mobility Spectrometry |
| DNA |
Deoxyribonucleic Acid |
| RNA |
Ribonucleic Acid |
| mRNA |
Messenger RNA |
| miRNA |
MicroRNA |
| lncRNA |
Long Non-Coding RNA |
| cfDNA |
Cell-Free DNA |
| AUC |
Area Under the Curve |
| NPV |
Negative Predictive Value |
| PPV |
Positive Predictive Value |
| ROC |
Receiver Operating Characteristic |
| EV |
Extracellular Vesicle |
| POC |
Point-of-Care |
References
- Filho, A.M.; Briganti, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol 2025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rumgay, H.; Li, M.; Yu, H.; Pan, H.; Ni, J. The global landscape of bladder cancer incidence and mortality in 2020 and projections to 2040. J. Glob. Health 2023, 13, 04109. [Google Scholar] [CrossRef] [PubMed]
- Farling, K.B. Bladder cancer: Risk factors, diagnosis, and management. Nurse Pract. 2017, 42, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Soloway, M.S. Bladder cancer: Lack of progress in bladder cancer--what are the obstacles? Nature reviews. Urology 2013, 10, 5–6. [Google Scholar] [CrossRef]
- Del Bene, G.; Calabrò, F.; Giannarelli, D.; Plimack, E.R.; Harshman, L.C.; Yu, E.Y.; Crabb, S.J.; Pal, S.K.; Alva, A.S.; Powles, T.; et al. Neoadjuvant vs. Adjuvant Chemotherapy in Muscle Invasive Bladder Cancer (MIBC): Analysis From the RISC Database. Front. Oncol. 2018, 8, 463. [Google Scholar] [CrossRef]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nature reviews. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef]
- Shariat, S.F.; Karam, J.A.; Lotan, Y.; Karakiewizc, P.I. Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev. Urol. 2008, 10, 120–135. [Google Scholar]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Linares Espinós, E.; Lorch, A.; Neuzillet, Y.; et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2021, 79, 82–104. [Google Scholar] [CrossRef]
- Winquist, E.; Kirchner, T.S.; Segal, R.; Chin, J.; Lukka, H. Neoadjuvant chemotherapy for transitional cell carcinoma of the bladder: A systematic review and meta-analysis. J. Urol. 2004, 171, 561–569. [Google Scholar] [CrossRef]
- Lin, C.C.; Hsu, C.H.; Huang, C.Y.; Cheng, A.L.; Vogelzang, N.J.; Pu, Y.S. Phase II trial of weekly paclitaxel, cisplatin plus infusional high dose 5-fluorouracil and leucovorin for metastatic urothelial carcinoma. J. Urol. 2007, 177, 84–89; discussion 89. [Google Scholar] [CrossRef]
- Flaig, T.W.; Spiess, P.E.; Abern, M.; Agarwal, N.; Bangs, R.; Buyyounouski, M.K.; Chan, K.; Chang, S.S.; Chang, P.; Friedlander, T.; et al. NCCN Guidelines® Insights: Bladder Cancer, Version 3.2024. J. Natl. Compr. Cancer Netw. JNCCN 2024, 22, 216–225. [Google Scholar] [CrossRef]
- Hahn, N.M.; Stadler, W.M.; Zon, R.T.; Waterhouse, D.; Picus, J.; Nattam, S.; Johnson, C.S.; Perkins, S.M.; Waddell, M.J.; Sweeney, C.J. Phase II trial of cisplatin, gemcitabine, and bevacizumab as first-line therapy for metastatic urothelial carcinoma: Hoosier Oncology Group GU 04-75. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1525–1530. [Google Scholar] [CrossRef]
- Jocham, D.; Stepp, H.; Waidelich, R. Photodynamic diagnosis in urology: State-of-the-art. Eur. Urol. 2008, 53, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; He, A.; Lv, T.; Xu, C.; Lin, L.; Lin, J. Overexpression of P4HB is correlated with poor prognosis in human clear cell renal cell carcinoma. Cancer Biomark. Sect. A Dis. Markers 2019, 26, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Siracusano, S.; Rizzetto, R.; Porcaro, A.B. Bladder cancer genomics. Urologia 2020, 87, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zhang, J.; Li, L.; Yang, Y.; Yuan, Y. Diagnostic accuracy of urinary survivin mRNA expression detected by RT-PCR compared with urine cytology in the detection of bladder cancer: A meta-analysis of diagnostic test accuracy in head-to-head studies. Oncol. Lett. 2020, 19, 1165–1174. [Google Scholar] [CrossRef]
- Bamias, A.; Moulopoulos, L.A.; Koutras, A.; Aravantinos, G.; Fountzilas, G.; Pectasides, D.; Kastritis, E.; Gika, D.; Skarlos, D.; Linardou, H.; et al. The combination of gemcitabine and carboplatin as first-line treatment in patients with advanced urothelial carcinoma. A Phase II study of the Hellenic Cooperative Oncology Group. Cancer 2006, 106, 297–303. [Google Scholar] [CrossRef]
- Feiertag, N.; Barry, E.; Abramson, M.; Park, J.Y.; Kovac, E.; Aboumohamed, A.; Watts, K.; Sankin, A. Urine Cytology Rarely Escalates Clinical Management in the Surveillance of Non-muscle-Invasive Bladder Cancer. Clin. Genitourin. Cancer 2023, 21, 258–264. [Google Scholar] [CrossRef]
- Tan, W.S.; Sarpong, R.; Khetrapal, P.; Rodney, S.; Mostafid, H.; Cresswell, J.; Watson, D.; Rane, A.; Hicks, J.; Hellawell, G.; et al. Does urinary cytology have a role in haematuria investigations? BJU Int. 2019, 123, 74–81. [Google Scholar] [CrossRef]
- Pan, T.; Lehman, E.; Raman, J.D. Performance characteristics of urinary cytology in patients presenting with gross and microscopic hematuria. Am. J. Clin. Exp. Urol. 2021, 9, 384–389. [Google Scholar]
- Allison, D.B.; Zhang, M.L.; Vohra, P.; VandenBussche, C.J. The Diagnostic Dilemma of Urothelial Tissue Fragments in Urinary Tract Cytology Specimens. Diagnostics 2022, 12, 931. [Google Scholar] [CrossRef]
- Velez Torres, J.M.; Gonzalez, M.L.; Duarte, E.M.; Zein-Sabatto, B.; Aron, M.; Gupta, N.S.; Kerr, D.A.; Netto, G.J.; Jorda, M.; Kryvenko, O.N. Urine Cytology Findings in Cases of Pseudocarcinomatous Urothelial Hyperplasia of the Bladder Often Represent a Diagnostic Challenge. Arch. Pathol. Lab. Med. 2023, 147, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Owens, C.L.; Lithgow, M.Y.; Jiang, Z.; Fischer, A.H. Causes of false-negative for high-grade urothelial carcinoma in urine cytology. Diagn. Cytopathol. 2016, 44, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Flores Monar, G.V.; Reynolds, T.; Gordon, M.; Moon, D.; Moon, C. Molecular Markers for Bladder Cancer Screening: An Insight into Bladder Cancer and FDA-Approved Biomarkers. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Li, H.X.; Wang, M.R.; Zhao, H.; Cao, J.; Li, C.L.; Pan, Q.J. Comparison of fluorescence in situ hybridization, NMP22 bladderchek, and urinary liquid-based cytology in the detection of bladder urothelial carcinoma. Diagn. Cytopathol. 2013, 41, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Zippe, C.; Pandrangi, L.; Agarwal, A. NMP22 is a sensitive, cost-effective test in patients at risk for bladder cancer. J. Urol. 1999, 161, 62–65. [Google Scholar] [CrossRef]
- Maas, M.; Todenhöfer, T.; Black, P.C. Urine biomarkers in bladder cancer - current status and future perspectives. Nature reviews. Urology 2023, 20, 597–614. [Google Scholar] [CrossRef]
- Chou, R.; Gore, J.L.; Buckley, D.; Fu, R.; Gustafson, K.; Griffin, J.C.; Grusing, S.; Selph, S. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Ann. Intern. Med. 2015, 163, 922–931. [Google Scholar] [CrossRef]
- Sarosdy, M.F. The use of the BTA Test in the detection of persistent or recurrent transitional-cell cancer of the bladder. World J. Urol. 1997, 15, 103–106. [Google Scholar] [CrossRef]
- Soorojebally, Y.; Neuzillet, Y.; Roumiguié, M.; Lamy, P.J.; Allory, Y.; Descotes, F.; Ferlicot, S.; Kassab-Chahmi, D.; Oudard, S.; Rébillard, X.; et al. Urinary biomarkers for bladder cancer diagnosis and NMIBC follow-up: A systematic review. World J. Urol. 2023, 41, 345–359. [Google Scholar] [CrossRef]
- Laukhtina, E.; Shim, S.R.; Mori, K.; D’Andrea, D.; Soria, F.; Rajwa, P.; Mostafaei, H.; Compérat, E.; Cimadamore, A.; Moschini, M.; et al. Diagnostic Accuracy of Novel Urinary Biomarker Tests in Non-muscle-invasive Bladder Cancer: A Systematic Review and Network Meta-analysis. Eur. Urol. Oncol. 2021, 4, 927–942. [Google Scholar] [CrossRef]
- Bhat, A.; Ritch, C.R. Urinary biomarkers in bladder cancer: Where do we stand? Curr. Opin. Urol. 2019, 29, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; He, L.; Liu, X.; Xiao, Y.; Hu, J.; Cai, S.; Cai, H.; Yu, B. Artificial intelligence application in the diagnosis and treatment of bladder cancer: Advance, challenges, and opportunities. Front. Oncol. 2024, 14, 1487676. [Google Scholar] [CrossRef] [PubMed]
- Katare, P.; Gorthi, S.S. Recent technical advances in whole slide imaging instrumentation. J. Microsc. 2021, 284, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Tsuji, K.; Masuda, K.; Ueno, K.; Henmi, K.; Nakagawa, S.; Fujita, R.; Suzuki, K.; Inoue, Y.; Teramukai, S.; et al. Urine cell image recognition using a deep-learning model for an automated slide evaluation system. BJU Int. 2022, 130, 235–243. [Google Scholar] [CrossRef]
- Sanghvi, A.B.; Allen, E.Z.; Callenberg, K.M.; Pantanowitz, L. Performance of an artificial intelligence algorithm for reporting urine cytopathology. Cancer Cytopathol. 2019, 127, 658–666. [Google Scholar] [CrossRef]
- Muralidaran, C.; Dey, P.; Nijhawan, R.; Kakkar, N. Artificial neural network in diagnosis of urothelial cell carcinoma in urine cytology. Diagn. Cytopathol. 2015, 43, 443–449. [Google Scholar] [CrossRef]
- Nojima, S.; Terayama, K.; Shimoura, S.; Hijiki, S.; Nonomura, N.; Morii, E.; Okuno, Y.; Fujita, K. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021, 129, 984–995. [Google Scholar] [CrossRef]
- Zhang, Z.; Fu, X.; Liu, J.; Huang, Z.; Liu, N.; Fang, F.; Rao, J. Developing a Machine Learning Algorithm for Identifying Abnormal Urothelial Cells: A Feasibility Study. Acta Cytol. 2021, 65, 335–341. [Google Scholar] [CrossRef]
- Yilmaz, E.C.; Belue, M.J.; Turkbey, B.; Reinhold, C.; Choyke, P.L. A Brief Review of Artificial Intelligence in Genitourinary Oncological Imaging. Can. Assoc. Radiol. J. = J. L’association Can. Des Radiol. 2023, 74, 534–547. [Google Scholar] [CrossRef]
- Chen, C.C.; Yen, T.H.; Li, J.R.; Chen, C.J.; Yang, C.S.; Lai, J.Y.; Lin, S.J.; Yeh, C.H.; Hsu, S.W.; Lin, M.Y.; et al. Artificial intelligence algorithms enhance urine cytology reporting confidence in postoperative follow-up for upper urinary tract urothelial carcinoma. Int. Urol. Nephrol 2024. [Google Scholar] [CrossRef]
- Liu, T.J.; Yang, W.C.; Huang, S.M.; Yang, W.L.; Wu, H.J.; Ho, H.W.; Hsu, S.W.; Yeh, C.H.; Lin, M.Y.; Hwang, Y.T.; et al. Evaluating artificial intelligence-enhanced digital urine cytology for bladder cancer diagnosis. Cancer Cytopathol. 2024, 132, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Falagario, U.G.; Barone, B.; Maggi, M.; Crocetto, F.; Busetto, G.M.; Giudice, F.D.; Terracciano, D.; Lucarelli, G.; Lasorsa, F.; et al. Artificial Intelligence in the Advanced Diagnosis of Bladder Cancer-Comprehensive Literature Review and Future Advancement. Diagnostics 2023, 13, 2308. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Du, S.; Zhang, Z.; Lv, F.; Hong, Q.; Lai, M. Artificial Neural Network Based on Memristive Circuit for High-Speed Equalization. IEEE Trans. Circuits Syst. I: Regul. Pap. 2024, 71, 1745–1756. [Google Scholar] [CrossRef]
- Chavlis, S.; Poirazi, P. Dendrites endow artificial neural networks with accurate, robust and parameter-efficient learning. Nat. Commun. 2025, 16, 943. [Google Scholar] [CrossRef]
- Roberts, D.; Yaida, S.; Hanin, B. The Principles of Deep Learning Theory: An Effective Theory Approach to Understanding Neural Networks; 2022. [Google Scholar]
- Indolia, S.; Goswami, A.K.; Mishra, S.P.; Asopa, P. Conceptual Understanding of Convolutional Neural Network- A Deep Learning Approach. Procedia Comput. Sci. 2018, 132, 679–688. [Google Scholar] [CrossRef]
- Wu, S.; Shen, R.; Hong, G.; Luo, Y.; Wan, H.; Feng, J.; Chen, Z.; Jiang, F.; Wang, Y.; Liao, C.; et al. Development and validation of an artificial intelligence-based model for detecting urothelial carcinoma using urine cytology images: A multicentre, diagnostic study with prospective validation. EClinicalMedicine 2024, 71, 102566. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, S.; Shen, Q.; Chang, L.; Fang, S.; Fan, Y.; Peng, H.; Yu, W. A Deep Learning System to Predict the Histopathological Results From Urine Cytopathological Images. Front. Oncol. 2022, 12, 901586. [Google Scholar] [CrossRef]
- Tsuji, K.; Kaneko, M.; Harada, Y.; Fujihara, A.; Ueno, K.; Nakanishi, M.; Konishi, E.; Takamatsu, T.; Horiguchi, G.; Teramukai, S.; et al. A Fully Automated Artificial Intelligence System to Assist Pathologists’ Diagnosis to Predict Histologically High-grade Urothelial Carcinoma from Digitized Urine Cytology Slides Using Deep Learning. Eur. Urol. Oncol. 2024, 7, 258–265. [Google Scholar] [CrossRef]
- Vorontsov, E.; Bozkurt, A.; Casson, A.; Shaikovski, G.; Zelechowski, M.; Severson, K.; Zimmermann, E.; Hall, J.; Tenenholtz, N.; Fusi, N.; et al. A foundation model for clinical-grade computational pathology and rare cancers detection. Nat. Med. 2024, 30, 2924–2935. [Google Scholar] [CrossRef]
- Lebret, T.; Pignot, G.; Colombel, M.; Guy, L.; Rebillard, X.; Savareux, L.; Roumigue, M.; Nivet, S.; Coutade Saidi, M.; Piaton, E.; et al. Artificial intelligence to improve cytology performances in bladder carcinoma detection: Results of the VisioCyt test. BJU Int. 2022, 129, 356–363. [Google Scholar] [CrossRef]
- Farahani, S.J.; Li, J.; Minder, B.; Vielh, P.; Glisic, M.; Muka, T. Impact of implementing the first edition of the Paris system for reporting: A systematic review and meta-analysis. Cytopathol. Off. J. Br. Soc. Clin. Cytol. 2024, 35, 616–633. [Google Scholar] [CrossRef]
- Ou, Y.C.; Tsao, T.Y.; Chang, M.C.; Lin, Y.S.; Yang, W.L.; Hang, J.F.; Li, C.B.; Lee, C.M.; Yeh, C.H.; Liu, T.J. Evaluation of an artificial intelligence algorithm for assisting the Paris System in reporting urinary cytology: A pilot study. Cancer Cytopathol. 2022, 130, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Liao, J.; Li, M.S.; Khoo, B.L. Urine biopsy technologies: Cancer and beyond. Theranostics 2020, 10, 7872–7888. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.H.T.; Jiang, P.; Tam, J.C.W.; Sun, X.; Lee, W.S.; Yu, S.C.Y.; Teoh, J.Y.C.; Chiu, P.K.F.; Ng, C.F.; Chow, K.M.; et al. Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin. Biochem. 2017, 50, 496–501. [Google Scholar] [CrossRef]
- Xu, Y.; Kim, Y.H.; Jeong, P.; Piao, X.M.; Byun, Y.J.; Kang, H.W.; Kim, W.T.; Lee, J.Y.; Kim, I.Y.; Moon, S.K.; et al. Diagnostic value of combined IQGAP3/BMP4 and IQGAP3/FAM107A expression ratios in urinary cell-free DNA for discriminating bladder cancer from hematuria. Urol. Oncol. 2019, 37, 86–96. [Google Scholar] [CrossRef]
- Xu, Y.; Kim, Y.H.; Jeong, P.; Piao, X.M.; Byun, Y.J.; Seo, S.P.; Kang, H.W.; Kim, W.T.; Lee, J.Y.; Ryu, D.H.; et al. Urinary Cell-Free DNA IQGAP3/BMP4 Ratio as a Prognostic Marker for Non-Muscle-Invasive Bladder Cancer. Clin. Genitourin. Cancer 2019, 17, e704–e711. [Google Scholar] [CrossRef]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Stasik, S.; Salomo, K.; Heberling, U.; Froehner, M.; Sommer, U.; Baretton, G.B.; Ehninger, G.; Wirth, M.P.; Thiede, C.; Fuessel, S. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin. Biochem. 2019, 64, 60–63. [Google Scholar] [CrossRef]
- Darwiche, F.; Parekh, D.J.; Gonzalgo, M.L. Biomarkers for non-muscle invasive bladder cancer: Current tests and future promise. Indian. J. Urol. IJU J. Urol. Soc. India 2015, 31, 273–282. [Google Scholar] [CrossRef]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; van der Keur, K.; van Kessel, K.; Zwarthoff, E.; Agerbæk, M.; Ørntoft, T.F.; Jensen, J.B.; et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, K.E.; Van Neste, L.; Lurkin, I.; Zwarthoff, E.C.; Van Criekinge, W. Evaluation of an Epigenetic Profile for the Detection of Bladder Cancer in Patients with Hematuria. J. Urol. 2016, 195, 601–607. [Google Scholar] [CrossRef] [PubMed]
- de Jong, F.C.; Iflé, I.G.; van der Made, A.C.; Kooper, D.; de Jong, J.J.; Franckena, M.; Zuiverloon, T.C.M.; van Criekinge, W.; Incrocci, L.; Zwarthoff, E.C.; et al. A Genomic Urine Assay for Surveillance of Patients with Bladder Cancer Treated with Radiotherapy. Eur. Urol. Open Sci. 2024, 62, 131–139. [Google Scholar] [CrossRef]
- Beukers, W.; van der Keur, K.A.; Kandimalla, R.; Vergouwe, Y.; Steyerberg, E.W.; Boormans, J.L.; Jensen, J.B.; Lorente, J.A.; Real, F.X.; Segersten, U.; et al. FGFR3, TERT and OTX1 as a Urinary Biomarker Combination for Surveillance of Patients with Bladder Cancer in a Large Prospective Multicenter Study. J. Urol. 2017, 197, 1410–1418. [Google Scholar] [CrossRef]
- Batista, R.; Vinagre, J.; Prazeres, H.; Sampaio, C.; Peralta, P.; Conceição, P.; Sismeiro, A.; Leão, R.; Gomes, A.; Furriel, F.; et al. Validation of a Novel, Sensitive, and Specific Urine-Based Test for Recurrence Surveillance of Patients With Non-Muscle-Invasive Bladder Cancer in a Comprehensive Multicenter Study. Front. Genet. 2019, 10, 1237. [Google Scholar] [CrossRef]
- Ou, Z.; Li, K.; Yang, T.; Dai, Y.; Chandra, M.; Ning, J.; Wang, Y.; Xu, R.; Gao, T.; Xie, Y.; et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin. Transl. Med. 2020, 9, 4. [Google Scholar] [CrossRef]
- Sieverink, C.A.; Batista, R.P.M.; Prazeres, H.J.M.; Vinagre, J.; Sampaio, C.; Leão, R.R.; Máximo, V.; Witjes, J.A.; Soares, P. Clinical Validation of a Urine Test (Uromonitor-V2(®)) for the Surveillance of Non-Muscle-Invasive Bladder Cancer Patients. Diagnostics 2020, 10, 745. [Google Scholar] [CrossRef]
- Azawi, N.; Vásquez, J.L.; Dreyer, T.; Guldhammer, C.S.; Saber Al-Juboori, R.M.; Nielsen, A.M.; Jensen, J.B. Surveillance of Low-Grade Non-Muscle Invasive Bladder Tumors Using Uromonitor: SOLUSION Trial. Cancers 2023, 15, 2341. [Google Scholar] [CrossRef]
- Marziali, A.; Akeson, M. New DNA sequencing methods. Annu. Rev. Biomed. Eng. 2001, 3, 195–223. [Google Scholar] [CrossRef]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Kok, J.; Buist, G.; Zomer, A.L.; van Hijum, S.A.; Kuipers, O.P. Comparative and functional genomics of lactococci. FEMS Microbiol. Rev. 2005, 29, 411–433. [Google Scholar] [CrossRef]
- Diamandis, M.; White, N.M.; Yousef, G.M. Personalized medicine: Marking a new epoch in cancer patient management. Mol. Cancer Res. MCR 2010, 8, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Springer, S.U.; Chen, C.H.; Rodriguez Pena, M.D.C.; Li, L.; Douville, C.; Wang, Y.; Cohen, J.D.; Taheri, D.; Silliman, N.; Schaefer, J.; et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Eich, M.L.; Rodriguez Pena, M.D.C.; Springer, S.U.; Taheri, D.; Tregnago, A.C.; Salles, D.C.; Bezerra, S.M.; Cunha, I.W.; Fujita, K.; Ertoy, D.; et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Modern pathology: An official journal of the United States and Canadian Academy of Pathology, Inc 2019, 32, 1544–1550. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Yu, D.W.; Zhou, X.; Vogler, A.P. Mitochondrial metagenomics: Letting the genes out of the bottle. GigaScience 2016, 5, 15. [Google Scholar] [CrossRef]
- Lau, T.K.; Cheung, S.W.; Lo, P.S.; Pursley, A.N.; Chan, M.K.; Jiang, F.; Zhang, H.; Wang, W.; Jong, L.F.; Yuen, O.K.; et al. Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: Review of 1982 consecutive cases in a single center. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2014, 43, 254–264. [Google Scholar] [CrossRef]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar] [CrossRef]
- Ge, G.; Peng, D.; Guan, B.; Zhou, Y.; Gong, Y.; Shi, Y.; Hao, X.; Xu, Z.; Qi, J.; Lu, H.; et al. Urothelial Carcinoma Detection Based on Copy Number Profiles of Urinary Cell-Free DNA by Shallow Whole-Genome Sequencing. Clin. Chem. 2020, 66, 188–198. [Google Scholar] [CrossRef]
- Wang, P.; Shi, Y.; Zhang, J.; Shou, J.; Zhang, M.; Zou, D.; Liang, Y.; Li, J.; Tan, Y.; Zhang, M.; et al. UCseek: Ultrasensitive early detection and recurrence monitoring of urothelial carcinoma by shallow-depth genome-wide bisulfite sequencing of urinary sediment DNA. EBioMedicine 2023, 89, 104437. [Google Scholar] [CrossRef]
- Zeng, S.; Ying, Y.; Xing, N.; Wang, B.; Qian, Z.; Zhou, Z.; Zhang, Z.; Xu, W.; Wang, H.; Dai, L.; et al. Noninvasive Detection of Urothelial Carcinoma by Cost-effective Low-coverage Whole-genome Sequencing from Urine-Exfoliated Cell DNA. Clin. Cancer Res. An Off. J. Am. Assoc. Cancer Res. 2020, 26, 5646–5654. [Google Scholar] [CrossRef]
- Schmidt, B.; Hildebrandt, A. Deep learning in next-generation sequencing. Drug Discov. Today 2021, 26, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Vyas, A.; Kumar, K.; Sharma, A.; Verma, D.; Bhatia, D.; Wahi, N.; Yadav, A.K. Advancing the frontier of artificial intelligence on emerging technologies to redefine cancer diagnosis and care. Comput. Biol. Med. 2025, 191, 110178. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Zhang, L. AI applications in HIV research: Advances and future directions. Front. Microbiol. 2025, 16, 1541942. [Google Scholar] [CrossRef]
- Paz, M.; Moratorio, G. Deep mutational scanning and CRISPR-engineered viruses: Tools for evolutionary and functional genomics studies. mSphere 2025, e0050824. [Google Scholar] [CrossRef]
- Rausch, T.; Marschall, T.; Korbel, J.O. The impact of long-read sequencing on human population-scale genomics. Genome Res. 2025, 35, 593–598. [Google Scholar] [CrossRef]
- Ward, D.G.; Baxter, L.; Ott, S.; Gordon, N.S.; Wang, J.; Patel, P.; Piechocki, K.; Silcock, L.; Sale, C.; Zeegers, M.P.; et al. Highly Sensitive and Specific Detection of Bladder Cancer via Targeted Ultra-deep Sequencing of Urinary DNA. Eur. Urol. Oncol. 2023, 6, 67–75. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Chen, K.; Babbra, R.K.; Feng, W.; Pejovic, N.; Nallicheri, A.; Harris, P.K.; Dienstbach, K.; Atkocius, A.; Maguire, L.; et al. Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: A cohort study. PLoS Med. 2021, 18, e1003732. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Shiang, A.; Alahi, I.; Sundby, R.T.; Feng, W.; Gungoren, B.; Nawaf, C.; Chen, K.; Babbra, R.K.; Harris, P.K.; et al. Urine cell-free DNA multi-omics to detect MRD and predict survival in bladder cancer patients. NPJ Precis. Oncol. 2023, 7, 6. [Google Scholar] [CrossRef]
- Lee, E.J.; Luo, J.; Wilson, J.M.; Shi, H. Analyzing the cancer methylome through targeted bisulfite sequencing. Cancer Lett. 2013, 340, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Mijnes, J.; Veeck, J.; Gaisa, N.T.; Burghardt, E.; de Ruijter, T.C.; Gostek, S.; Dahl, E.; Pfister, D.; Schmid, S.C.; Knüchel, R.; et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin. Epigenetics 2018, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, D.; Kaufmann, M.; Hippe, J.; Gajda, M.; Grimm, M.O. High Detection Rate for Non-Muscle-Invasive Bladder Cancer Using an Approved DNA Methylation Signature Test. Clin. Genitourin. Cancer 2020, 18, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Chao, H.; Deng, H.; Yu, Z.; Zhao, R.; Huang, L.; Gong, Y.; Zhu, Y.; Wang, Q.; Li, F.; et al. A novel and sensitive DNA methylation marker for the urine-based liquid biopsies to detect bladder cancer. BMC Cancer 2022, 22, 510. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, G.; Zhang, N.; Liu, S.; Lin, X.; Perschon, C.; Zheng, S.L.; Ding, Q.; Wang, X.; Na, R.; et al. HOXA9, PCDH17, POU4F2, and ONECUT2 as a Urinary Biomarker Combination for the Detection of Bladder Cancer in Chinese Patients with Hematuria. Eur. Urol. Focus 2020, 6, 284–291. [Google Scholar] [CrossRef]
- Ruan, W.; Chen, X.; Huang, M.; Wang, H.; Chen, J.; Liang, Z.; Zhang, J.; Yu, Y.; Chen, S.; Xu, S.; et al. A urine-based DNA methylation assay to facilitate early detection and risk stratification of bladder cancer. Clin. Epigenetics 2021, 13, 91. [Google Scholar] [CrossRef]
- van Kessel, K.E.; Beukers, W.; Lurkin, I.; Ziel-van der Made, A.; van der Keur, K.A.; Boormans, J.L.; Dyrskjøt, L.; Márquez, M.; Ørntoft, T.F.; Real, F.X.; et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J. Urol. 2017, 197, 590–595. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, R.; Lu, Q.; Xu, Z.; Liu, J.; Li, P.; Zhang, Y.; Zhou, C.; Luo, L.; Tang, W.; et al. A Novel Methylation Marker NRN1 plus TERT and FGFR3 Mutation Using Urine Sediment Enables the Detection of Urothelial Bladder Carcinoma. Cancers 2023, 15. [Google Scholar] [CrossRef]
- Pierconti, F.; Martini, M.; Cenci, T.; Fiorentino, V.; Gianfrancesco, L.D.; Ragonese, M.; Bientinesi, R.; Rossi, E.; Larocca, L.M.; Racioppi, M.; et al. The bladder epicheck test and cytology in the follow-up of patients with non-muscle-invasive high grade bladder carcinoma. Urol. Oncol. 2022, 40, e119–e108-108.e125. [Google Scholar] [CrossRef]
- Pierconti, F.; Rossi, E.D.; Cenci, T.; Carlino, A.; Fiorentino, V.; Totaro, A.; Sacco, E.; Palermo, G.; Iacovelli, R.; Larocca, L.M.; et al. DNA methylation analysis in urinary samples: A useful method to predict the risk of neoplastic recurrence in patients with urothelial carcinoma of the bladder in the high-risk group. Cancer Cytopathol. 2023, 131, 158–164. [Google Scholar] [CrossRef]
- Pierconti, F.; Martini, M.; Cenci, T.; Fiorentino, V.; Sacco, E.; Bientinesi, R.; Pugliese, D.; Iacovelli, R.; Schinzari, G.; Larocca, L.M.; et al. Methylation study of the Paris system for reporting urinary (TPS) categories. J. Clin. Pathol. 2021, 74, 102–105. [Google Scholar] [CrossRef]
- Pierconti, F.; Martini, M.; Fiorentino, V.; Cenci, T.; Capodimonti, S.; Straccia, P.; Sacco, E.; Pugliese, D.; Cindolo, L.; Larocca, L.M.; et al. The combination cytology/epichek test in non muscle invasive bladder carcinoma follow-up: Effective tool or useless expence? Urol. Oncol. 2021, 39, e117–e131-131.e121. [Google Scholar] [CrossRef] [PubMed]
- Trenti, E.; D’Elia, C.; Mian, C.; Schwienbacher, C.; Hanspeter, E.; Pycha, A.; Kafka, M.; Degener, S.; Danuser, H.; Roth, S.; et al. Diagnostic predictive value of the Bladder EpiCheck test in the follow-up of patients with non-muscle-invasive bladder cancer. Cancer Cytopathol. 2019, 127, 465–469. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, D.; Soria, F.; Zehetmayer, S.; Gust, K.M.; Korn, S.; Witjes, J.A.; Shariat, S.F. Diagnostic accuracy, clinical utility and influence on decision-making of a methylation urine biomarker test in the surveillance of non-muscle-invasive bladder cancer. BJU Int. 2019, 123, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Witjes, J.A.; Morote, J.; Cornel, E.B.; Gakis, G.; van Valenberg, F.J.P.; Lozano, F.; Sternberg, I.A.; Willemsen, E.; Hegemann, M.L.; Paitan, Y.; et al. Performance of the Bladder EpiCheck™ Methylation Test for Patients Under Surveillance for Non-muscle-invasive Bladder Cancer: Results of a Multicenter, Prospective, Blinded Clinical Trial. Eur. Urol. Oncol. 2018, 1, 307–313. [Google Scholar] [CrossRef]
- Su, S.F.; de Castro Abreu, A.L.; Chihara, Y.; Tsai, Y.; Andreu-Vieyra, C.; Daneshmand, S.; Skinner, E.C.; Jones, P.A.; Siegmund, K.D.; Liang, G. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin. Cancer Res. An Off. J. Am. Assoc. Cancer Res. 2014, 20, 1978–1989. [Google Scholar] [CrossRef]
- Varol, N.; Keles, İ.; Yildiz, H.; Karaosmanoglu, C.; Karalar, M.; Zengin, K.; Sarici, H.; Tokyol, C. Methylation analysis of histone 4-related gene HIST1H4F and its effect on gene expression in bladder cancer. Gene 2023, 866, 147352. [Google Scholar] [CrossRef]
- de Jong, J.J.; Pijpers, O.M.; van Kessel, K.E.M.; Boormans, J.L.; Van Criekinge, W.; Zwarthoff, E.C.; Lotan, Y. A Urine-based Genomic Assay Improves Risk Stratification for Patients with High-risk Hematuria Stratified According to the American Urological Association Guidelines. Eur. Urol. Oncol. 2023, 6, 183–189. [Google Scholar] [CrossRef]
- Olkhov-Mitsel, E.; Savio, A.J.; Kron, K.J.; Pethe, V.V.; Hermanns, T.; Fleshner, N.E.; van Rhijn, B.W.; van der Kwast, T.H.; Zlotta, A.R.; Bapat, B. Epigenome-Wide DNA Methylation Profiling Identifies Differential Methylation Biomarkers in High-Grade Bladder Cancer. Transl. Oncol. 2017, 10, 168–177. [Google Scholar] [CrossRef]
- Xu, Z.; Ge, G.; Guan, B.; Lei, Z.; Hao, X.; Zhou, Y.; Shi, Y.; Lu, H.; Wang, J.; Peng, D.; et al. Noninvasive Detection and Localization of Genitourinary Cancers Using Urinary Sediment DNA Methylomes and Copy Number Profiles. Eur. Urol. 2020, 77, 288–290. [Google Scholar] [CrossRef]
- Feber, A.; Dhami, P.; Dong, L.; de Winter, P.; Tan, W.S.; Martínez-Fernández, M.; Paul, D.S.; Hynes-Allen, A.; Rezaee, S.; Gurung, P.; et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin. Epigenetics 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kang, G.; Jiang, P.; Qiao, R.; Lam, W.K.J.; Yu, S.C.Y.; Ma, M.L.; Ji, L.; Cheng, S.H.; Gai, W.; et al. Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proc. Natl. Acad. Sci. USA 2022, 119, e2209852119. [Google Scholar] [CrossRef] [PubMed]
- Hanke, M.; Kausch, I.; Dahmen, G.; Jocham, D.; Warnecke, J.M. Detailed technical analysis of urine RNA-based tumor diagnostics reveals ETS2/urokinase plasminogen activator to be a novel marker for bladder cancer. Clin. Chem. 2007, 53, 2070–2077. [Google Scholar] [CrossRef] [PubMed]
- Ribal, M.J.; Mengual, L.; Lozano, J.J.; Ingelmo-Torres, M.; Palou, J.; Rodríguez-Faba, O.; Witjes, J.A.; Van der Heijden, A.G.; Medina, R.; Conde, J.M.; et al. Gene expression test for the non-invasive diagnosis of bladder cancer: A prospective, blinded, international and multicenter validation study. Eur. J. Cancer 2016, 54, 131–138. [Google Scholar] [CrossRef]
- Malentacchi, F.; Vinci, S.; Melina, A.D.; Kuncova, J.; Villari, D.; Nesi, G.; Selli, C.; Orlando, C.; Pazzagli, M.; Pinzani, P. Urinary carbonic anhydrase IX splicing messenger RNA variants in urogenital cancers. Urol. Oncol. 2016, 34, e299–e292-292.e216. [Google Scholar] [CrossRef]
- Zhang, M.; Ren, B.; Li, Z.; Niu, W.; Wang, Y. Expression of N-Myc Downstream-Regulated Gene 2 in Bladder Cancer and Its Potential Utility as a Urinary Diagnostic Biomarker. Med. Sci. Monit. 2017, 23, 4644–4649. [Google Scholar] [CrossRef]
- Lotan, Y.; OʼSullivan, P.; Raman, J.D.; Shariat, S.F.; Kavalieris, L.; Frampton, C.; Guilford, P.; Luxmanan, C.; Suttie, J.; Crist, H.; et al. Clinical comparison of noninvasive urine tests for ruling out recurrent urothelial carcinoma. Urol. Oncol. 2017, 35, e515–e531-531.e522. [Google Scholar] [CrossRef]
- Montalbo, R.; Lozano, J.J.; Izquierdo, L.; Ingelmo-Torres, M.; BaÑos, C.; Palou, J.; Van der Heijden, A.G.; Medina, R.; Schmidbauer, J.; Prat, A.; et al. Ability of a urine gene expression classifier to reduce the number of follow-up cystoscopies in bladder cancer patients. Transl. Res. 2019, 208, 73–84. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, L.; Ma, W.; Meng, Z.; Li, P.; Zhang, X.; Wang, Y.; Lu, Y.; Sun, Y.; Wu, Y.; et al. UriBLAD: A Urine-Based Gene Expression Assay for Noninvasive Detection of Bladder Cancer. J. Mol. Diagn. 2021, 23, 61–70. [Google Scholar] [CrossRef]
- Ratert, N.; Meyer, H.A.; Jung, M.; Lioudmer, P.; Mollenkopf, H.J.; Wagner, I.; Miller, K.; Kilic, E.; Erbersdobler, A.; Weikert, S.; et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J. Mol. Diagn. 2013, 15, 695–705. [Google Scholar] [CrossRef]
- Tölle, A.; Jung, M.; Rabenhorst, S.; Kilic, E.; Jung, K.; Weikert, S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol. Rep. 2013, 30, 1949–1956. [Google Scholar] [CrossRef]
- Pardini, B.; Cordero, F.; Naccarati, A.; Viberti, C.; Birolo, G.; Oderda, M.; Di Gaetano, C.; Arigoni, M.; Martina, F.; Calogero, R.A.; et al. microRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget 2018, 9, 20658–20669. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zhang, Z.; Zheng, F.; Wang, L.; Guo, J.; Zhang, T.; Dai, X.; Zhang, S.; Yang, D.; Kuang, R.; et al. Combining Protein and miRNA Quantification for Bladder Cancer Analysis. ACS Appl. Mater. Interfaces 2017, 9, 23420–23427. [Google Scholar] [CrossRef] [PubMed]
- Moisoiu, T.; Dragomir, M.P.; Iancu, S.D.; Schallenberg, S.; Birolo, G.; Ferrero, G.; Burghelea, D.; Stefancu, A.; Cozan, R.G.; Licarete, E.; et al. Combined miRNA and SERS urine liquid biopsy for the point-of-care diagnosis and molecular stratification of bladder cancer. Mol. Med. 2022, 28, 39. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Fujita, K.; Jingushi, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Ueda, Y.; Tanigawa, G.; Yoshioka, I.; Ueda, K.; et al. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 2017, 8, 24668–24678. [Google Scholar] [CrossRef]
- Hofbauer, S.L.; de Martino, M.; Lucca, I.; Haitel, A.; Susani, M.; Shariat, S.F.; Klatte, T. A urinary microRNA (miR) signature for diagnosis of bladder cancer. Urol. Oncol. 2018, 36, e531–e531-531.e538. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, J.; Xu, D.; Liu, Y.; Zhang, D.; Ye, J.; Li, H. Diagnostic performance of urine and blood microRNAs for bladder cancer: A meta-analysis. Expert. Rev. Anticancer. Ther. 2022, 22, 1357–1369. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Essawy, N.O.; Shehta, M.; Kotb, Y.M. Rapid detection of urinary long non-coding RNA urothelial carcinoma associated one using a PCR-free nanoparticle-based assay. Biomarkers 2015, 20, 212–217. [Google Scholar] [CrossRef]
- Du, L.; Duan, W.; Jiang, X.; Zhao, L.; Li, J.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J. Cell Mol. Med. 2018, 22, 2838–2845. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Y.; Wang, X.; Gao, Y.; Li, L.; Zhang, J.; Zhang, L.; Che, F. Circulating lncRNAs as noninvasive biomarkers in bladder cancer: A diagnostic meta-analysis based on 15 published articles. Int. J. Biol. Markers 2020, 35, 40–48. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, Z.Y.; Zhang, Z.; Xiao, X.Y.; Gao, S.L.; Lu, C.; Zuo, L.; Zhang, L.F. Prediction of bladder cancer outcome by identifying and validating a mutation-derived genomic instability-associated long noncoding RNA (lncRNA) signature. Bioengineered 2021, 12, 1725–1738. [Google Scholar] [CrossRef]
- Ecke, T.H.; Meisl, C.J.; Schlomm, T.P.; Rabien, A.; Labonté, F.; Rong, D.; Hofbauer, S.; Friedersdorff, F.P.; Sommerfeldt, L.; Gagel, N.; et al. BTA stat®, NMP22® BladderChek®, UBC® Rapid Test, and CancerCheck® UBC® rapid VISUAL as urinary marker for bladder cancer: Final results of a German multicenter study. Urol. Oncol. 2023, 41, e417–e484-484.e426. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Xu, J.; Zhang, Q. Sandwich ELISA for detecting urinary Survivin in bladder cancer. Chin. J. Cancer Res. = Chung-Kuo Yen Cheng Yen Chiu 2013, 25, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Karpiuk, K.R.; Młynarczyk, G.; Matowicka-Karna, J.; Darewicz, B. Emerging Biomarkers in Urological Cancers: Angiogenesis and Damage-Associated Molecular Pattern Signaling. Int. J. Mol. Sci. 2025, 26. [Google Scholar] [CrossRef] [PubMed]
- Glas, A.S.; Roos, D.; Deutekom, M.; Zwinderman, A.H.; Bossuyt, P.M.; Kurth, K.H. Tumor markers in the diagnosis of primary bladder cancer. A systematic review. J. Urol. 2003, 169, 1975–1982. [Google Scholar] [CrossRef] [PubMed]
- Atsü, N.; Ekici, S.; Oge, O.O.; Ergen, A.; Hasçelik, G.; Ozen, H. False-positive results of the NMP22 test due to hematuria. J. Urol. 2002, 167, 555–558. [Google Scholar] [CrossRef]
- Joung, J.Y.; Park, S.; Yoon, H.; Kwon, W.A.; Cho, I.C.; Seo, H.K.; Chung, J.; Hwang, S.H.; Lee, C.W.; Lee, K.H. Overestimation of nuclear matrix protein 22 in concentrated urine. Urology 2013, 82, 1059–1064. [Google Scholar] [CrossRef]
- Shariat, S.F.; Marberger, M.J.; Lotan, Y.; Sanchez-Carbayo, M.; Zippe, C.; Lüdecke, G.; Boman, H.; Sawczuk, I.; Friedrich, M.G.; Casella, R.; et al. Variability in the performance of nuclear matrix protein 22 for the detection of bladder cancer. J. Urol. 2006, 176, 919–926; discussion 926. [Google Scholar] [CrossRef]
- Chen, L.M.; Chang, M.; Dai, Y.; Chai, K.X.; Dyrskjøt, L.; Sanchez-Carbayo, M.; Szarvas, T.; Zwarthoff, E.C.; Lokeshwar, V.; Jeronimo, C.; et al. External validation of a multiplex urinary protein panel for the detection of bladder cancer in a multicenter cohort. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1804–1812. [Google Scholar] [CrossRef]
- Furuya, H.; Sakatani, T.; Tanaka, S.; Murakami, K.; Waldron, R.T.; Hogrefe, W.; Rosser, C.J. Bladder cancer risk stratification with the Oncuria 10-plex bead-based urinalysis assay using three different Luminex xMAP instrumentation platforms. Res Sq 2023. [Google Scholar] [CrossRef]
- Guo, H.; Jin, B.; Zhu, Z.; Dai, X.; Wang, M.; Xie, Y.; Xu, C.; Wang, Z.; Liu, Y.; Tan, W. Nanoparticle-Protein Corona Boosted Cancer Diagnosis with Proteomic Transfer Learning. ACS Nano 2025, 19, 23592–23605. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhu, S.; Cui, X.; Xu, W.; Kong, C.; Zhang, Z.; Qian, W. Identifying non-muscle-invasive and muscle-invasive bladder cancer based on blood serum surface-enhanced Raman spectroscopy. Biomed. Opt. Express 2019, 10, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jue, M.; Lee, K.; Paulson, B.; Oh, J.; Cho, M.; Kim, J.K. Early-stage diagnosis of bladder cancer using surface-enhanced Raman spectroscopy combined with machine learning algorithms in a rat model. Biosens. Bioelectron. 2024, 246, 115915. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, J.; Bi, X.; Qian, H.; Pan, J.; Ye, J. Non-invasive and rapid diagnosis of low-grade bladder cancer via SERSomes of urine. Nanoscale 2025, 17, 7303–7312. [Google Scholar] [CrossRef]
- Zhong, Q.; Shao, L.; Yao, Y.; Chen, S.; Lv, X.; Liu, Z.; Zhu, S.; Yan, Z. Urine-based SERS and multivariate statistical analysis for identification of non-muscle-invasive bladder cancer and muscle-invasive bladder cancer. Anal. Bioanal. Chem. 2024, 416, 6973–6984. [Google Scholar] [CrossRef]
- Keum, C.; Yeom, H.; Noh, T.I.; Yi, S.Y.; Jin, S.; Kim, C.; Shim, J.S.; Yoon, S.G.; Kim, H.; Lee, K.H.; et al. Diagnosis of early-stage bladder cancer via unprocessed urine samples at the point of care. Nat. Biomed. Eng. 2025, 9, 1026–1038. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Issaq, H.J.; Nativ, O.; Waybright, T.; Luke, B.; Veenstra, T.D.; Issaq, E.J.; Kravstov, A.; Mullerad, M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J. Urol. 2008, 179, 2422–2426. [Google Scholar] [CrossRef]
- Li, J.; Cheng, B.; Xie, H.; Zhan, C.; Li, S.; Bai, P. Bladder cancer biomarker screening based on non-targeted urine metabolomics. Int. Urol. Nephrol. 2022, 54, 23–29. [Google Scholar] [CrossRef]
- Oto, J.; Fernández-Pardo, Á.; Roca, M.; Plana, E.; Cana, F.; Herranz, R.; Pérez-Ardavín, J.; Vera-Donoso, C.D.; Martínez-Sarmiento, M.; Medina, P. LC-MS metabolomics of urine reveals distinct profiles for non-muscle-invasive and muscle-invasive bladder cancer. World J. Urol. 2022, 40, 2387–2398. [Google Scholar] [CrossRef]
- Wang, R.; Kang, H.; Zhang, X.; Nie, Q.; Wang, H.; Wang, C.; Zhou, S. Urinary metabolomics for discovering metabolic biomarkers of bladder cancer by UPLC-MS. BMC Cancer 2022, 22, 214. [Google Scholar] [CrossRef]
- Lett, L.; George, M.; Slater, R.; De Lacy Costello, B.; Ratcliffe, N.; García-Fiñana, M.; Lazarowicz, H.; Probert, C. Investigation of urinary volatile organic compounds as novel diagnostic and surveillance biomarkers of bladder cancer. Br. J. Cancer 2022, 127, 329–336. [Google Scholar] [CrossRef]
- Mengual, L.; Frantzi, M.; Mokou, M.; Ingelmo-Torres, M.; Vlaming, M.; Merseburger, A.S.; Roesch, M.C.; Culig, Z.; Alcaraz, A.; Vlahou, A.; et al. Multicentric validation of diagnostic tests based on BC-116 and BC-106 urine peptide biomarkers for bladder cancer in two prospective cohorts of patients. Br. J. Cancer 2022, 127, 2043–2051. [Google Scholar] [CrossRef]
- Shapiro, A.; Gofrit, O.N.; Pizov, G.; Cohen, J.K.; Maier, J. Raman molecular imaging: A novel spectroscopic technique for diagnosis of bladder cancer in urine specimens. Eur. Urol. 2011, 59, 106–112. [Google Scholar] [CrossRef]
- Yosef, H.K.; Krauß, S.D.; Lechtonen, T.; Jütte, H.; Tannapfel, A.; Käfferlein, H.U.; Brüning, T.; Roghmann, F.; Noldus, J.; Mosig, A.; et al. Noninvasive Diagnosis of High-Grade Urothelial Carcinoma in Urine by Raman Spectral Imaging. Anal. Chem. 2017, 89, 6893–6899. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).