1. Introduction
The integration of artificial intelligence (AI) into scientific research has progressed from a
methodological enhancement to a fundamental paradigm shift (Tang et al. 2025; Zhang et al. 2024b).
Early AI models primarily functioned as computational tools, facilitating low-level analytical tasks,
such as pattern extraction and representation learning (Ye et al. 2025a; Zhang et al. 2024a). More recently,
advances in AI, particularly large language models (LLMs), have introduced stronger capabilities for
reasoning (Ma et al. 2024; Yao et al. 2023), exerting a transformative effect on the scientific discovery
process. For example, some AI systems even demonstrate the ability to plan and execute experiments
autonomously (Jansen et al. 2025; Pratiush et al. 2024; Zheng et al. 2025).
Despite the rapid development of AI, scientific discovery remains a fundamentally creative
and complex process that requires significant human involvement. Especially in high-stakes and
resource-intensive scientific domains (e.g., medicine, chemistry, and genomics) where errors can be
costly or irreversible, human scientists are still expected to continuously monitor AI outcomes and
make critical research decisions. However, existing surveys predominantly focus on the technical
capabilities of AI models, often overlooking the role of human scientists in the discovery process.
For instance, Zheng et al. (2025) reviews LLM-based systems for scientific discovery and proposes
a three-level autonomy taxonomy (i.e., Tool, Analyst, Scientist). Reddy and Shojaee (2025) provides
a survey of generative AI for scientific tasks and summarizes challenges in building AI systems for
scientific discovery. Consequently, our theoretical understanding of how humans and AI can effectively
collaborate together throughout the scientific research process remains limited. Although surveys on human-NLP cooperation (Huang et al. 2025) and general human-AI interaction discuss relevant
interaction principles (Mohanty et al. 2025; Rajashekar et al. 2024), they often overlook the specific
context of scientific discovery. Therefore, there is a lack of a structured framework for understanding
the human-AI partnership in scientific discovery.
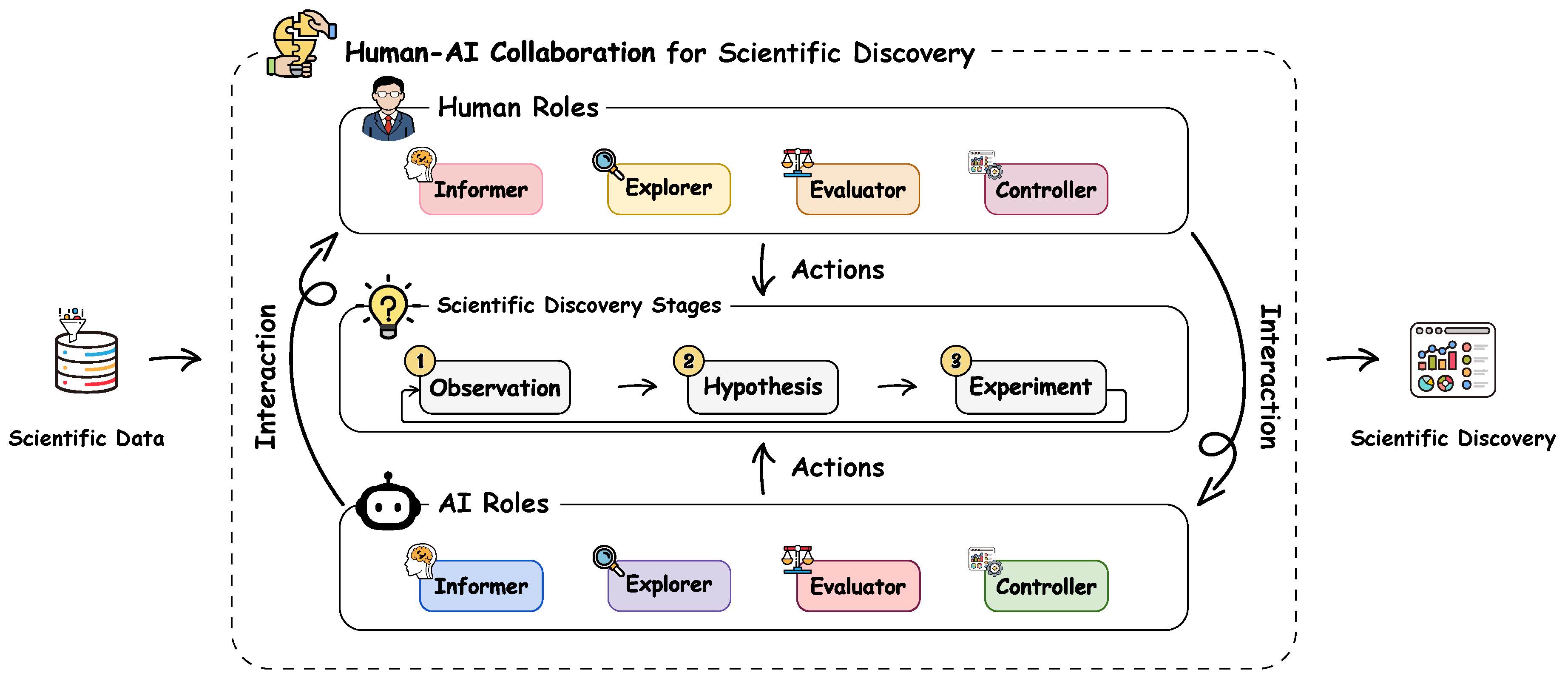
Figure 1.
Our taxonomy characterizes research on human-AI collaborative scientific discovery from four roles of human and AI across the three stages of scientific discovery.
Figure 1.
Our taxonomy characterizes research on human-AI collaborative scientific discovery from four roles of human and AI across the three stages of scientific discovery.
To bridge this gap, we presented a systematic taxonomy for human-AI collaboration in scientific discovery. First, we identified four roles of human and AI based on a systematic review of 51 papers and anchored our analysis in the established three stages of scientific discovery (Wang et al. 2023a) (i.e., observation, hypothesis, and experiment). This established a unified framework that organized collaborative dynamics into consistent and comparable units. Building upon this taxonomic framework, we analyzed the specific role allocation between human and AI partners, identifying and discussing their common collaboration patterns in scientific discovery and their differences across the three stages. We conclude by outlining five open challenges and future directions for establishing efficient and trustworthy human–AI partnerships. The main contributions of this paper are summarized as follows:
We presented a comprehensive survey that systematically reviews human-AI collaboration specifically in scientific discovery.
We introduced a novel taxonomy that defines the four roles of human and AI and characterized their distinct collaboration patterns across the three stages of scientific discovery.
We identified critical challenges and future pathways for building human-AI partnerships in the scientific discovery process.
2. Methodology
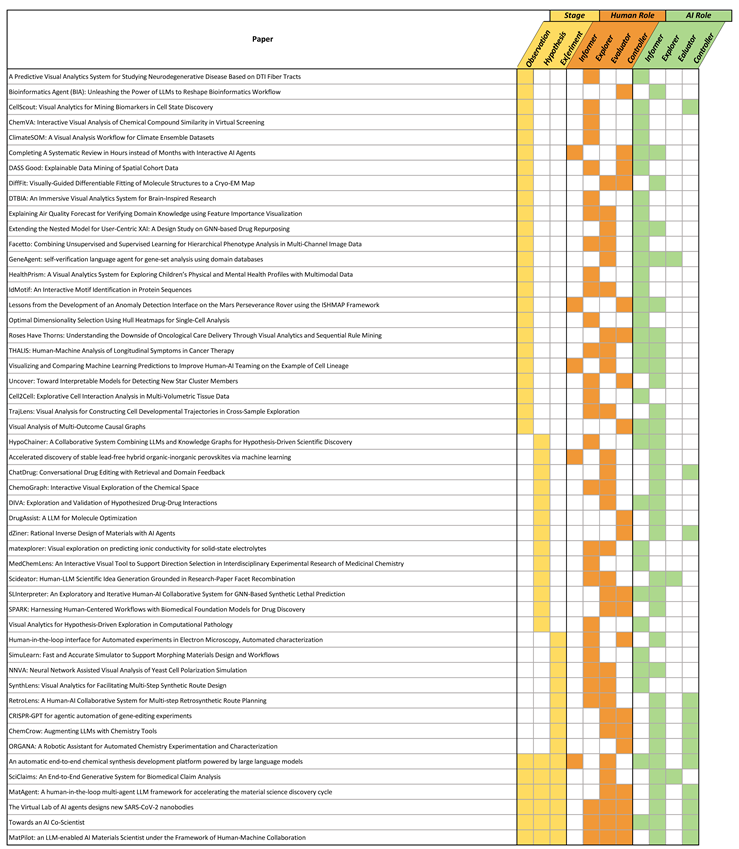
In this section, we outline the methodology for collecting a corpus of 51 papers on human-AI collaboration for scientific discovery and the coding process used to identify the roles of human and AI.
2.1. Paper Collection
To assemble a high-quality corpus focused on human-AI collaboration for scientific discovery, we implemented a systematic selection process encompassing research published between 2015 and 2025. We began by identifying seed papers from authoritative surveys on AI for science (Reddy and
Shojaee 2025; Zheng et al. 2025). Using these papers as a baseline, we then conducted an iterative snowballing procedure, examining references and citations to identify relevant work until no further relevant studies emerged.
To ensure relevance, we applied several screening criteria to select papers for inclusion in our corpus. We began by reviewing the abstracts, and if necessary, examined other sections. Each paper had to present an interactive system or workflow explicitly designed to facilitate scientific discovery. As a result, we excluded papers whose contribution was the development of a fully automated algorithm. Additionally, we excluded studies focused solely on data labeling tasks (e.g., SciDaSynth (Wang et al.
2025d)), even if the paper suggested that the dataset could contribute to future scientific discovery, as these works were considered too preliminary. To maintain high quality, we included only published papers or preprints with more than 100 citations. The final corpus consisted of 51 papers from a variety of reputable venues, such as Nature, ACL, EMNLP, CHI, and TVCG.
2.2. Paper Coding
Initially, six co-authors independently coded a subset of the corpus to derive the roles of humans and AI. Through weekly discussions, they resolved conflicts and unified the coding results, ultimately identifying four distinct roles. To better analyze their functions in scientific discovery, we followed a commonly used three-stage decomposition of the scientific process (i.e., observation, hypothesis, experiment) (Wang et al. 2023a), coding each paper according to which stage it belonged to. Specifically, the observation stage involves collecting and examining data or phenomena to identify patterns, anomalies, or open questions that require explanation. Based on these observations and prior knowledge, researchers formulate hypotheses—tentative, testable explanations or predictions that guide inquiry. The experiment stage then designs and conducts controlled studies or analyses to test these hypotheses, using the results to validate, refine, or reject them, often leading to new observations and continuing the discovery cycle.
3. Taxonomy
In this section, we first introduce four roles that humans or AI can play in the scientific discovery process. Built upon the definitions of these roles, we then elaborate on common human–AI collaboration patterns at the three stages of scientific discovery. Finally, we analyze how these roles differ across the three distinct stages.
3.1. Roles of Human and AI
Based on our systematic analysis of the corpus, we identify four roles of human and AI in scientific discovery: Informer, Explorer, Evaluator, and Controller. To clarify agency, we apply “human-” or “AI-” prefixes before these roles (e.g., human-Informer, AI-Informer).
⋄Informer. The Informer synthesizes, distills, or articulates key information, insights, or constraints from raw data or intermediate analyses to guide the actions of other roles. For instance, the AI-Informer in THALIS extracts temporal patterns from longitudinal symptom records in cancer therapy (Floricel et al. 2022). This provides a summarized trajectory view for experts to analyze patient responses to treatment. Similarly, in ISHMAP for Mars rover operations (Wright et al. 2023), the human-Informer marks instrument states and operational events on the telemetry timeline. The AI uses these annotations to reduce false alarms during state changes and to highlight unexpected signals.
⋄Explorer. The Explorer operates within the space of data patterns, hypotheses, or experimental designs to explore promising candidates or directions. Compared with Informer, whose output provides low-level data insights, the Explorer directly generates candidates tailored to the specific needs of each stage in the scientific discovery process. For instance, in the hAE interface (Pratiush
et al. 2024), the AI-Explorer searches the parameter space to select the next experimental conditions for electron microscopy. ChemVA (Sabando et al. 2021) enables the human-Explorer to interactively navigate a projected chemical space to identify molecular targets.
⋄Evaluator. Once artifacts are proposed, their scientific merit must be rigorously evaluated and even refined. The Evaluator assesses observed patterns, hypotheses, or experimental designs based on predefined criteria, evidence, and domain constraints, revising them as necessary to meet quality standards. For instance, in RetroLens (Shi et al. 2023a), chemists serve as the human-Evaluator. Specifically, they can assess AI-predicted synthetic routes for chemical feasibility or refine the synthetic steps by themselves.
⋄Controller. The Controller oversees the scientific discovery workflow to ensure correct and constraint-compliant execution, intervening when necessary to adjust procedures and handle runtime exceptions. This role is central to BIA (Xin et al. 2024), where the AI-Controller orchestrates the execution of complex bioinformatics toolchains, dynamically handling errors and modifying the workflow logic to ensure successful completion.
3.2. Common Collaboration Patterns Within Each Stage of Scientific Discovery
In this section, we introduce common human–AI collaboration patterns at each of the three stages (i.e., observation, hypothesis, and experiment) of the scientific discovery process. Note that although many papers in our corpus involve multiple human or AI roles, we only focus on roles that actively participate in human-AI collaboration and derive collaboration patterns from them to ensure that our analysis reflects meaningful human–AI collaboration rather than mere role co-existence
3.2.1. Observation Stage
The observation stage involves collecting and analyzing data to identify patterns and anomalies that warrant further investigation. During this stage, humans and AI collaborate to organize large datasets, highlight potential patterns, and verify them against the raw data to support hypothesis generation.
AI-Informer & Human-Explorer. A common human-AI collaboration pattern in the observation
stage involves an AI-Informer transforming raw data into structured representations (e.g., embeddings
or feature importance maps), supporting a human-Explorer to efficiently identify clusters, trends,
and outliers in scientific data. For example, some AI-Informers map high-dimensional data into a
two-dimensional view, allowing human-Explorers to observe how patterns change across different
conditions (Jeong et al. 2025; Kawakami et al. 2025). In biological domains, AI-Informers visualize tissue
interactions (Mörth et al. 2025), pediatric health profiles (Jiang et al. 2024), neural connections (Yao
et al. 2025), and cell trajectories (Wang et al. 2025b). Human-Explorers follow these visual pathways
to uncover disease trends or developmental stages. Other tools organize items by similarity. The
AI-Informers can also cluster compounds or highlight sequence motifs, allowing human-Explorers to
search for promising chemicals (Sabando et al. 2021), biomarkers (Sheng et al. 2025b), or protein
functions (Park et al. 2024). Furthermore, the AI-Informers can rank the influence of features
on predictions. The Human-Explorers investigate these cues to understand air pollution drivers
(Palaniyappan Velumani et al. 2022), explore raw fiber tracts (Xu et al. 2023), and analyze phenotype
images (Krueger et al. 2020).
AI-Explorer & Human-Evaluator. When models can automatically suggest candidates, the
collaboration often follows an examination by humans. The AI-Explorer suggests candidate patterns
and displays the primary evidence it used, such as highlighted inputs, matched records, or similar
past cases. The human-Evaluator reviews these candidates to determine whether they are correct and
meaningful. For instance, in drug research, the AI-Explorer highlights connections among different
drugs for repurposing, enabling the human Evaluator to assess and verify the underlying biological
rationale (Wang et al. 2023b). In structural biology, the AI-Explorer can generate 3D atomic models to
match density maps, allowing the human-Evaluator to examine the structural configuration (Luo et al.
2025). For longitudinal records in medicine, the AI-Explorers find distinct treatment pathways or care
rules. The human-evaluators review these specific sequences to validate symptom progression (Floricel
et al. 2022) or hospital protocols (Floricel et al. 2024). In plant embryo lineage analysis, the AI-Explorer
can generate classification results from multiple models. The human-Evaluator then assesses these
outputs to identify the correct cell type based on consensus (Hong et al. 2024). Additionally, the
AI-Explorer can retrieve and explore the functional roles of gene groups, while the human Evaluator
reviews this information to validate their biological relevance (Wang et al. 2025e).
AI-Explorer & Human-Controller. With autonomous tools, observation follows an iterative search loop. The human-Controller directs the search process, while the AI-Explorer scans the data. For literature discovery, the human-Controllers guide the search direction, while the AI-Explorers navigate databases to summarize relevant papers (Qiu et al. 2025; Xin et al. 2024). To identify star clusters
or spatial groups, the human-Controller adjust the detection parameters. The AI-Explorer scans
astronomical surveys to locate new star clusters (Ratzenböck et al. 2023) or analyzes spatial population
data to find notable spatial groups (Wentzel et al. 2023). In causal analysis, the human-Controller
refines the search constraints, enabling the AI-Explorer to explore cause-and-effect relationships (Fan
et al. 2025). Additionally, for Mars rover operations, the human-Controller sets anomaly criteria to
enable the AI-Explorer to detect signal anomalies (Wright et al. 2023).
3.2.2. Hypothesis Stage
Hypothesis generation involves proposing explanations or solutions for observed phenomena. Collaborative efforts during this phase focus on retrieving background knowledge and developing candidate theories or designs to guide subsequent testing.
AI-Explorer & Human-Evaluator. A common collaboration pattern involves the AI-Explorer
generating the initial hypothesis draft, while the human-Evaluator performs the final scientific
review. For instance, AI-Explorers generate candidate drug structures or material compositions,
while human-Evaluators assess whether these designs are chemically feasible (Liu et al. 2024; Lu et al.
2018; Ni et al. 2024). Similarly, AI-Explorers scan vast chemical or protein spaces to find promising
candidates. Human-Evaluators review the list to select the best options for testing (Kale et al. 2023;
Swanson et al. 2025). In scientific idea generation, AI-Explorers combine concepts from the literature
to propose new research directions or claims. Human-Evaluators validate these proposals against
domain knowledge (Kakar et al. 2019; Ortega and Gomez-Perez 2025; Wang et al. 2025a).
AI-Explorer & Human-Controller. In interactive design tasks, the human-Controller guides the
optimization process, while the AI-Explorer generates candidate solutions. For example, in material
and drug design, the human-Controller directs the design process by updating the requirements. The
AI-Explorer then generates a new batch of molecules based on this guidance (Ansari et al. 2024; Ye
et al. 2025b). For drug discovery, the human-Controller guides the search toward a target protein and
sets the property limits. The AI-Explorer generates and ranks candidate molecules to meet these goals
(Kwon et al. 2024). Furthermore, in gene analysis, the human-Controller prioritizes which biological
relationships are important for the search. The AI-Explorer can then use these relationship patterns to
predict gene pairs that cause cell death (Jiang et al. 2025b).
AI-Informer & Human-Explorer. The AI-Informer can gather evidence or predictions, while the
human-Explorer analyzes them to propose new candidates. For example, in biomedical research,
the AI-Informer integrates dispersed findings from the literature, helping the human-Explorer infer
potential hypotheses about relationships among biological factors (Jiang et al. 2025a). Additionally,
some AI-Informers can assist in retrieving relevant information from large volumes of data. For
instance, this can enable human-Explorers to explore promising compound candidates (Shi et al. 2023b)
or find similar image patches that support diagnostic theories (Corvò et al. 2021). In materials science,
the AI-Informer forecasts physical properties such as conductivity, allowing the human-Explorer to
combine these predictions to explore stable electrolytes for batteries (Pu et al. 2022).
3.2.3. Experiment Stage
The experiment stage involves designing and conducting tests to validate proposed hypotheses. In this phase, humans and AI collaborate to plan procedures and manage physical or computational processes to collect data.
AI-Controller & Human-Controller. During physical execution, the workflow functions as a shared control loop in which the AI and human manage different levels of the process. The AI-Controller handles immediate machine tasks, while the human-Controller directs the overall strategy. For instance, in robotic laboratories, the AI-Controller performs physical actions such as manipulating chemical samples. Human-Controller supervises the operation and updates targets based on real-time observations (Darvish et al. 2025). In materials laboratories, AI-Controller automates major steps from material preparation to characterization, while human-Controller provides oversight and adjusts actions based on the results (Ni et al. 2024). Similarly, in electron microscopy, the AI-Controller manages instrument settings to optimize data collection. Human-Controller monitors the live stream and directs the beam to explore relevant sample areas (Pratiush et al. 2024).
AI-Explorer & Human-Evaluator. When an experiment involves multiple steps, the AI-Explorer
drafts a step-by-step plan. The human-Evaluator then reviews the final plan, corrects any errors,
and determines whether it is usable. For example, some AI-Explorers draft chemistry workflows
by calling external chemistry tools during the planning phase, then return a complete procedure for
human-Evaluators to review (Boiko et al. 2023; Bran et al. 2024). In gene editing, an AI-Explorer
can generate an experimental plan, including suggested guide choices and key setup steps, with
a human-Evaluator reviewing the final output before execution (Qu et al. 2025). For multi-step
synthesis planning, the AI-Explorer proposes a full reaction route, and the human-Evaluator edits or
replaces problematic steps in the route before laboratory work begins (Shi et al. 2023a). Furthermore,
some AI-Explorers return a small set of candidate procedures along with a brief test plan, allowing
human-Evaluators to edit the selected option and finalize what will be executed (Bazgir et al. 2025;
Gottweis et al. 2025; Swanson et al. 2025).
AI-Informer & Human-Explorer. Before conducting physical experiments, the AI-Informer quickly
forecasts potential results. The human-Explorer navigates these predictions to find the optimal
experimental conditions. For instance, in material design, the AI-Informer predicts how structures
deform, while the human-Explorer searches the design space to drive configurations that achieve the
desired shape changes (Yang et al. 2020). For biological simulations, the AI-Informer predicts yeast cell
polarization, allowing the human-Explorer to navigate the parameter space and explore settings that
match real-world observations (Hazarika et al. 2020). In chemical synthesis, the AI-Informer maps out
alternative reaction pathways and provides risk estimates. Then the human-Explorer explores these
pathways to develop a practical plan for laboratory execution (Wang et al. 2025c).
3.2.4. Role Differences Across Three Stages
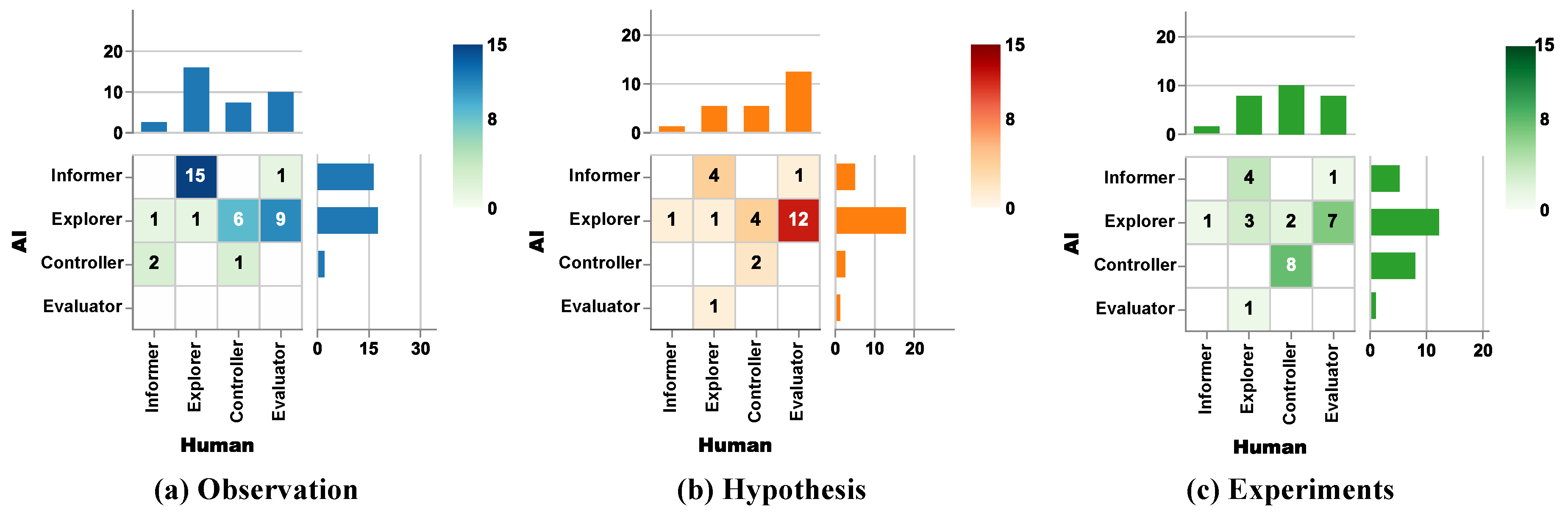
Figure 2 illustrates a distinct imbalance in the distribution of human and AI roles across the three stages of scientific discovery. The observation stage frequently features the combination of AI-Informer and human-Explorer. In contrast, the hypothesis stage shows a significant shift toward the pairing of AI-Explorer and human-Evaluator. The experiment stage reveals a trend in which AI-Controller and human-Controller collaborate in executing protocols.
This imbalance reflects the differing cognitive and operational demands across discovery stages. The observation stage is inherently open-ended, relying heavily on human insight to interpret phenomena and identify meaningful patterns, with AI primarily acting as an Informer that aggregates and summarizes data. As inquiry advances to the hypothesis stage, the problem space becomes more structured, allowing AI to systematically explore candidate hypotheses, while humans increasingly assume the Evaluator role to assess plausibility. In the experiment stage, the focus shifts to procedural execution, where requirements for correctness, safety, and reproducibility motivate a shared Controller role, with AI supporting automation and parameter control under human oversight.
These patterns indicate that human–AI role allocation in scientific discovery dynamically reconfigures as epistemic uncertainty decreases and task structure increases. Humans dominate stages that demand sensemaking under ambiguity and normative judgment, whereas AI gains prominence as tasks become formalizable and computationally searchable. Notably, the experiment stage reflects a convergence rather than a transfer of control, highlighting the need to preserve meaningful human authority even in highly automated settings. This suggests that effective human–AI collaboration should adapt role assignments across discovery stages, rather than imposing static responsibilities throughout the workflow.
4. Discussion
Built upon the key findings from our systematic survey, this section discusses several significant challenges and potential future research directions for human-AI collaboration in scientific discovery.
4.1. From Asymmetric Growth to Symbiotic Evolution
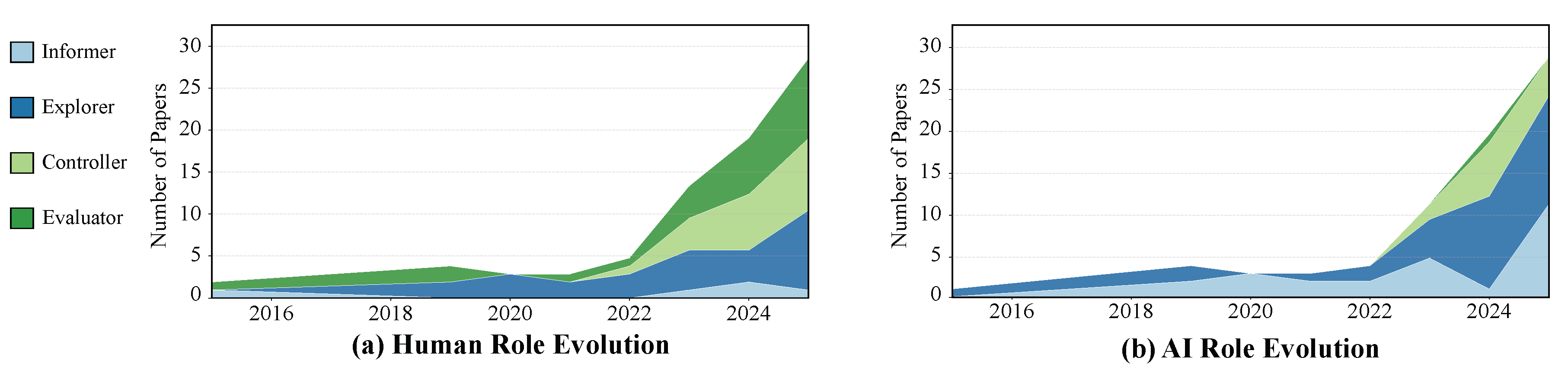
From
Figure 3, we can observe that the evolution of roles reveals a complementary pattern: AI’s development is specialized in computational tasks, while human involvement remains concentrated on high-judgment functions. Specifically,
Figure 3a indicates that humans are least used as Informers but dominate as Evaluators and Controllers, with a strong presence as Explorers. This confirms that humans anchor the process in critical judgment, contextual reasoning, creative thinking, and ethical decision-making—areas requiring deep expertise and accountability. In contrast,
Figure 3b shows that AI is predominantly deployed as an Informer and Explorer, with steady growth as a Controller. The Evaluator role remains minimal. This reflects a rational deployment of AI for its core strengths: processing data at scale, exploring solution spaces, and automating procedural workflows.
The scarcity of the AI-Evaluator is the most pronounced example of this lag, stemming
from fundamental technical gaps. Scientific evaluation requires: (1) Calibrated uncertainty
quantification, whereas current models often provide overconfident point estimates, risking unreliable
conclusions Heo et al. (2025); Xie et al. (2025). (2) Causal and mechanistic reasoning, beyond surface
pattern recognition; without understanding the underlying “why”, evaluations may be misled by
spurious correlations Chen et al. (2024). (3) Contextual and normative judgment, aligning with tacit
scientific standards–a challenge reflected in AI’s difficulty with complex, value-laden trade-offs Rezaei
et al. (2025). These challenges in reliability, depth, and contextual alignment currently limit AI’s role as
a primary evaluator. Notably, the AI-Controller role also depends on robust reasoning and trustworthy
autonomy. This shared need for reliability explains why its development, while progressing, remains
more cautious than that of the well-established Informer and Explorer roles.
To evolve from the current functional division into a deeper, symbiotic partnership, future research must address these core limitations across roles. One potential direction is the development of AI-Controllers that go beyond basic workflow execution toward verifiable robustness. This necessitates benchmarks for failure recovery and methods for explainable workflow logic, ensuring systems can be audited and trusted in dynamic environments. As for AI-Evaluators, the focus should shift from automation to calibrated assistance. The immediate goal should be to develop tools that provide uncertainty-quantified assessments and evidence-attributed rationales, augmenting rather than replacing human judgment. In addition, effective collaborative design demands interfaces that formally position the human as a strategic supervisor. These systems should streamline the oversight of AI-generated options, making the human’s role in guidance, interpretation, and final validation more efficient.
4.2. Generative Interfaces for Supporting Human Involvement
Most existing interfaces for human–AI collaboration in scientific discovery are highly customized, often tailored to specific domains or discovery tasks. While such designs enable deep integration with domain workflows, they limit reusability and hinder generalization to other contexts. Recent advances in generative AI for user interface generation offer new opportunities to address this limitation (Chen
et al. 2025). Rather than designing fixed, task-specific interfaces, future systems could dynamically generate interfaces that adapt to human responsibilities, such as exploration, evaluation, or control, throughout the scientific discovery process. Such context-aware interfaces have the potential to both enhance human oversight and maintain flexibility in exploration, paving the way for more effective human-AI collaboration.
However, there are still several challenges. First, generative UI can implicitly constrain human exploration pathways. In scientific discovery, research directions emerge through iterative choices about variables, comparisons, and analytical operations rather than being predefined. When a UI is generated dynamically, these choices are partially delegated to the interface, which determines what controls, views, and exploration paths are available. Consequently, valid lines of inquiry may remain unexplored—not due to lack of scientific merit, but because they are interactionally unavailable. Therefore, it is critical to consider how to design generative interfaces that preserve open-ended exploration without inadvertently narrowing or biasing scientific discovery. Second, hallucinations in generative UIs pose heightened risks. Scientific data are often heterogeneous and multimodal, and even governed by domain-specific constraints Sheng et al. (2025a); Wang et al. (2025c), which can further exacerbate hallucinations in generative AI. Future work should therefore focus on developing domain-aware generative models, benchmarks, and evaluation protocols that explicitly test whether generated interfaces respect data compatibility, experimental assumptions, and validity constraints.
4.3. Adaptive Role Assignments Between Human and AI
Existing human-AI collaborative research typically predefines the roles of human and AI based on their abilities and limitations to solve scientific discovery tasks (Hong et al. 2024; Jiang et al.
2024). However, scientific research is an inherently creative, exploratory, and non-linear process, in which research goals, hypotheses, and methods often evolve based on intermediate findings. Such static designs fail to adapt to the uncertainty in the discovery process. Fixed role assignments may overlook scenarios where AI demonstrates unexpected proficiency in non-traditional tasks or where human intervention becomes necessary due to contextual judgments that exceed the predefined scope of automation. These limitations necessitate a paradigm shift toward dynamic and adaptive role assignment between humans and AI. Rather than framing human–AI collaboration as a predefined division of labor, future research should conceptualize it as a problem of dynamic coordination. The roles of human and AI and their task allocation can be continuously negotiated based on contextual signals such as task difficulty, model confidence, experimental risk, and human cognitive load.
4.4. Empowering Embodied AI in Scientific Experiments
The papers in our corpus that address the experiment stage of scientific discovery primarily emphasize data analysis, while only a few investigate AI support for the practical execution of experimental processes. However, in domains like biology, chemistry, and medicine, scientific discovery fundamentally relies on a large number of physical laboratory experiments rather than data analysis alone (Luro et al. 2020; Wright et al. 2014). Embodied AI offers the potential to bridge this gap by converting AI models’ planning capabilities into concrete experimental actions and operating directly at the laboratory bench (Pratiush et al. 2024).It tightens the coupling between scientific research decision-making and execution, allowing errors or uncertainties in AI-generated plans to directly affect physical experiments. This raises the stakes of human oversight and requires more continuous, real-time engagement across both conceptual and operational levels. These shifts highlight the need to carefully consider how humans can be more effectively integrated into the loop.
4.5. Long-Term Implications for Leveraging AI in Scientific Discovery
As AI becomes increasingly powerful, the emergence of large language models has enabled systems to handle the entire research process, from hypothesis generation to experimental design and even paper writing (Schmidgall et al. 2025). In this context, it is increasingly important to consider the long-term implications of integrating AI into scientific workflows.
First, future work should establish standardized, auditable protocols for tracing AI involvement throughout the research process, rather than merely declaring AI usage. This can include the queries posed to AI systems, the full set of alternatives they generate, and the points at which human researchers intervene, modify, or reject AI suggestions. Such protocols enable process-level reproducibility, accountability, and responsible attribution of human and AI contributions, thereby supporting the long-term sustainability of the AI-augmented scientific ecosystem.
Second, the widespread use of AI in hypothesis exploration and decision-making raises important epistemic questions about how scientific reasoning may be reshaped over time. By mediating which hypotheses are generated, prioritized, or discarded, AI systems may systematically influence scientists’ exploration strategies and cognitive trajectories. Future research should empirically identify the stages at which reliance on AI may gradually reshape key forms of human judgment (e.g., intuition, value-based reasoning, or cross-domain insight) and assess whether such shifts introduce systematic bias or ethical risk.
5. Conclusion
In this work, we presented a systematic review of human-AI collaboration in scientific discovery, focusing on the roles of humans and AI across the stages of observation, hypothesis, and experiment. By introducing a novel taxonomy of four roles of human-AI collaboration (i.e., Informer, Explorer, Evaluator, and Controller), we provided a framework to better understand how AI and humans interact and contribute throughout the discovery process. Through our analysis, we identified key collaboration patterns and highlighted critical gaps, including challenges related to role coordination, validation, and transparency. Finally, we outlined a research agenda for developing more adaptive, trustworthy, and efficient human-AI systems.
Limitations
One limitation of this study is that we only included papers that explicitly address specific problems in the three stages of scientific discovery, while excluding more general-purpose papers, such as those aimed at assisting humans with tasks like writing or data collection. This choice was made because such tools operate outside the three main stages we focus on, serving more preparatory or subsequent roles. Nevertheless, these tools remain important, and future work could explore how they can be integrated into a broader framework of scientific discovery. Another limitation is that our analysis focuses exclusively on the natural sciences. This is because methodologies in the social sciences differ substantially, which may result in human–AI collaboration approaches that do not directly align with those observed in the natural sciences. Future work could explore whether our taxonomy can be adapted or extended to the social sciences.
Appendix A. Coding Results
References
- Mehrad Ansari, Jeffrey Watchorn, Carla E. Brown, and Joseph S Brown. 2024. dZiner: Rational Inverse Design of Materials with AI Agents. In AI for Accelerated Materials Design - NeurIPS 2024.
- Adib Bazgir, Rama chandra Praneeth Madugula, and Yuwen Zhang. 2025. MatAgent: A human-in-the-loop multi-agent LLM framework for accelerating the material science discovery cycle. In AI for Accelerated Materials Design - ICLR 2025.
- Daniil A. Boiko, Robert MacKnight, Ben Kline, and Gabe Gomes. 2023. Autonomous chemical research with large language models. Nature, 624(7992):570–578. [CrossRef]
- Andres M. Bran, Sam Cox, Oliver Schilter, Carlo Baldassari, Andrew D. White, and Philippe Schwaller. 2024. Augmenting large language models with chemistry tools. Nature Machine Intelligence, 6(5):525–535. [CrossRef]
- Jiaqi Chen, Yanzhe Zhang, Yutong Zhang, Yijia Shao, and Diyi Yang. 2025. Generative interfaces for language models. arXiv preprint arXiv:2508.19227.
- Yiming Chen, Chen Zhang, Danqing Luo, Luis Fernando D’Haro, Robby Tan, and Haizhou Li. 2024. Unveiling the Achilles’ Heel of NLG Evaluators: A Unified Adversarial Framework Driven by Large Language Models. In Findings of the Association for Computational Linguistics: ACL 2024, pages 1359–1375.
- A. Corvò, H. S. Garcia Caballero, M. A. Westenberg, M. A. van Driel, and J. J. van Wijk. 2021. Visual Analytics for Hypothesis-Driven Exploration in Computational Pathology. IEEE Transactions on Visualization and Computer Graphics, 27(10):3851–3866. [CrossRef]
- Kourosh Darvish, Marta Skreta, Yuchi Zhao, Naruki Yoshikawa, Sagnik Som, Miroslav Bogdanovic, Yang Cao, Han Hao, Haoping Xu, Alán Aspuru-Guzik, Animesh Garg, and Florian Shkurti. 2025. ORGANA: A robotic assistant for automated chemistry experimentation and characterization. Matter, 8(2):101897. [CrossRef]
- Mengjie Fan, Jinlu Yu, Daniel Weiskopf, Nan Cao, Huai-Yu Wang, and Liang Zhou. 2025. Visual Analysis of Multi-Outcome Causal Graphs. IEEE Transactions on Visualization and Computer Graphics, 31(1):656–666. [CrossRef]
- Carla Floricel, Nafiul Nipu, Mikayla Biggs, Andrew Wentzel, Guadalupe Canahuate, Lisanne Van Dijk, Abdallah Mohamed, C.David Fuller, and G.Elisabeta Marai. 2022. THALIS: Human-Machine Analysis of Longitudinal Symptoms in Cancer Therapy. IEEE Transactions on Visualization and Computer Graphics, 28(1):151–161.
- Carla Floricel, Andrew Wentzel, Abdallah Mohamed, C.David Fuller, Guadalupe Canahuate, and G.Elisabeta Marai. 2024. Roses Have Thorns: Understanding the Downside of Oncological Care Delivery Through Visual Analytics and Sequential Rule Mining. IEEE Transactions on Visualization and Computer Graphics, 30(1):1227–1237. [CrossRef]
- Juraj Gottweis, Wei-Hung Weng, Alexander Daryin, Tao Tu, Anil Palepu, Petar Sirkovic, Artiom Myaskovsky, Felix Weissenberger, Keran Rong, Ryutaro Tanno, and 1 others. 2025. Towards an AI Co-Scientist. arXiv preprint arXiv:2502.18864.
- Subhashis Hazarika, Haoyu Li, Ko-Chih Wang, Han-Wei Shen, and Ching-Shan Chou. 2020. NNVA: Neural Network Assisted Visual Analysis of Yeast Cell Polarization Simulation. IEEE Transactions on Visualization and Computer Graphics, 26(1):34–44.
- Juyeon Heo, Miao Xiong, Christina Heinze-Deml, and Jaya Narain. 2025. Do LLMs estimate uncertainty well in instruction-following? In The Thirteenth International Conference on Learning Representations.
- Jiayi Hong, Ross Maciejewski, Alain Trubuil, and Tobias Isenberg. 2024. Visualizing and Comparing Machine Learning Predictions to Improve Human-AI Teaming on the Example of Cell Lineage. IEEE Transactions on Visualization and Computer Graphics, 30(4):1956–1969. [CrossRef]
- Chen Huang, Yang Deng, Wenqiang Lei, Jiancheng Lv, Tat-Seng Chua, and Jimmy Huang. 2025. How to Enable Effective Cooperation Between Humans and NLP Models: A Survey of Principles, Formalizations, and Beyond. In Proceedings of the 63rd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 466–488.
- Peter Jansen, Oyvind Tafjord, Marissa Radensky, Pao Siangliulue, Tom Hope, Bhavana Dalvi Mishra, Bodhisattwa Prasad Majumder, Daniel S Weld, and Peter Clark. 2025. CodeScientist: End-to-end semi-automated scientific discovery with code-based experimentation. pages 13370–13467.
- Haejin Jeong, Hyoung-oh Jeong, Semin Lee, and Won-Ki Jeong. 2025. Optimal Dimensionality Selection Using Hull Heatmaps for Single-Cell Analysis. Computer Graphics Forum, 44(6):e70151.
- Haoran Jiang, Shaohan Shi, Yunjie Yao, Chang Jiang, and Quan Li. 2025a. HypoChainer: a Collaborative System Combining LLMs and Knowledge Graphs for Hypothesis-Driven Scientific Discovery. IEEE Transactions on Visualization and Computer Graphics, pages 1–11.
- Haoran Jiang, Shaohan Shi, Shuhao Zhang, Jie Zheng, and Quan Li. 2025b. SLInterpreter: An Exploratory and Iterative Human-AI Collaborative System for GNN-Based Synthetic Lethal Prediction. IEEE Transactions on Visualization and Computer Graphics, 31(1):919–929. [CrossRef]
- Zhihan Jiang, Handi Chen, Rui Zhou, Jing Deng, Xinchen Zhang, Running Zhao, Cong Xie, Yifang Wang, and Edith C.H. Ngai. 2024. HealthPrism: A Visual Analytics System for Exploring Children’s Physical and Mental Health Profiles with Multimodal Data . IEEE Transactions on Visualization & Computer Graphics, 30(01):1205–1215. [CrossRef]
- Tabassum Kakar, Xiao Qin, Elke A. Rundensteiner, Lane Harrison, Sanjay K. Sahoo, and Suranjan De. 2019. DIVA: Exploration and Validation of Hypothesized Drug-Drug Interactions. In Computer Graphics Forum, volume 38, pages 95–106. [CrossRef]
- Bharat Kale, Austin Clyde, Maoyuan Sun, Arvind Ramanathan, Rick Stevens, and Michael E. Papka. 2023. ChemoGraph: Interactive Visual Exploration of the Chemical Space. volume 42, pages 13–24. [CrossRef]
- Yuya Kawakami, Daniel Cayan, Dongyu Liu, and Kwan-Liu Ma. 2025. ClimateSOM: a Visual Analysis Workflow for Climate Ensemble Datasets. IEEE Transactions on Visualization and Computer Graphics, pages 1–11.
- Robert Krueger, Johanna Beyer, Won-Dong Jang, Nam Wook Kim, Artem Sokolov, Peter K. Sorger, and Hanspeter Pfister. 2020. Facetto: Combining Unsupervised and Supervised Learning for Hierarchical Phenotype Analysis in Multi-Channel Image Data. IEEE Transactions on Visualization and Computer Graphics, 26(1):227–237. [CrossRef]
- Bum Chul Kwon, Simona Rabinovici-Cohen, Beldine Moturi, Ruth Mwaura, Kezia Wahome, Oliver Njeru, Miguel Shinyenyi, Catherine Wanjiru, Sekou Remy, William Ogallo, Itai Guez, Partha Suryanarayanan, Joseph Morrone, Shreyans Sethi, Seung-Gu Kang, Tien Huynh, Kenney Ng, Diwakar Mahajan, Hongyang Li, and 4 others. 2024. SPARK: harnessing human-centered workflows with biomedical foundation models for drug discovery. IJCAI ’24.
- Shengchao Liu, Jiongxiao Wang, Yijin Yang, Chengpeng Wang, Ling Liu, Hongyu Guo, and Chaowei Xiao. 2024. Conversational Drug Editing Using Retrieval and Domain Feedback. In The Twelfth International Conference on Learning Representations (ICLR).
- Shuaihua Lu, Qionghua Zhou, Yixin Ouyang, Yilv Guo, Qiang Li, and Jinlan Wang. 2018. Accelerated Discovery of Stable Lead-Free Hybrid Organic-Inorganic Perovskites via Machine Learning. Nature Communications, 9(1):3405.
- Deng Luo, Zainab Alsuwaykit, Dawar Khan, Ondřej Strnad, Tobias Isenberg, and Ivan Viola. 2025. DiffFit: Visually-Guided Differentiable Fitting of Molecule Structures to a Cryo-EM Map. IEEE Transactions on Visualization and Computer Graphics, 31(1):558–568. [CrossRef]
- Stéphane Luro, Laurie Potvin-Trottier, Burak Okumus, and Johan Paulsson. 2020. Isolating live cells after high-throughput, long-term time-lapse microscopy. Nature Methods, 17:93–100. [CrossRef]
- Yubo Ma, Zhibin Gou, Junheng Hao, Ruochen Xu, Shuohang Wang, Liangming Pan, Yujiu Yang, Yixin Cao, and Aixin Sun. 2024. SciAgent: Tool-augmented language models for scientific reasoning. In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, pages 15701–15736.
- Vikram Mohanty, Jude Lim, and Kurt Luther. 2025. What Lies Beneath? Exploring the Impact of Underlying AI Model Updates in AI-Infused Systems. In Proceedings of the 2025 CHI Conference on Human Factors in Computing Systems, CHI ’25.
- Eric Mörth, Kevin Sidak, Zoltan Maliga, Torsten Möller, Nils Gehlenborg, Peter Sorger, Hanspeter Pfister, Johanna Beyer, and Robert Krüger. 2025. Cell2Cell: Explorative Cell Interaction Analysis in Multi-Volumetric Tissue Data. IEEE Transactions on Visualization and Computer Graphics, 31(1):569–579. [CrossRef]
- Ziqi Ni, Yahao Li, Kaijia Hu, Kunyuan Han, Ming Xu, Xingyu Chen, Fengqi Liu, Yicong Ye, and Shuxin Bai. 2024. MatPilot: An LLM-Enabled AI Materials Scientist Under the Framework of Human-Machine Collaboration. arXiv preprint arXiv:2411.08063.
- Raúl Ortega and Jose Manuel Gomez-Perez. 2025. SciClaims: An end-to-end generative system for biomedical claim analysis. In Proceedings of the 2025 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, pages 141–154.
- Reshika Palaniyappan Velumani, Meng Xia, Jun Han, Chaoli Wang, ALEXIS K LAU, and Huamin Qu. 2022. AQX: Explaining Air Quality Forecast for Verifying Domain Knowledge using Feature Importance Visualization. In Proceedings of the 27th International Conference on Intelligent User Interfaces, page 720–733.
- Ji Hwan Park, Vikash Prasad, Sydney Newsom, Fares Najar, and Rakhi Rajan. 2024. IdMotif: An Interactive Motif Identification in Protein Sequences. IEEE Computer Graphics and Applications, 44(3):114–125. [CrossRef]
- Utkarsh Pratiush, Gerd Duscher, and Sergei Kalinin. 2024. Human-in-the-loop interface for Automated experiments in Electron Microscopy, Automated characterization. In AI for Accelerated Materials Design - NeurIPS 2024.
- Jiansu Pu, Hui Shao, Boyang Gao, Zhengguo Zhu, Yanlin Zhu, Yunbo Rao, and Yong Xiang. 2022. matExplorer: Visual Exploration on Predicting Ionic Conductivity for Solid-state Electrolytes. IEEE Transactions on Visualization and Computer Graphics, 28(1):65–75. [CrossRef]
- Rui Qiu, Shijie Chen, Yu Su, Po-Yin Yen, and Han Wei Shen. 2025. Completing a systematic review in hours instead of months with interactive AI agents. In Proceedings of the 63rd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 31559–31593.
- Yuanhao Qu, Kaixuan Huang, Ming Yin, Kanghong Zhan, Dyllan Liu, Di Yin, Henry C. Cousins, William A. Johnson, Xiaotong Wang, Mihir Shah, and 1 others. 2025. CRISPR-GPT for agentic automation of gene-editing experiments. Nature Biomedical Engineering.
- Niroop Channa Rajashekar, Yeo Eun Shin, Yuan Pu, Sunny Chung, Kisung You, Mauro Giuffre, Colleen E Chan, Theo Saarinen, Allen Hsiao, Jasjeet Sekhon, Ambrose H Wong, Leigh V Evans, Rene F. Kizilcec, Loren Laine, Terika Mccall, and Dennis Shung. 2024. Human-Algorithmic Interaction Using a Large Language Model-Augmented Artificial Intelligence Clinical Decision Support System. In Proceedings of the 2024 CHI Conference on Human Factors in Computing Systems, CHI ’24.
- Sebastian Ratzenböck, Verena Obermüller, Torsten Möller, João Alves, and Immanuel M. Bomze. 2023. Uncover: Toward Interpretable Models for Detecting New Star Cluster Members. IEEE Transactions on Visualization and Computer Graphics, 29(9):3855–3872.
- Chandan K. Reddy and Parshin Shojaee. 2025. Towards scientific discovery with generative AI: progress, opportunities, and challenges. In Proceedings of the Thirty-Ninth AAAI Conference on Artificial Intelligence. AAAI Press. [CrossRef]
- MohammadHossein Rezaei, Yicheng Fu, Phil Cuvin, Caleb Ziems, Yanzhe Zhang, Hao Zhu, and Diyi Yang. 2025. EgoNormia: Benchmarking Physical-Social Norm Understanding. In Findings of the Association for Computational Linguistics: ACL 2025, pages 19256–19283.
- María Virginia Sabando, Pavol Ulbrich, Matías Selzer, Jan Byška, Jan Mičan, Ignacio Ponzoni, Axel J. Soto, María Luján Ganuza, and Barbora Kozlíková. 2021. ChemVA: Interactive Visual Analysis of Chemical Compound Similarity in Virtual Screening. IEEE Transactions on Visualization and Computer Graphics, 27(2):891–901. [CrossRef]
- Samuel Schmidgall, Yusheng Su, Ze Wang, Ximeng Sun, Jialian Wu, Xiaodong Yu, Jiang Liu, Michael Moor, Zicheng Liu, and Emad Barsoum. 2025. Agent laboratory: Using LLM agents as research assistants. In Findings of the Association for Computational Linguistics: EMNLP 2025, pages 5977–6043.
- Rui Sheng, Xingbo Wang, Jiachen Wang, Xiaofu Jin, Zhonghua Sheng, Zhenxing Xu, Suraj Rajendran, Huamin Qu, and Fei Wang. 2025a. Trialcompass: Visual analytics for enhancing the eligibility criteria design of clinical trials. IEEE Transactions on Visualization and Computer Graphics, pages 1–11.
- Rui Sheng, Zelin Zang, Jiachen Wang, Yan Luo, Zixin Chen, Yan Zhou, Shaolun Ruan, and Huamin Qu. 2025b. CellScout: Visual Analytics for Mining Biomarkers in Cell State Discovery. IEEE Transactions on Visualization and Computer Graphics, pages 1–16.
- Chuhan Shi, Yicheng Hu, Shenan Wang, Shuai Ma, Chengbo Zheng, Xiaojuan Ma, and Qiong Luo. 2023a. RetroLens: A Human-AI Collaborative System for Multi-step Retrosynthetic Route Planning.
- Chuhan Shi, Fei Nie, Yicheng Hu, Yige Xu, Lei Chen, Xiaojuan Ma, and Qiong Luo. 2023b. MedChemLens: An Interactive Visual Tool to Support Direction Selection in Interdisciplinary Experimental Research of Medicinal Chemistry. IEEE Transactions on Visualization and Computer Graphics, 29(1):63–73.
- Kyle Swanson, Wesley Wu, Nash L Bulaong, John E Pak, and James Zou. 2025. The Virtual Lab of AI agents designs new SARS-CoV-2 nanobodies. Nature, 646(8085):716–723.
- Jiabin Tang, Lianghao Xia, Zhonghang Li, and Chao Huang. 2025. AI-researcher: Autonomous scientific innovation. In The Thirty-ninth Annual Conference on Neural Information Processing Systems.
- Hanchen Wang, Tianfan Fu, Yuanqi Du, Wenhao Gao, Kexin Huang, Ziming Liu, Payal Chandak, Shengchao Liu, Peter Van Katwyk, Andreea Deac, and 1 others. 2023a. Scientific discovery in the age of artificial intelligence. Nature, 620(7972):47–60. [CrossRef]
- Qianwen Wang, Kexin Huang, Payal Chandak, Marinka Zitnik, and Nils Gehlenborg. 2023b. Extending the Nested Model for User-Centric XAI: A Design Study on GNN-based Drug Repurposing. IEEE Transactions on Visualization and Computer Graphics, 29(1):1266–1276. [CrossRef]
- Qingyun Wang, Rylan Schaeffer, Fereshte Khani, Filip Ilievski, and 1 others. 2025a. Scideator: Human-LLM Scientific Idea Generation Grounded in Research-Paper Facet Recombination. Transactions on Machine Learning Research.
- Qipeng Wang, Shaolun Ruan, Rui Sheng, Yong Wang, Min Zhu, and Huamin Qu. 2025b. Trajlens: Visual analysis for constructing cell developmental trajectories in cross-sample exploration. IEEE Transactions on Visualization and Computer Graphics, pages 1–11.
- Qipeng Wang, Rui Sheng, Shaolun Ruan, Xiaofu Jin, Chuhan Shi, and Min Zhu. 2025c. SynthLens: Visual Analytics for Facilitating Multi-Step Synthetic Route Design. IEEE Transactions on Visualization and Computer Graphics, 31(10):7647–7660.
- Xingbo Wang, Samantha L. Huey, Rui Sheng, Saurabh Mehta, and Fei Wang. 2025d. SciDaSynth: Interactive Structured Data Extraction From Scientific Literature With Large Language Model. Campbell Systematic Reviews, 21(1):e70073.
- Zhizheng Wang, Qiao Jin, Chih-Hsuan Wei, Shubo Tian, Po-Ting Lai, Qingqing Zhu, Chi-Ping Day, Christina Ross, Robert Leaman, and Zhiyong Lu. 2025e. GeneAgent: Self-verification language agent for gene-set analysis using domain databases. Nature Methods, 22(8):1677–1685. [CrossRef]
- Andrew Wentzel, Carla Floricel, Guadalupe Canahuate, Mohamed A. Naser, Abdallah S. Mohamed, Clifton D. Fuller, Lisanne van Dijk, and G. Elisabeta Marai. 2023. DASS Good: Explainable Data Mining of Spatial Cohort Data. Computer Graphics Forum, 42(3):283–295. [CrossRef]
- Austin P Wright, Peter Nemere, Adrian Galvin, Duen Horng Chau, and Scott Davidoff. 2023. Lessons from the development of an anomaly detection interface on the mars perseverance rover using the ishmap framework. In Proceedings of the 28th International Conference on Intelligent User Interfaces, page 91–105.
- Peter M. Wright, Ian B. Seiple, and Andrew G. Myers. 2014. The evolving role of chemical synthesis in antibacterial drug discovery. Angewandte Chemie International Edition, 53(34):8840–8869. [CrossRef]
- Qiujie Xie, Qingqiu Li, Zhuohao Yu, Yuejie Zhang, Yue Zhang, and Linyi Yang. 2025. An Empirical Analysis of Uncertainty in Large Language Model Evaluations. In The Thirteenth International Conference on Learning Representations.
- Qi Xin, Quyu Kong, Hongyi Ji, Yue Shen, Yuqi Liu, Yan Sun, Zhilin Zhang, Zhaorong Li, Xunlong Xia, Bing Deng, and Yinqi Bai. 2024. BioInformatics Agent (BIA): Unleashing the Power of Large Language Models to Reshape Bioinformatics Workflow. bioRxiv.
- Chaoqing Xu, Tyson Neuroth, Takanori Fujiwara, Ronghua Liang, and Kwan-Liu Ma. 2023. A Predictive Visual Analytics System for Studying Neurodegenerative Disease Based on DTI Fiber Tracts. IEEE Transactions on Visualization and Computer Graphics, 29(4):2020–2035. [CrossRef]
- Humphrey Yang, Kuanren Qian, Haolin Liu, Yuxuan Yu, Jianzhe Gu, Matthew McGehee, Yongjie Jessica Zhang, and Lining Yao. 2020. SimuLearn: Fast and Accurate Simulator to Support Morphing Materials Design and Workflows. In Proceedings of the 33rd Annual ACM Symposium on User Interface Software and Technology, UIST ’20, page 71–84.
- Jun-Hsiang Yao, Mingzheng Li, Jiayi Liu, Yuxiao Li, Jielin Feng, Jun Han, Qibao Zheng, Jianfeng Feng, and Siming Chen. 2025. DTBIA: An Immersive Visual Analytics System for Brain-Inspired Research. IEEE Transactions on Visualization and Computer Graphics, 31(6):3796–3808.
- Shunyu Yao, Jeffrey Zhao, Dian Yu, Nan Du, Izhak Shafran, Karthik Narasimhan, and Yuan Cao. 2023. ReAct: Synergizing Reasoning and Acting in Language Models. In International Conference on Learning Representations. [CrossRef]
- Fei Ye, Zaixiang Zheng, Dongyu Xue, Yuning Shen, Lihao Wang, Yiming Ma, Yan Wang, Xinyou Wang, Xiangxin Zhou, and Quanquan Gu. 2025a. Proteinbench: A holistic evaluation of protein foundation models. In International Conference on Representation Learning, volume 2025, pages 29857–29891.
- Geyan Ye, Xibao Cai, Houtim Lai, Xing Wang, Junhong Huang, Longyue Wang, Wei Liu, and Xiangxiang Zeng. 2025b. DrugAssist: a large language model for molecule optimization. Briefings in Bioinformatics, 26(1):bbae693.
- Yang Zhang, Zhewei Wei, Ye Yuan, Chongxuan Li, and Wenbing Huang. 2024a. EquiPocket: an e(3)-equivariant geometric graph neural network for ligand binding site prediction. In Proceedings of the 41st International Conference on Machine Learning, volume 235, pages 60021–60039.
- Yu Zhang, Xiusi Chen, Bowen Jin, Sheng Wang, Shuiwang Ji, Wei Wang, and Jiawei Han. 2024b. A Comprehensive Survey of Scientific Large Language Models and Their Applications in Scientific Discovery. In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, pages 8783–8817.
- Tianshi Zheng, Zheye Deng, Hong Ting Tsang, Weiqi Wang, Jiaxin Bai, Zihao Wang, and Yangqiu Song. 2025. From Automation to Autonomy: A Survey on Large Language Models in Scientific Discovery. pages 17744–17761.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).