Submitted:
31 December 2025
Posted:
01 January 2026
You are already at the latest version
Abstract
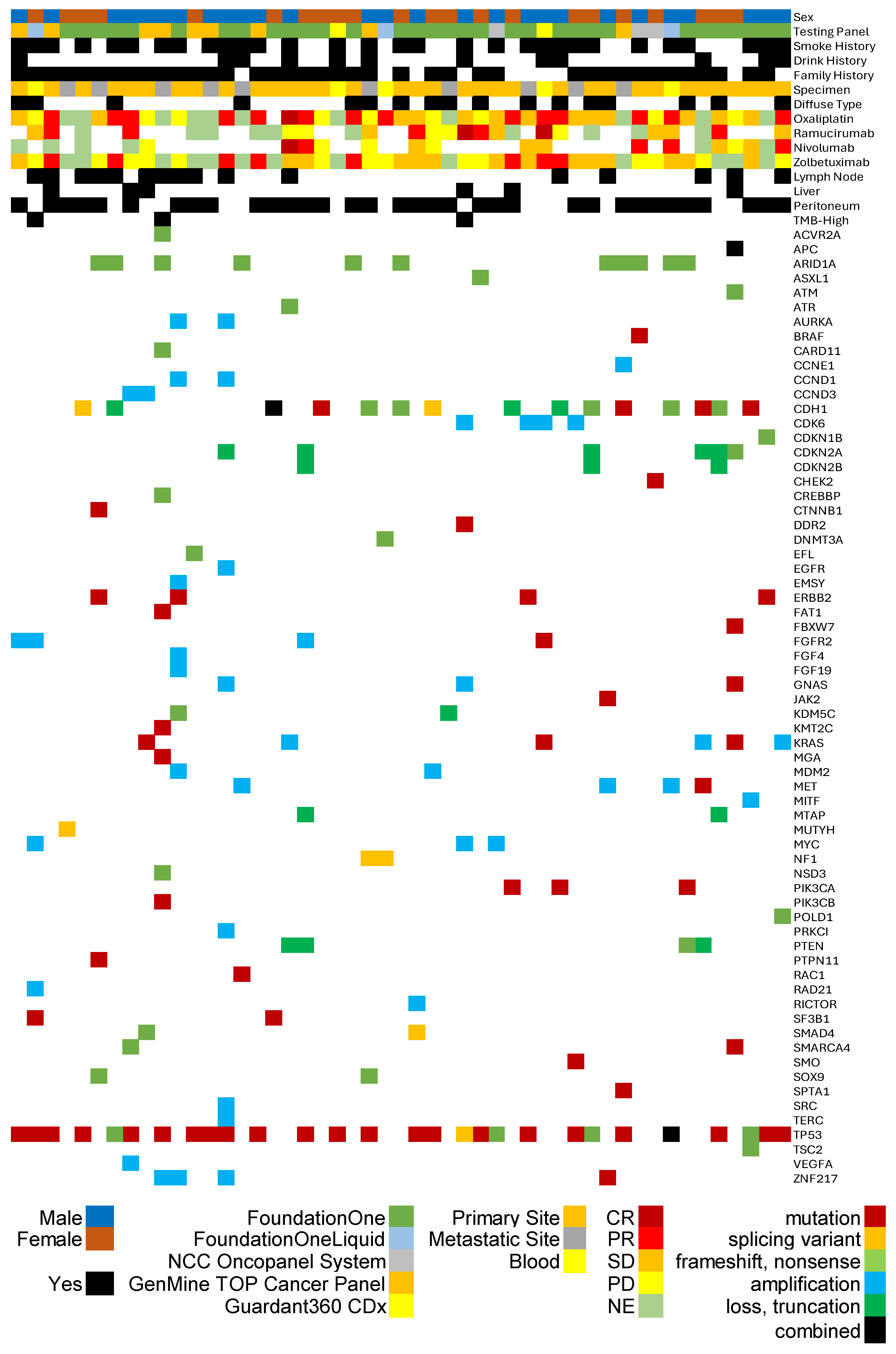
Background/Objectives: The anti-CLDN18.2 antibody zolbetuximab has emerged as a novel therapeutic option for advanced gastric adenocarcinoma. However, robust predictive biomarkers for its efficacy remain an unmet need. Methods: Utilizing the Japanese Center for Cancer Genomics and Advanced Therapeutics database, we retrospectively analyzed the clinical and genomic profiles of 49 patients with gastric adenocarcinoma who received zolbetuximab-containing regimens. Due to Japanese health insurance regulations, these patients were deemed to have CLDN18.2-positive tumors. We explored the association between objective response rate (ORR) and concurrent genomic alterations, focusing on tumor mutational burden (TMB) and major mutations (TP53, ARID1A, CDH1). Results: The ORR to zolbetuximab-based therapy in this cohort was 22.2%. Statistical analysis revealed a trend toward higher clinical response in patients with lower TMB (median 1.82 in responders vs. 4.0 in non-responders; p=0.050). Furthermore, patients without a CDH1 single nucleotide variant also showed a suggestive trend toward better response (p=0.086). No significant associations were found with TP53 or ARID1A alterations (p=0.787 and p=0.239, respectively). Conclusions: Our findings suggest that low TMB and the absence of CDH1 variants may serve as potential predictive biomarkers for response to zolbetuximab in CLDN18.2-positive gastric cancer. Prospective validation is warranted to maximize patient selection for this targeted therapy.
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Overview of C-CAT Registered Cases and Gastric Cancer Cohort
3.2. Patient and Genomic Characteristics of Zolbetuximab-Treated Cohort

3.3. Assessment of Treatment Efficacy and Genomic Predictors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GC | Gastric Cancer |
| ICI | Immune Checkpoint Inhibitor |
| CLDN18.2 | Claudin 18.2 |
| ADCC | Antibody-Dependent Cell-mediated Cytotoxicity |
| C-CAT | Center for Cancer Genomics and Advanced Therapeutics |
| TMB | Tumor Mutational Burden |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| CR | Complete Response |
| PR | Partial Response |
| SD | Stable Disease |
| PD | Progressive Disease |
| NE | Not Evaluated |
| ORR | Objective Response Rate |
| SNV | Single Nucleotide Variant |
| TCGA | The Cancer Gnome Atlas |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J Clin 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Chen, L.T.; Ryu, M.H.; Oh, D.Y.; Oh, S.C.; Chung, H.C.; Lee, K.W.; Omori, T.; Shitara, K.; Sakuramoto, S.; et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022, 23, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Kawazoe, A.; Bai, Y.; Xu, J.; Lonardi, S.; Metges, J.P.; Yanez, P.; Wyrwicz, L.S.; Shen, L.; Ostapenko, Y.; et al. Pembrolizumab plus trastuzumab and chemotherapy for HER2-positive gastric or gastro-oesophageal junction adenocarcinoma: interim analyses from the phase 3 KEYNOTE-811 randomised placebo-controlled trial. Lancet 2023, 402, 2197–2208. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Z.; Jiang, W.; Zhang, Y.; Meng, Z.; Niu, Y.; Sheng, Z.; Chen, C.; Liu, X.; Chen, X.; et al. CLDN18.2 and 4-1BB bispecific antibody givastomig exerts antitumor activity through CLDN18.2-expressing tumor-directed T-cell activation. J Immunother Cancer 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Shitara, K. Zolbetuximab for Claudin18.2-positive gastric or gastroesophageal junction cancer. Ther Adv Med Oncol 2024, 16, 17588359231217967. [Google Scholar] [CrossRef] [PubMed]
- Türeci; Mitnacht-Kraus, R.; Wöll, S.; Yamada, T.; Sahin, U. Characterization of zolbetuximab in pancreatic cancer models. Oncoimmunology 2019, 8, e1523096. [Google Scholar] [CrossRef] [PubMed]
- Shitara, K.; Lordick, F.; Bang, Y.J.; Enzinger, P.; Ilson, D.; Shah, M.A.; Van Cutsem, E.; Xu, R.H.; Aprile, G.; Xu, J.; et al. Zolbetuximab plus mFOLFOX6 in patients with CLDN18.2-positive, HER2-negative, untreated, locally advanced unresectable or metastatic gastric or gastro-oesophageal junction adenocarcinoma (SPOTLIGHT): a multicentre, randomised, double-blind, phase 3 trial. Lancet 2023, 401, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Shitara, K.; Ajani, J.A.; Bang, Y.J.; Enzinger, P.; Ilson, D.; Lordick, F.; Van Cutsem, E.; Gallego Plazas, J.; Huang, J.; et al. Zolbetuximab plus CAPOX in CLDN18.2-positive gastric or gastroesophageal junction adenocarcinoma: the randomized, phase 3 GLOW trial. Nat Med 2023, 29, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Kato, M.; Kohsaka, S.; Sudo, T.; Tamai, I.; Shiraishi, Y.; Okuma, Y.; Ogasawara, D.; Suzuki, T.; Yoshida, T.; et al. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer Discov 2022, 12, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Saito, Y. Genomic Analysis of Advanced Phyllodes Tumors Using Next-Generation Sequencing and Their Chemotherapy Response: A Retrospective Study Using the C-CAT Database. Medicina (Kaunas) 2024, 60. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Seino, M.; Sato, H.; Saito, Y.; Saito, K.; Yamada, Y.; Takahashi, K.; Kumanishi, R.; Fukui, T. Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database. Curr Issues Mol Biol 2025, 47. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020, 21, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Corso, G.; Comelli, G.; Veronesi, P.; Bianchi, B.; Petitto, S.; Polizzi, A.; Girardi, A.; Cioffi, A.; La Vecchia, C.; Bagnardi, V.; et al. Germline CDH1 variants in hereditary diffuse gastric cancer syndrome with focus on younger women. J Cancer Res Clin Oncol 2023, 149, 16147–16155. [Google Scholar] [CrossRef] [PubMed]
- Lobo, S.; Dias, A.; Pedro, A.M.; Ferreira, M.; Pinto-Oliveira, A.; São José, C.; Herrera-Mullar, J.; Pinto, N.; Colas, C.; Hüneburg, R.; et al. Hereditary diffuse gastric cancer spectrum associated with germline CTNNA1 loss of function revealed by clinical and molecular data from 351 carrier families and over 37 000 non-carrier controls. Gut 2025. [Google Scholar] [CrossRef] [PubMed]
| Features | FoundationOne CDx |
FoundationOne Liquid CDx |
NCC Oncopanel System |
Guardant360 CDx | GenMine TOP Cancer Panel |
|---|---|---|---|---|---|
| Sample Type | FFPE Tissue | Blood | FFPE Tissue and Blood |
Blood | FFPE Tissue and Blood |
| Number of Genes | 324 | 324 | 124 | 74 | 723 |
| MSI Testing | Yes | Yes | Yes * | Yes | Yes * |
| TMB Assesment | Yes | Yes | Yes | No | Yes |
| Minimum Tumor Content Required |
20% | N/A | 20% | N/A | 20% |
| Required DNA Input | 50 ng | 2 tubes | 50 ng | 2 tubes** | 50 ng |
| Total Cases (n = 110,125) | |||
|---|---|---|---|
| Primary Site | Cancer GenomicsTest | ||
| Pancreas | 17789 (16.2%) | FoundationOne CDx | 74409 (67.6%) |
| Colorectal | 17595 (16.0%) | FoundationOne Liquid CDx | 16492 (15.0%) |
| Bile Duct | 9679 (8.8%) | NCC Oncopanel System | 10078 (9.2%) |
| Breast | 8846 (8.0%) | GenMineTM TOP Cancer Panel | 6132 (5.6%) |
| Lung | 6646 (6.0%) | Guardant360 CDx | 2744 (2.5%) |
| Esophagus/Stomach | 6593 (6.0%) | ||
| Prostate | 6585 (6.0%) | Age Group (years) | |
| Ovary/Fallopian Tube | 5822 (5.3%) | 70-79 | 32843 (29.8%) |
| Soft Tissue | 4151 (3.8%) | 60-69 | 30868 (28.0%) |
| Uterus | 3606 (3.3%) | 50-59 | 23017 (20.9%) |
| Others | 22843 (20.7%) | 40-49 | 11212 (10.2%) |
| 80-89 | 5338 (4.8%) | ||
| Sex | 30-29 | 3700 (3.6%) | |
| Male | 55272 (50.2%) | 20-29 | 1235 (1.1%) |
| Female | 54878 (49.8%) | 10-19 | 1047 (1.0%) |
| Unknown | 5 (0.0%) | 0-9 | 901 (0.8%) |
| 90- | 81 (0.1%) | ||
| Gastric Cancer Cases (n=2,483) | |||
|---|---|---|---|
| Cancer GenomicsTest | Treatment Response to Oxaliplatin* | ||
| FoundationOne CDx | 1736 (69.9%) | Complete Response | 28 (1.5%) |
| FoundationOne Liquid CDx | 316 (12.7%) | Partial Response | 689 (37.8%) |
| NCC Oncopanel System | 250 (10.1%) | Stable Disease | 662 (36.4%) |
| GenMineTM TOP Cancer Panel | 121 (4.9%) | Progressive Disease | 442 (24.3%) |
| Guardant360 CDx | 60 (2.4%) | Not Evaluated | 336 |
| Sex | Treatment Response toNivolumab* | ||
| Male | 1650 (66.5%) | Complete Response | 19 (1.3%) |
| Female | 833 (33.5%) | Partial Response | 388 (26.0%) |
| Stable Disease | 515 (34.5%) | ||
| Age Group (years) | Progressive Disease | 573 (38.3%) | |
| 70-79 | 902 (36.3%) | Not Evaluated | 325 |
| 60-69 | 704 (28.4%) | ||
| 50-59 | 374 (15.1%) | Treatment Response to Zolbetuximab* | |
| 40-49 | 230 (9.3%) | Complete Response | 0 |
| 80-89 | 125 (5.0%) | Partial Response | 8 (22.2%) |
| 30-39 | 111 (4.5%) | Stable Disease | 14 (38.9%) |
| 20-29 | 30 (1.2%) | Progressive Disease | 14 (38.9%) |
| 10-19 | 5 (0.2%) | Not Evaluated | 13 |
| 90- | 2 (0.1%) | ||
| Pathological Classification | |||
| Diffuse | 511 (20.6%) | ||
| Tubular | 503 (20.3%) | ||
| Intestinal | 232 (9.3%) | ||
| Mucinous | 33 (1.3%) | ||
| Papillary | 17 (0.7%) | ||
| Not Other Specified or Unknown |
1187 (47.8%) | ||
| Gastric Cancer Cases Treated with Zolbetuximab-based regimens (N=49) | |||
|---|---|---|---|
| Age Group (years; median 62) | Metastatic Sites | ||
| 60-69 | 14 (28.6%) | Peritoneum | 34 (69.4%) |
| 70-79 | 12 (24.5%) | Lymph Node | 16 (32.7%) |
| 50-59 | 8 (16.3%) | Liver | 6 (12.2%) |
| 30-39 | 7 (14.3%) | ||
| 40-49 | 7 (14.3%) | Treatment Response to Oxaliplatin* | |
| 80-89 | 1 (2.0%) | Complete Response | 1 (2.7%) |
| Partial Response | 14 (37.8%) | ||
| Sex | Stable Disease | 13 (35.1%) | |
| Male | 27 (55.1%) | Progressive Disease | 9 (24.3%) |
| Female | 22 (44.9%) | Not Evaluated | 11 |
| Smoking History | Treatment Response toNivolumab* | ||
| No | 30 (61.2%) | Complete Response | 1 (4.8%) |
| Yes | 18 (36.7%) | Partial Response | 4 (19.0%) |
| Unknown | 1 (2.0%) | Stable Disease | 5 (23.8%) |
| Progressive Disease | 5 (23.8%) | ||
| Drinking History | Not Evaluated | 6 | |
| No | 35 (71.4%) | ||
| Yes | 12 (36.7%) | Treatment Response to Zolbetuximab* | |
| Unknown | 2 (4.1%) | Complete Response | 0 |
| Partial Response | 8 (22.2%) | ||
| CancerTestingPanel | Stable Disease | 14 (38.9%) | |
| FoundationOne CDx | 32 (65.3%) | Progressive Disease | 14 (38.9%) |
| FoundationOne Liquid CDx | 3 (6.1%) | Not Evaluated | 13 |
| NCC Oncopanel System | 3 (6.1%) | ||
| GenMineTM TOP Cancer Panel | 9 (18.4%) | Treatment Line of Zolbetuximab* | |
| Guardant360 CDx | 2 (4.1%) | 1st line | 23 |
| 2nd line | 9 | ||
| Pathological Classification | 3rd line | 5 | |
| Diffuse | 17 (34.7%) | 4th line | 3 |
| Tubular | 7 (14.3%) | 5th line or Later | 8 |
| Intestinal | 1 (2.0%) | unknown | 1 |
| Not Other Specified or Unknown |
22 (44.5%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





