Submitted:
18 December 2025
Posted:
19 December 2025
You are already at the latest version
Abstract
Brain health monitoring is increasingly essential as modern cognitive load, stress, and lifestyle pressures contribute to widespread neural instability. The paper introduces BrainTwin, a next-generation cognitive digital twin that integrates advanced MRI analytics for comprehensive neuro-oncological assessment with real-time EEG–based brain health intelligence.Structural analysis is driven by an Enhanced Vision Transformer (ViT++), which improves spatial representation and boundary localization, achieving more accurate tumor prediction than conventional models. Extracted tumor volume forms the baseline for short-horizon tumor progression modeling. Parallel to MRI analysis, continuous EEG signals are captured through an in-house wearable skullcap, preprocessed using Edge AI on a Hailo Toolkit–enabled Raspberry Pi 5 for low-latency denoising and secure cloud transmission. Pre-processed EEG packets are authenticated at the fog layer ensuring secure and reliable cloud transfer, enabling significant load reduction in the edge and cloud nodes. In the digital twin, EEG characteristics offer real-time functional monitoring through dynamic brain-wave analysis,while a BiLSTM classifier distinguishes relaxed, stress, and fatigue states. Unlike static MRI imaging, EEG provides real-time brain health monitoring. The Brain-Twin performs EEG–MRI fusion, co-relating functional EEG metrics with ViT++ structural embeddings to produce a single risk score that can be interpreted by clinicians to determine brain vulnerability to future diseases. Explainable artificial intelligence (XAI) provides clinical interpretability through Gradient weighted class activation mapping (Grad-CAM) heatmaps, which are used to interpret ViT++ decisions and are visualized on a 3D interactive brain model to allow more in-depth inspection of spatial details. The evaluation metrics demonstrate a BiLSTM macro-F1 of 0.94 (Precision/ Recall/ F1: Relaxed 0.96, Stress 0.93, Fatigue 0.92) and ViT++ MRI accuracy of 96% outperforming baseline architectures. These results demonstrate BrainTwin’s reliability, interpretability, and clinical utility as an integrated digital companion for tumor assessment and real-time functional brain monitoring.
Keywords:
1. Introduction
2. Related Works
| ARCHITECTURE | NOVELTY | EVALUATION METRICS | DRAWBACKS | BRAINTWIN SOLUTION | REFERENCE |
|---|---|---|---|---|---|
| Attention-based Residual Network-152V2 (ResNet-152V2) + PCA + DCGAN | Integrates attention mechanisms for focused hemorrhage feature extraction, PCA for dimensionality reduction, and DCGAN-based synthesis to compensate for minority intracranial hemorrhage subtypes in digital twin applications. | Accuracy: 99.2% (Epidural), 97.1% (Intraparenchymal) |
Heavy reliance on synthetic images increases overfitting risk; lacks cross-dataset generalization; no explainable AI, limiting clinical interpretability. | Our digital twin enables MRI–EEG fusion in real time removing the dependence on synthetic augmentation. It incorporates Vision Transformer++ with Grad-CAM explainability, ensuring transparency, and uses edge–fog processing for robust generalization to diverse clinical conditions. | Aftab Hussain et al. [1] |
| Transfer Learning on Tangent Space with SVM (TL-TSS) + Riemannian Manifold EEG Analysis | Employs cognitive computing and Riemannian geometry to extract robust EEG features, enabling high-accuracy motor imagery decoding for BCI-driven digital twins. | Accuracy: up to 97.88%; High kappa & transfer accuracy across datasets | Limited to motor imagery tasks; no MRI or multimodal integration; lacks transformer-based scalability and edge deployment. | Our model overcomes this by integrating functional EEG and structural MRI which has facilitated broader neurological coverage than motor imagery. Enhanced ViT++ and edge preprocessing on Raspberry Pi ensures scalability and applicability in real-time. | Zhihan Lv et al. [2] |
| Deep CNN without Batch Normalization + Adaptive MRI–PET/SPECT Fusion | Introduces a customized loss function to allow for convergence stability; excludes batch normalization layers, improving model’s training time; and develops a novel adaptive method that preserves the integrity of both clinical information and spatial information through multimodal fusion. | PSNR: 34.11 dB; SSIM: 85.24% | Offline-only processing; no real-time inference; lacks EEG functional integration and digital-twin updating mechanisms. | Our digital twin performs continuous real-time MRI–EEG monitoring, supports dynamic twin updates, and provides 3D visualization of tissue-level states with real-time inference, addressing the limitations of offline processing. | Jinxia Wang et al. [3] |
| DTBIA — Digital Twin-Based Brain-Inspired Analytics (VR Interface) | Provides an immersive VR-driven visualization engine enabling exploration of BOLD and DTI signals at voxel and regional resolutions. | Qualitative user feedback; no quantitative metrics reported | Requires expensive VR/GPU hardware; lacks predictive modeling, EEG integration, and real-time physiological inputs. | Our system uses lightweight 3D brain visualization without VR, integrates MRI+EEG, and includes a Tumor Kinetics Engine for forecasting, achieving predictive modeling and real-time functionality without costly hardware. | Yao et al. [4] |
| RF Backscatter Sensing + Stacked Autoencoder + Fine-Tuned KNN Classifier | Uses wearable ultra-wideband RF sensors and machine learning for portable, real-time stroke monitoring within a lightweight digital twin framework. | Binary Accuracy: 93.4%; Multiclass Accuracy: 92.3% | Not validated on clinical EEG; reactive rather than predictive; lacks explainability, multimodal fusion, and visualization. | Our digital twin incorporates predictive tumor/stroke progression modeling, XAI-based visual explanations, and 3D neuro-visualization, enabling proactive monitoring validated on real multimodal data. | Sagheer Khan et al. [5] |
| Blockchain-Enabled Digital Twin + Logistic Regression | Introduces a secure, decentralized twin architecture for stroke prediction using blockchain for tamper-proof data exchange and logistic regression for classification. | Overall Accuracy: 98.28% | Works only with static datasets; no imaging or EEG support; no real-time streaming or visualization. | Our model supports real-time continuous data flow, integrates MRI and EEG, performs dynamic updates at the edge and fog layers, and includes interactive 3D visualization, addressing all missing components. | Upadrista et al. [6] |
| Neuro-Symbolic Reasoning with GRU-Based Neural Translator | Enables voice-based interaction with digital twins by translating natural language into symbolic logic executed on annotated 3D models. | BLEU: 0.989; Translation Accuracy: 96.2%; Failure Rate: 0.2% | Not healthcare-specific; lacks multimodal physiological integration; no real-time clinical data ingestion. | Our twin incorporates multimodal MRI–EEG streaming, enabling real-time physiological analysis. It also supports explainability and forecasting, going far beyond symbolic interaction alone. | Siyaev et al. [7] |
| IoT-Enabled MRI Pipeline + CNN/SVM/ELM with PSO Feature Selection | Uses IoT-based data acquisition with PSO for optimal MRI feature selection and evaluates CNN, SVM, and ELM for tumor classification in a cloud-based digital twin. | CNN achieved highest performance; training and execution times reported | No explainable AI (Grad-CAM/SHAP); cloud latency issues; no EEG integration; not multimodal or real-time. | Our edge–fog–cloud architecture minimizes latency, integrates both MRI and EEG, provides Grad-CAM explainability, and ensures real-time digital-twin responsiveness. | Sultanpure et al. [8] |
| S3VM + Graph-Based Similarity Learning + Improved AlexNet | Combines semi-supervised learning and graph-based similarity to exploit both labeled and unlabeled MRI data; modifies AlexNet pooling and normalization for improved segmentation. | Accuracy: 92.52%; DSC: 75.58%; Jaccard: 79.55%; RMSE: 4.91%; MAE: 5.59% | Requires manual hyperparameter tuning; no real-time streaming; lacks EEG integration, XAI, and dynamic visualization. | Our model automates feature extraction via ViT++, integrates EEG functional data, supports explainability, and provides real-time 3D visualization, surpassing static semi-supervised approaches. | Wan et al. [9] |
| MARS + Mixed Spline Regression (B-Spline Basis + Toeplitz Covariance) | Models digital twins of brain aging to detect thalamic atrophy in MS years before clinical onset; constructs disease-specific aging curves using multi-cohort MRI. | Onset Detection: 5–6 years earlier; Repeated Measure Correlation: 0.88 | Requires large longitudinal datasets; no real-time inference; lacks multimodal or functional tracking. | Our digital twin tracks MRI volume changes and EEG cognitive patterns in real time, enabling multimodal functional-structural monitoring without requiring massive longitudinal datasets. | Steven Cen et al. [10] |
| BTSC-TNAS — Nested U-Shape CNN + Transformer with NAS-Searched Blocks | Joint segmentation-classification architecture using neural architecture search to optimize transformer and CNN feature extraction for brain tumors. | Dice: 80.9% (Tumor), 87.1% (Abnormal); Accuracy: 0.941 | Structural-only MRI; no functional data; no real-time processing capability. | Our twin integrates multimodal MRI–EEG, offers real-time edge processing, and provides explainable tumor insights not possible with structural-only offline architectures. | Liu et al. [11] |
| CKD-TransBTS — Hybrid CNN–Transformer with Modality Pairing (MCCA + TCFC) | Introduces clinically informed modality grouping (T1+T1Gd, T2+T2FLAIR), Modality-Correlated Cross-Attention (MCCA), and feature calibration via TCFC for efficient multimodal MRI segmentation. | Dice (BraTS2021): ET = 0.8850, TC = 0.9066, WT > 0.92; HD95: 5.93–7.60 mm | Works offline only; structural imaging only; lacks explainability and dynamic digital-twin updates. | Our digital twin uses multimodal integration, explainable ViT++, and dynamic fog-layer updates to provide transparent, continuously updated predictions. | Lin et al. [12] |
| PBViT — Patch-Based Vision Transformer + DenseNet Blocks + Custom CNN Kernel | Uses spatial patch tokenization with transformer encoders and DenseNet connections to enhance representation learning; includes ablation studies on patch size and encoder depth. | Accuracy: 95.8%; Precision: 95.3%; Recall: 93.2%; F1: 92% | No multimodal data; lacks real-time operation; no predictive modeling or XAI. | Our system fuses MRI with EEG cognitive information, enabling predictive modeling via a Tumor Kinetics Engine and delivering real-time, clinically actionable insights. | Chauhan et al. [13] |
| LTSpice-Modeled EEG Acquisition Twin + Random Forest/ANN Denoising | Creates a digital twin of the EEG acquisition chain by modeling electrode–skin–amplifier dynamics in LTSpice and applying supervised ML to denoise EEG signals, improving electrophysiological fidelity. | Metrics: R², MSE, RMSE, MAE (Random Forest outperforms ANN across all metrics) | Operates on simulated data only; restricted dataset; no MRI integration; lacks real-time clinical applicability and multimodal fusion. | Our digital twin incorporates real clinical MRI–EEG data, supports real-time preprocessing on Raspberry Pi, and improves signal quality using transformer-based embeddings. | Massaro [16] |
|
GCN + LSTM Hybrid Architecture for Seizure Prediction |
Converts multichannel EEG into Pearson-correlation graphs for spatial modeling using GCN and captures temporal seizure dynamics with LSTMs; delivers near-perfect seizure prediction performance. | Binary Accuracy: 99.39%; Ternary Accuracy: 98.69%; Sensitivity: 99.12%; Specificity: 95.72%; AUC: ≈1.0 | EEG-only framework; limited to CHB-MIT dataset; no multimodal MRI integration; lacks real-time deployment or XAI support. | Our model integrates MRI structural context with EEG seizure dynamics, adds real-time inference, provides XAI visualizations, and offers predictive twin behavior via a Tumor Kinetics Engine. | Kuang et al. [17] |
| Paper | Vision Transformer | Multi-modal (MRI + EEG) | XAI | Tumor Growth Prediction | Edge Computing | 3D Brain Visualization | Real-Time Monitoring | Wearable Skull Cap |
|---|---|---|---|---|---|---|---|---|
| Aftab Hussain et al. [1] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Zhihan Lv et al. [2] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Jinxia Wang et al. [3] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Yao et al. [4] | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ | ❌ |
| Sagheer Khan et al. [5] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ |
| Upadrista et al. [6] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Siyaev et al. [7] | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ | ❌ |
| Sultanpure et al. [8] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Wan et al. [9] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Cen et al. [10] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Liu et al. [11] | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Lin et al. [12] | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Chauhan et al. [13] | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Massaro [16] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Kuang et al. [17] | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
3. Dataset Description
3.1. In-House EEG Dataset
- Sampling Rate: 500 Hz
- Channels: 8 (including EOG)
- Participants: 500 Medically supervised human subjects.
- Demographics:Ages 20–75 years (mean 47.2 ± 12.5); 280 males, 220 females
| Cognitive State | Number of Segments |
|---|---|
| Relaxed | 1,200 |
| Stress | 1,200 |
| Fatigue | 1,200 |
| Total | 3,600 |
3.2. Clinically Acquired MRI Dataset
- Modalities Captured: T1-weighted, T2-weighted, and contrast-enhanced (T1-Gd) sequences
- Resolution: High-resolution Gray-Scale MRI Scans
- Size of MRI Scans: 600 x 600 pixels
- Format: NIfTI (.nii) or DICOM, later standardized for model ingestion
3.3. Data Splitting and Validation
- Training/Validation Set (70%): 350 patients (2,520 EEG Segments + 350 MRI volumes)
- Testing Set (30%): 150 patients (1,080 EEG Segments + 150 MRI volumes)
4. Proposed Model
4.1. System Architecture and Overview
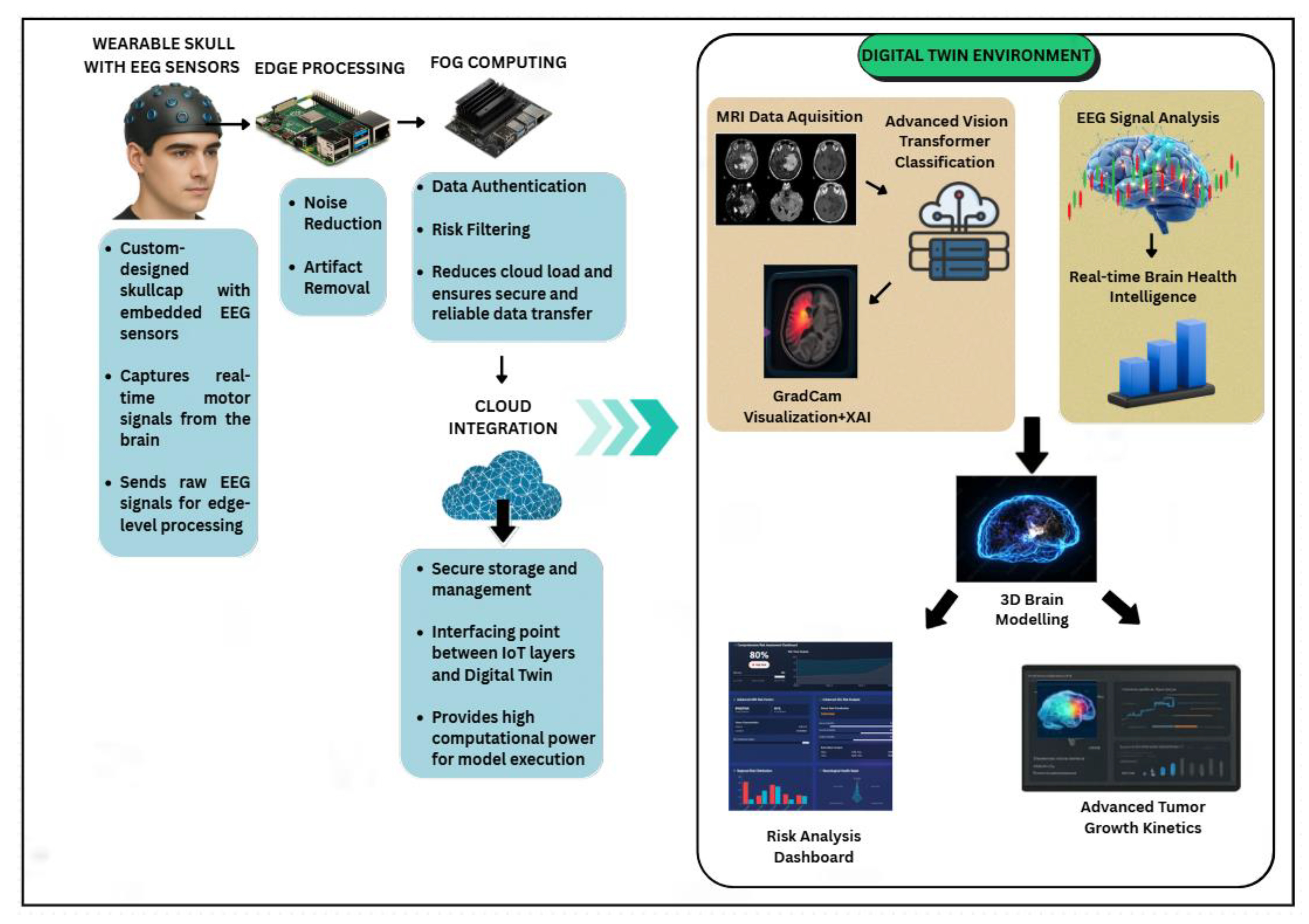
4.2. EEG Signal Acquisition Through Wearable Skullcap
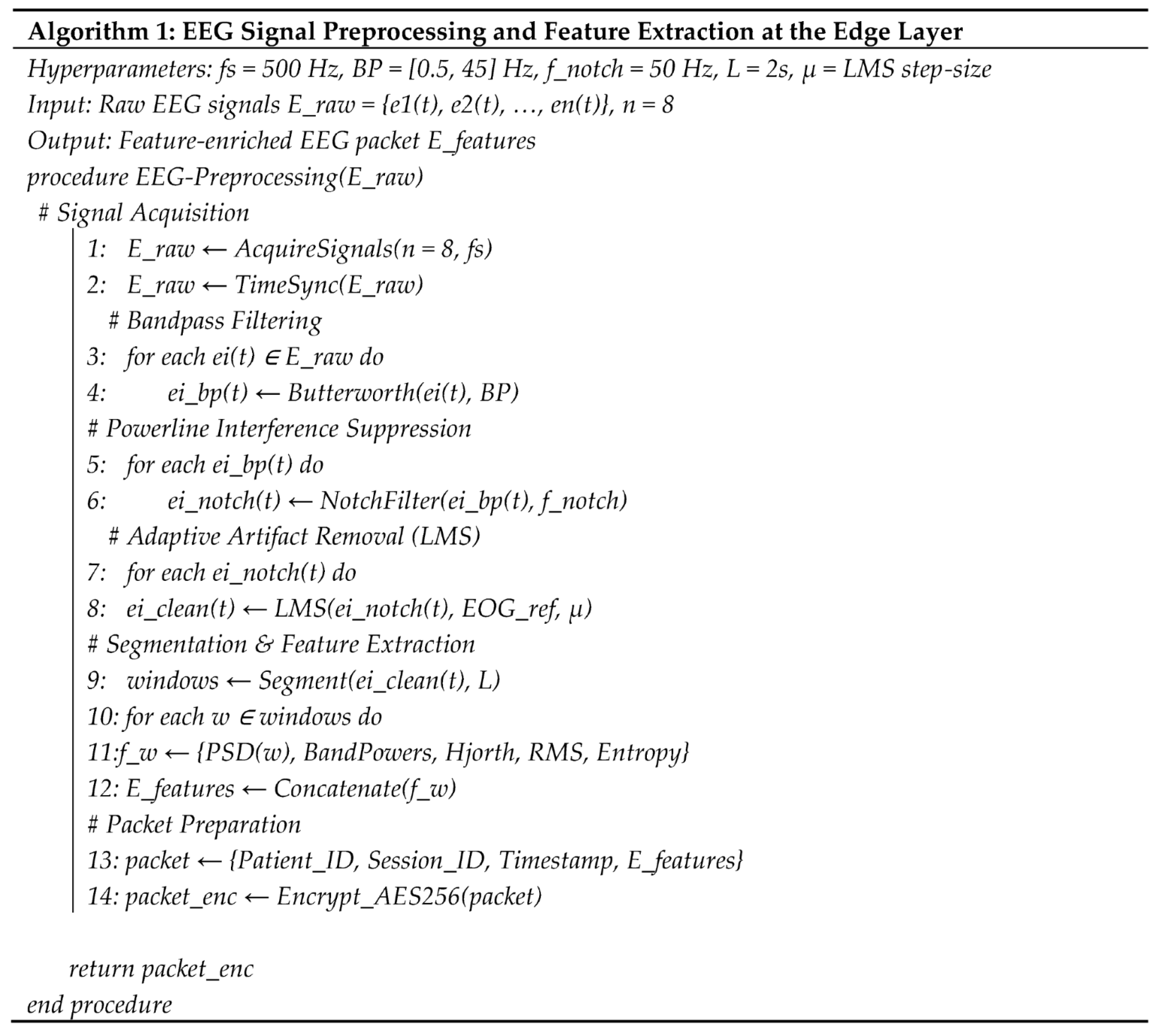
4.3. Edge Processing Using Raspberry Pi
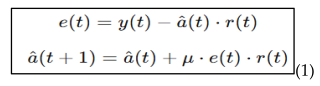
4.4. Fog Layer Authentication and Threshold Based Filtering
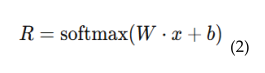
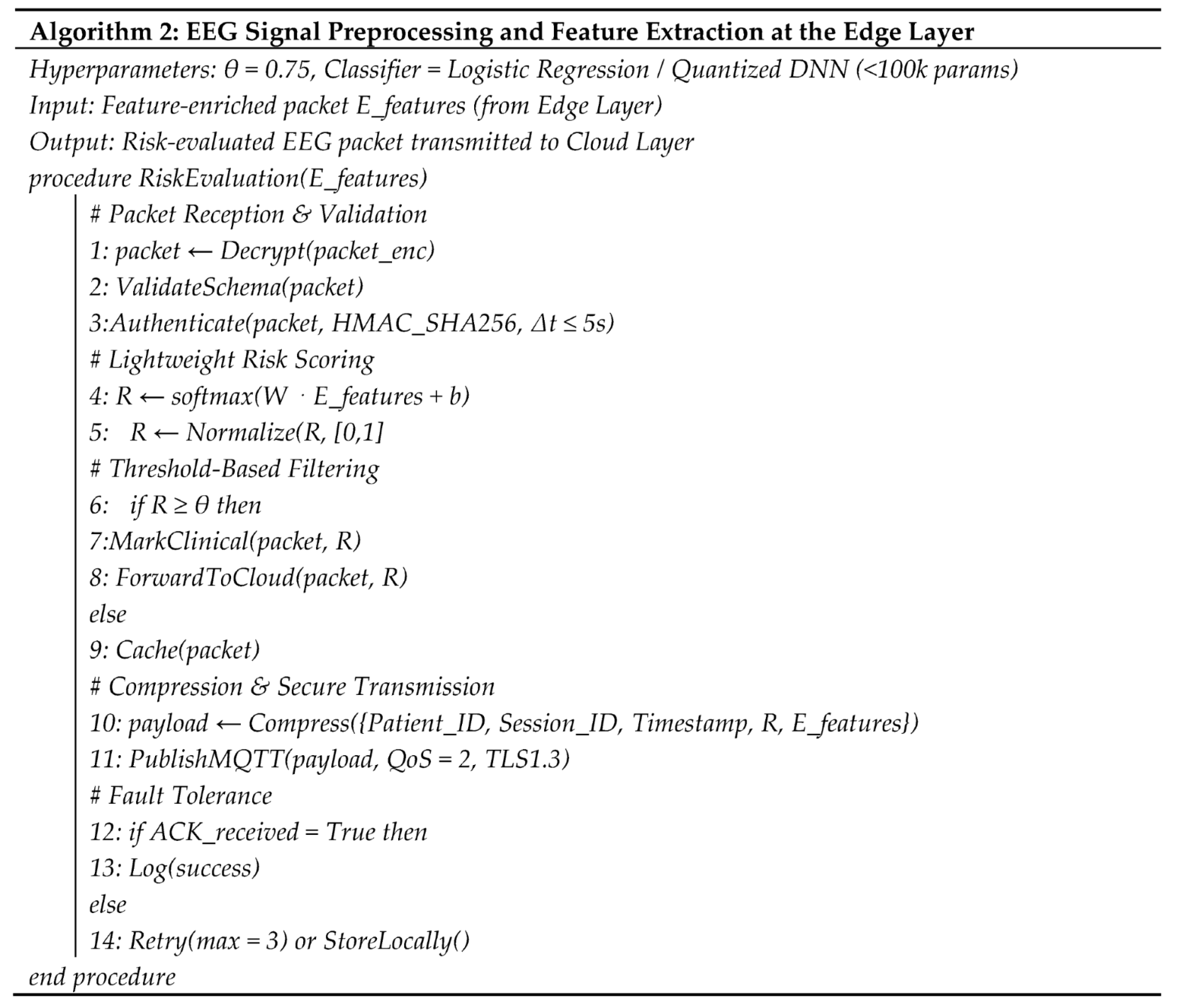
4.5. Cloud Integrated Digital Twin Environment
4.5.1. Patch-Level Attention Regularization (PLAR)

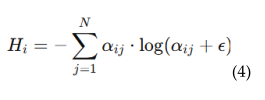
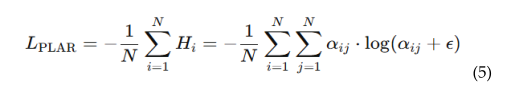
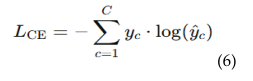
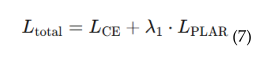
4.5.2. Adaptive Threshold Mechanism
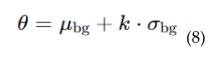

4.6. EEG Based Real-Time Brain Health Monitoring
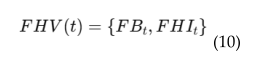
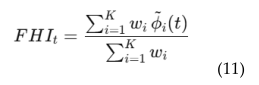
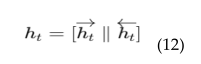
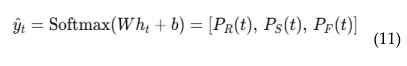
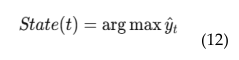
4.6. EEG-MRI Fusion
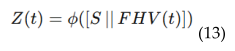
- Risk scoring and anomaly detection
- Interactive visualization.
4.7. XAI Based Grad-Cam Visualization
4.8. 3D Brain Interface
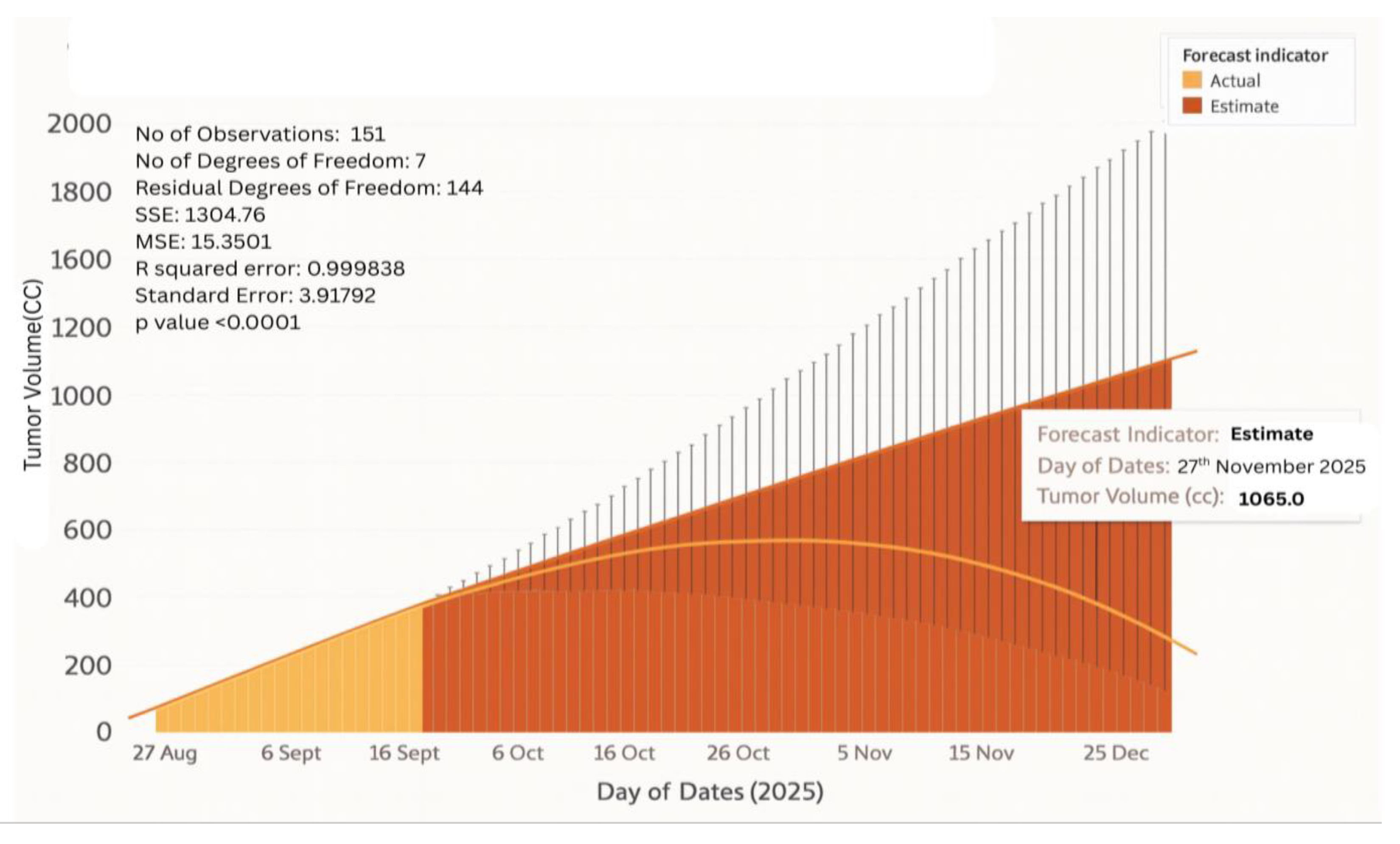
4.9. Tumor Kinetics Prediction Engine
5. Methodology
5.1. Clinical Scenarios and Applications
- A neurosurgeon evaluating a glioblastoma case can use the digital twin to visualize tumor boundaries in 3D, along with functional EEG overlays. This enables:
- Precision mapping of tumor location relative to eloquent cortex (motor, speech areas).
- Risk minimization by identifying functional regions likely to be affected during resection.
- Better patient counseling with visual models that explain risks and expected outcomes.
- Most of the patients with brain tumors present with seizures. Conventional EEG in hospitals is episodic and can fail to detect intermittent abnormalities. Using the EEG skullcap and edge-fog of wearable, continuous data is recorded in the daily activities.
- Clinicians get immediate alerts in case of abnormal brain signals. Treatment (e.g., anti-epileptic drugs) can be dynamically customized and this minimises unnecessary dose.
- When the patient is undergoing chemotherapy or radiotherapy, it is usually difficult to know the way that a tumor will react. The tumor kinetics engine provides predictive modelling of growth or shrinkage in various treatment regimes as a result, resistance is identified early enough, and the oncologists can change therapies before clinical failure.
- Continuous monitoring of EEGs correlates structural shrinkage with enhanced brain functionality providing a complete measure of treatment efficacy.
5.2. Clinical Advantages
5.3. Scalability and System Security
5.3.1. Minimization of Data & Local Processing
- EEG acquisition units transfers raw data directly into edge devices (Raspberry Pi, Jetson Nano).
- On-device preprocessing modules handle artifact removal (EEG filtering, ICA) and denoising
- A local pipeline script converts raw signals into compact feature vectors or masks.
- Only these processed, anonymized features are pushed to the fog layer. This ensures raw signals (which are identified as low risk EEG) are confined to the acquisition site.
5.3.2. Secure Transmission & Storage
- Data packets are published via MQTT brokers with TLS 1.3 encryption, so only authenticated fog/cloud subscribers can receive them.
- At the fog node, incoming data is stored in an encrypted database (AES-256).
- Cloud servers mirror this practice: all storage volumes are encrypted (AES-256, managed keys).
- Together, this ensures no unencrypted feature data exists outside the acquisition site.
5.3.3. Anonymization & Pseudonymization
- A local anonymization service strips identifiers (patient name, ID, DICOM headers in MRI files) before the data leaves the hospital.
- A pseudonymization script assigns a unique case ID (e.g., P1024/S0425) to each patient.
- Mapping tables (real ID ↔ pseudonym) are stored in a secure, local-only database, accessible only to hospital IT/admins.
- In the fog/cloud layers, models and dashboards only ever see pseudonymized IDs — never real patient identifiers.
5.3.4. Access Control & Auditability
- The system uses Role-Based Access Control (RBAC) with OAuth 2.0/JWT authentication.
- Clinicians can view diagnostic dashboards, researchers can analyze unidentified datasets, administrators can configure nodes — all restricted by roles.
- Every action (querying a case, running an inference, exporting results) is logged in immutable audit trails using blockchain-style append-only logs and secure logging frameworks.
- These logs allow retrospective tracking of who accessed what, when, and why — satisfying GDPR’s “accountability” requirement.

6.1. In-House Dataset Validation
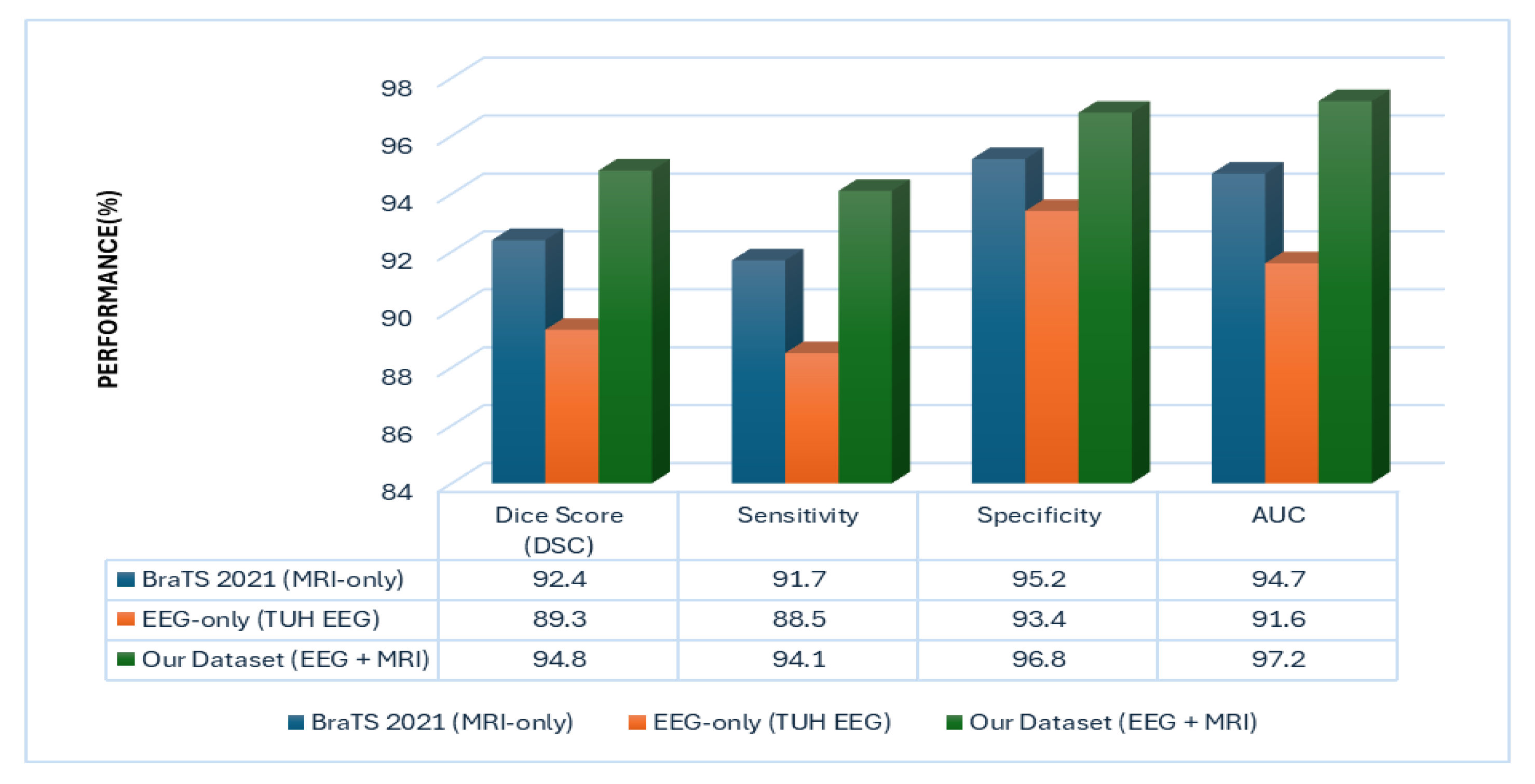
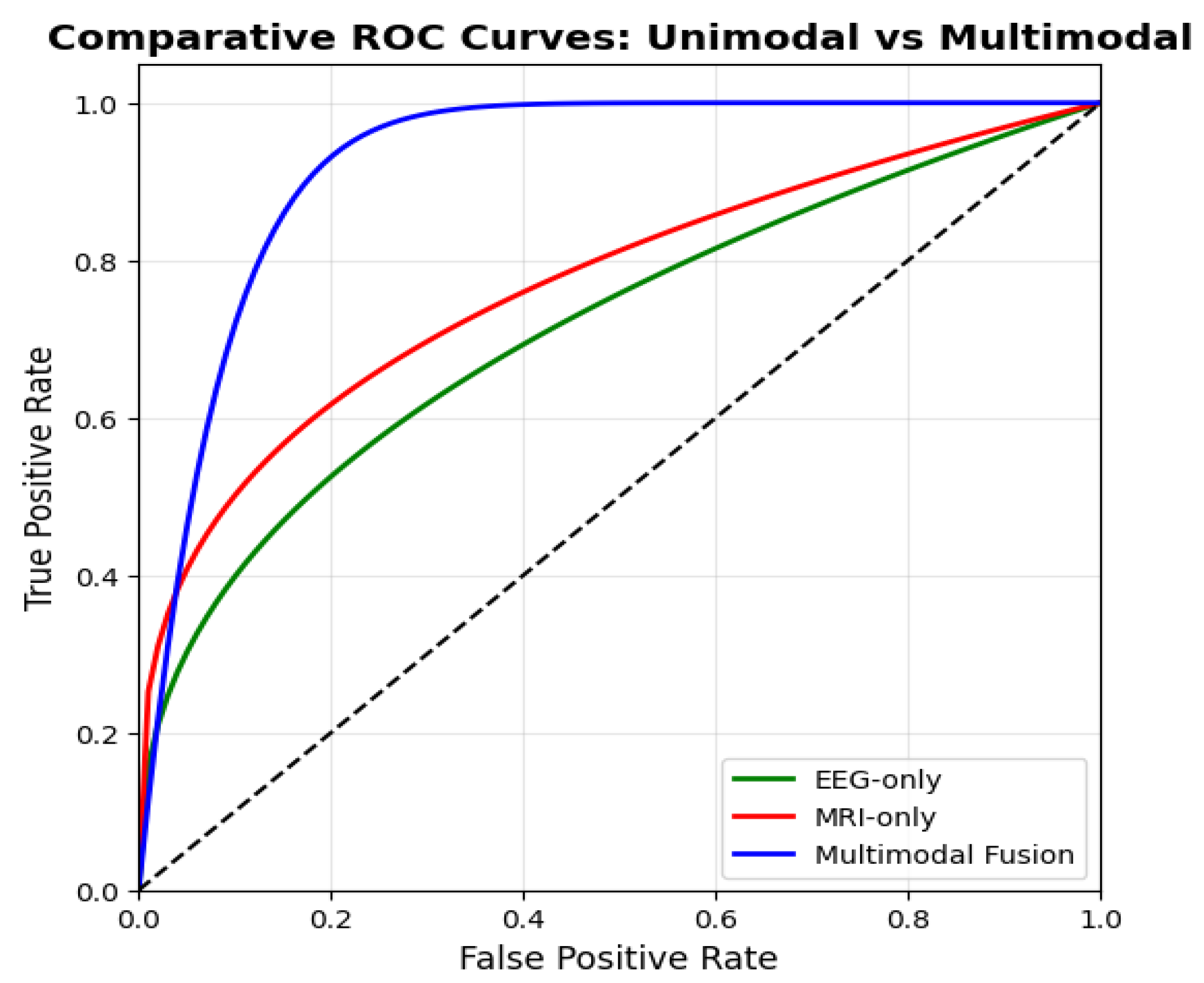
6.2. Multi-Modal Superiority over Unimodal Baselines
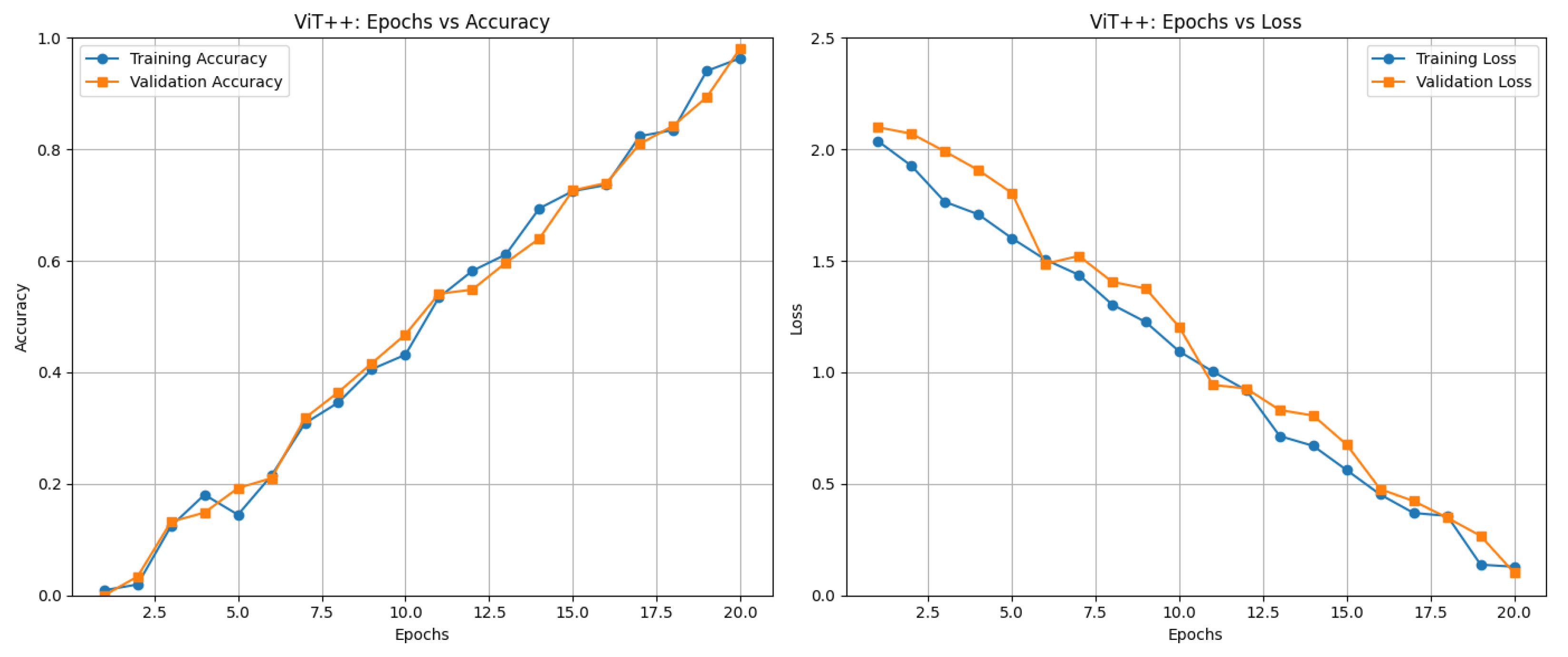
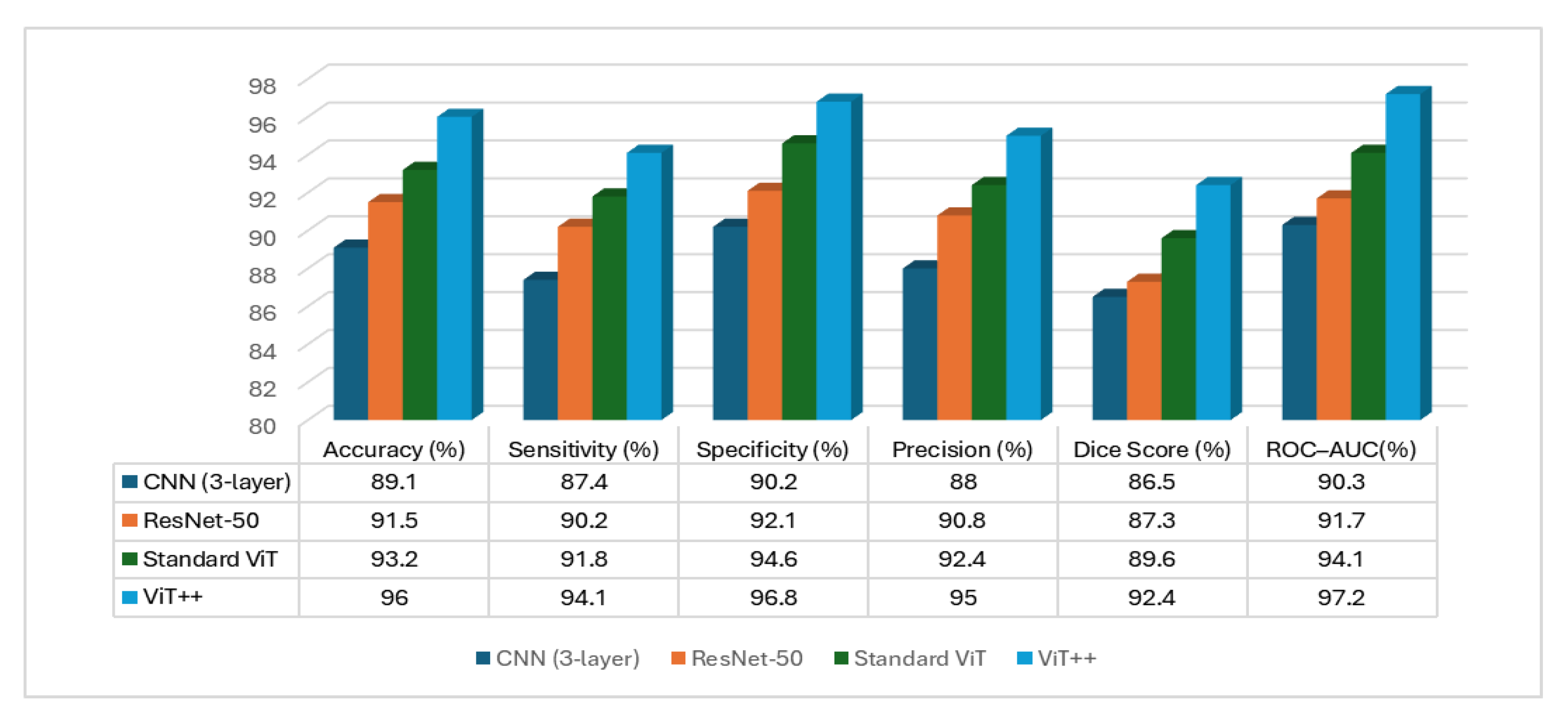
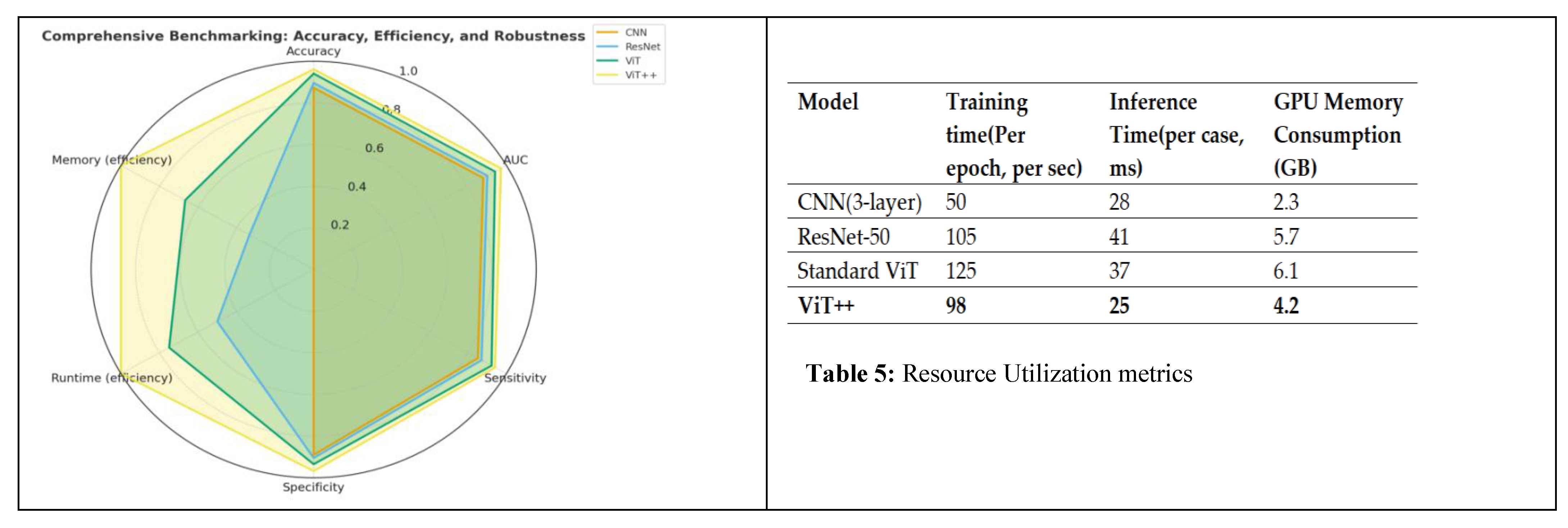
6.3. Performance Evaluation
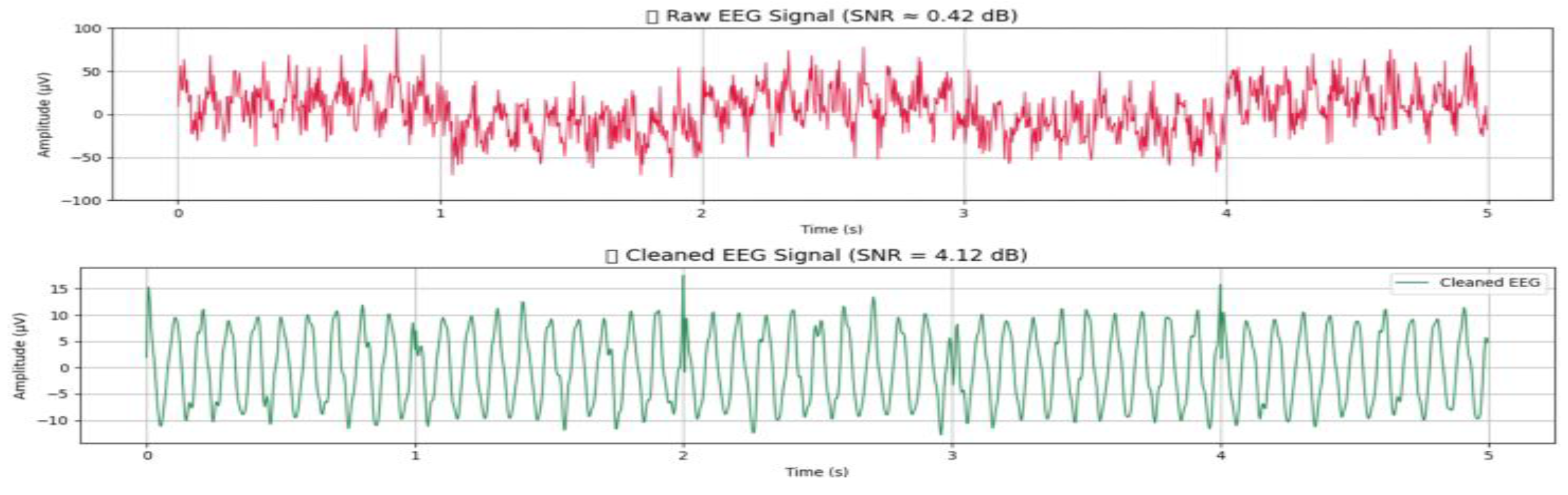
6.4. Denoising and SNR Enhancement
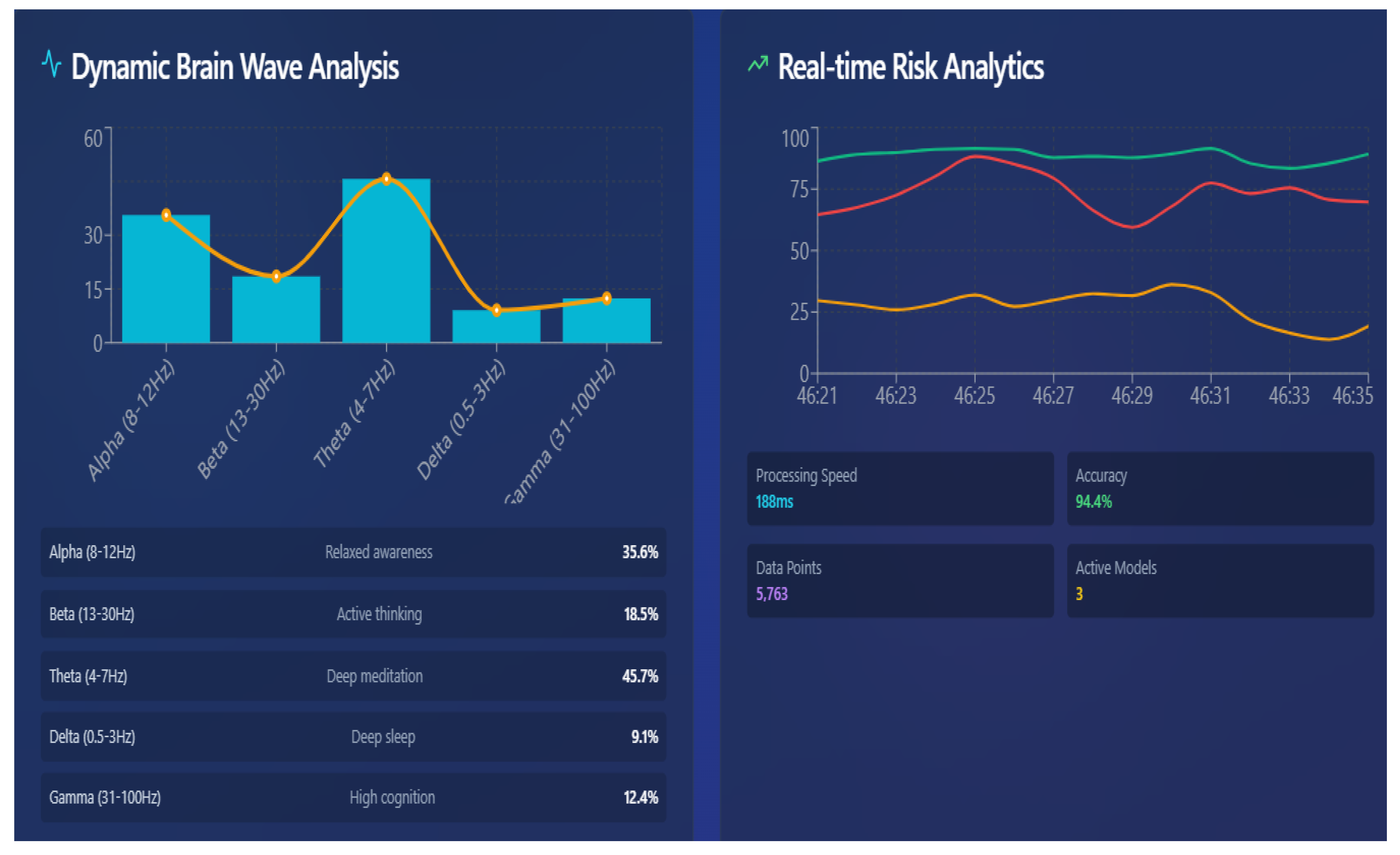
6.4. EEG-Driven Real-Time Brain Wave Analysis
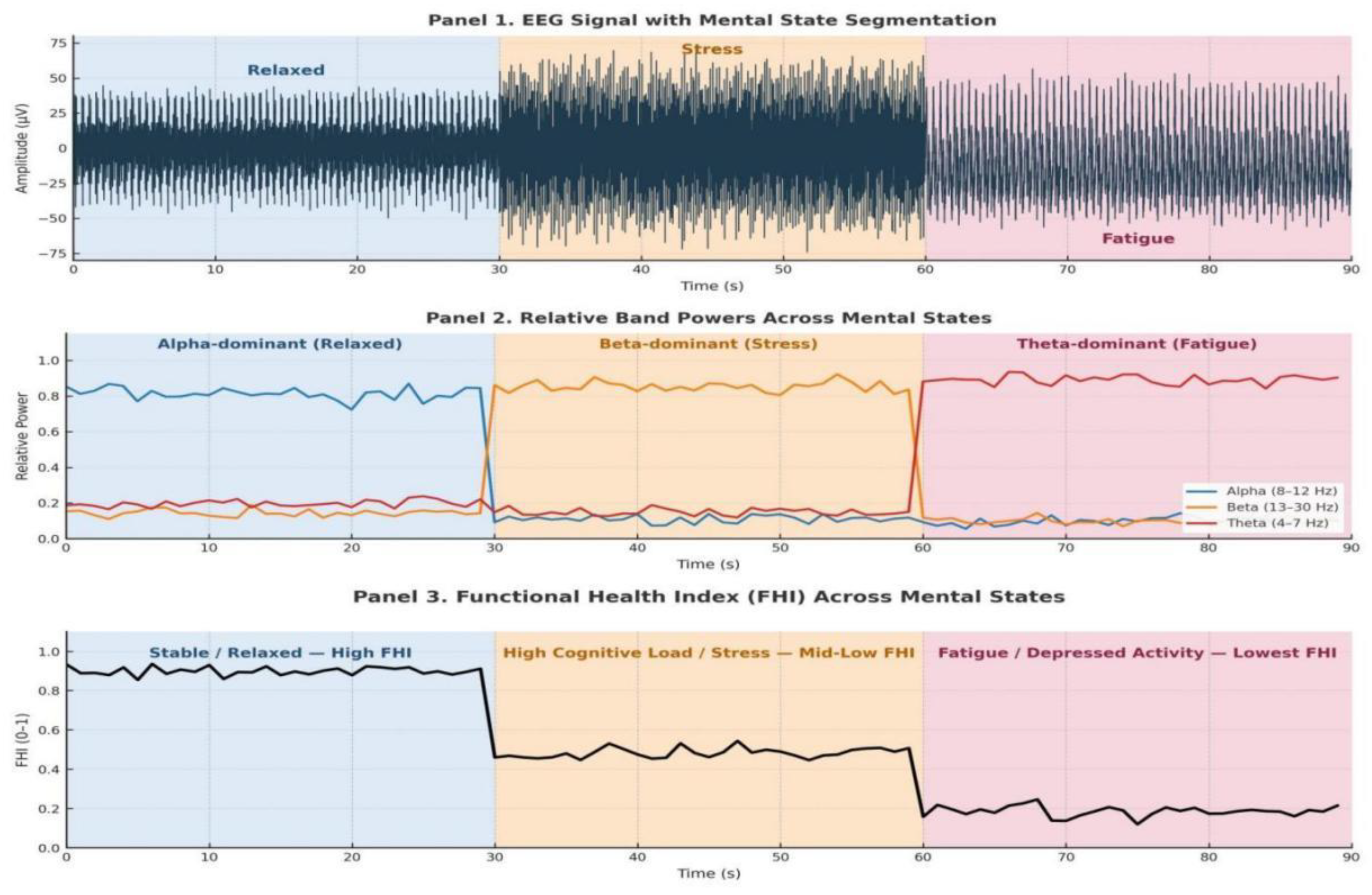
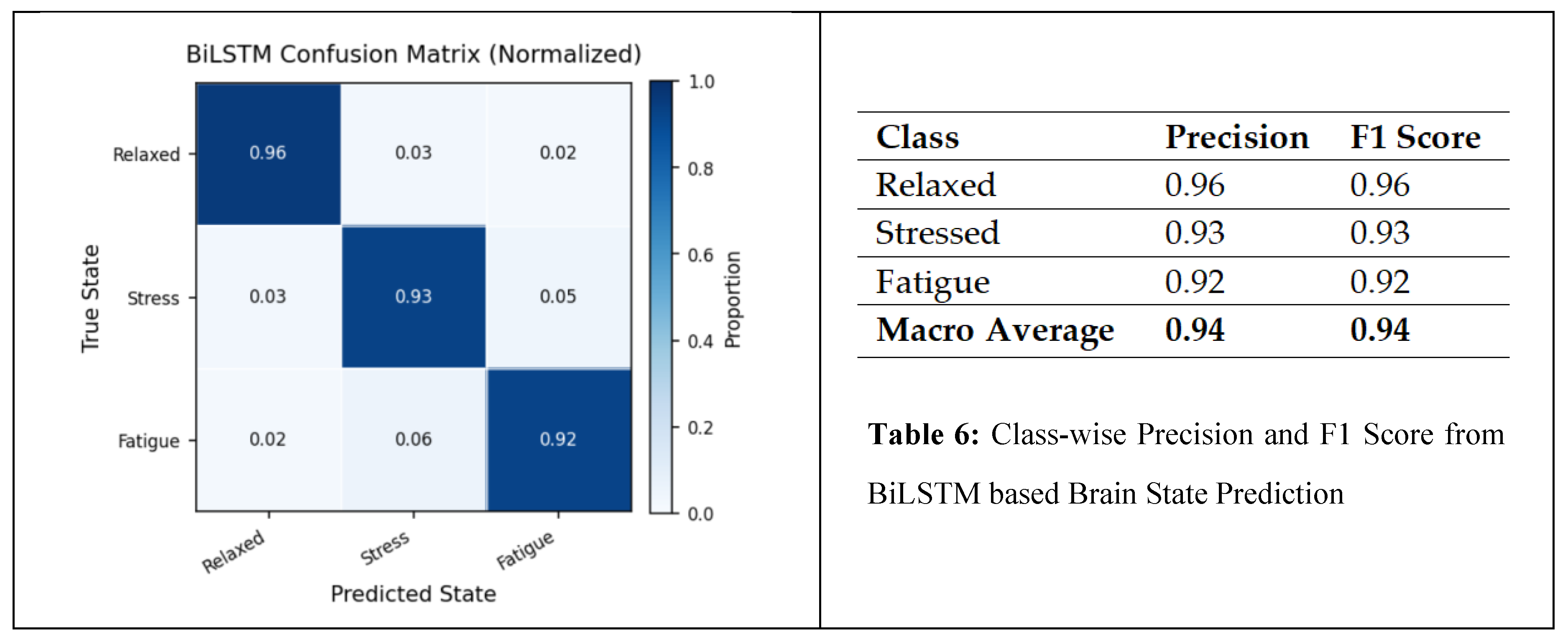
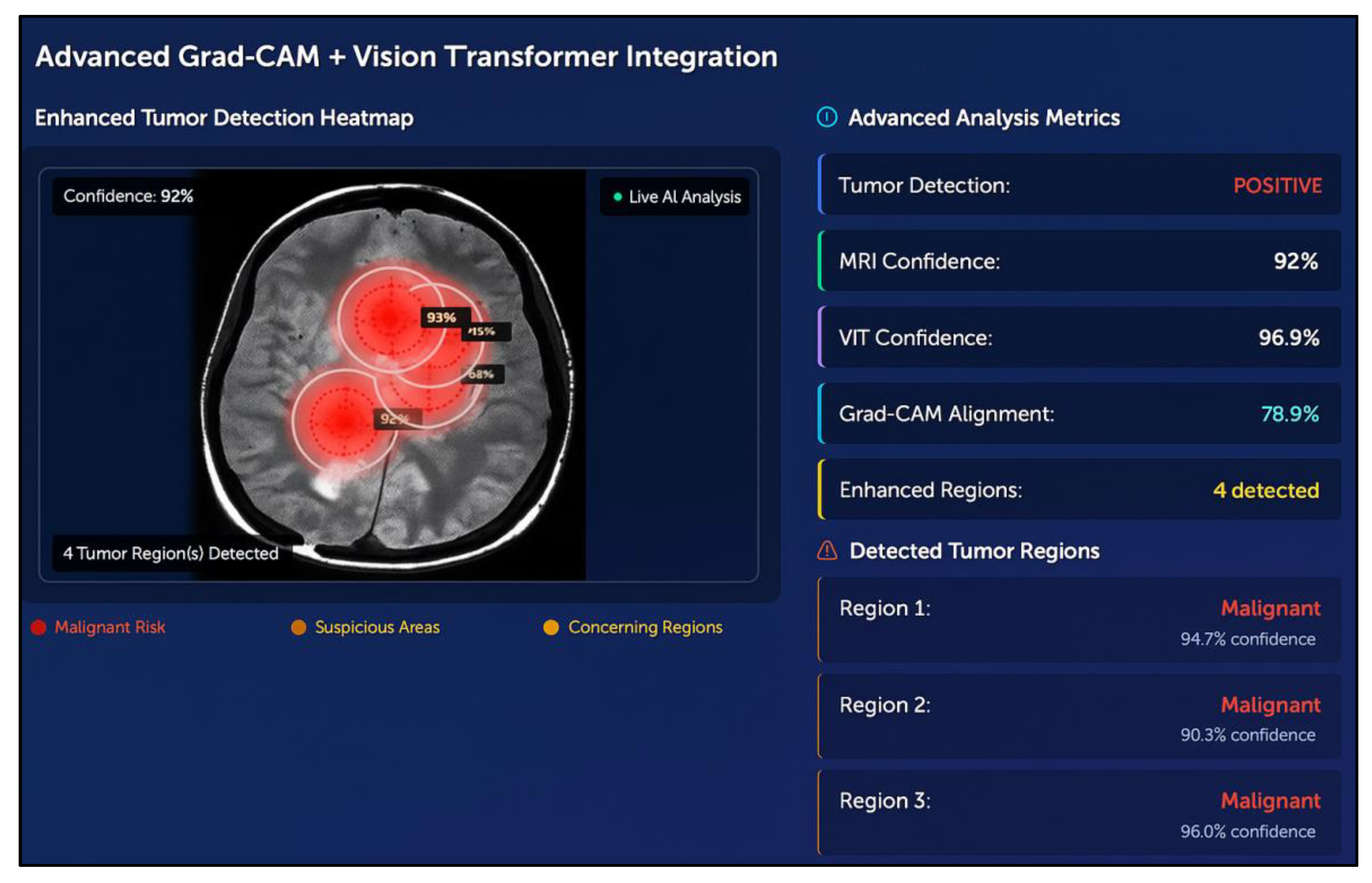
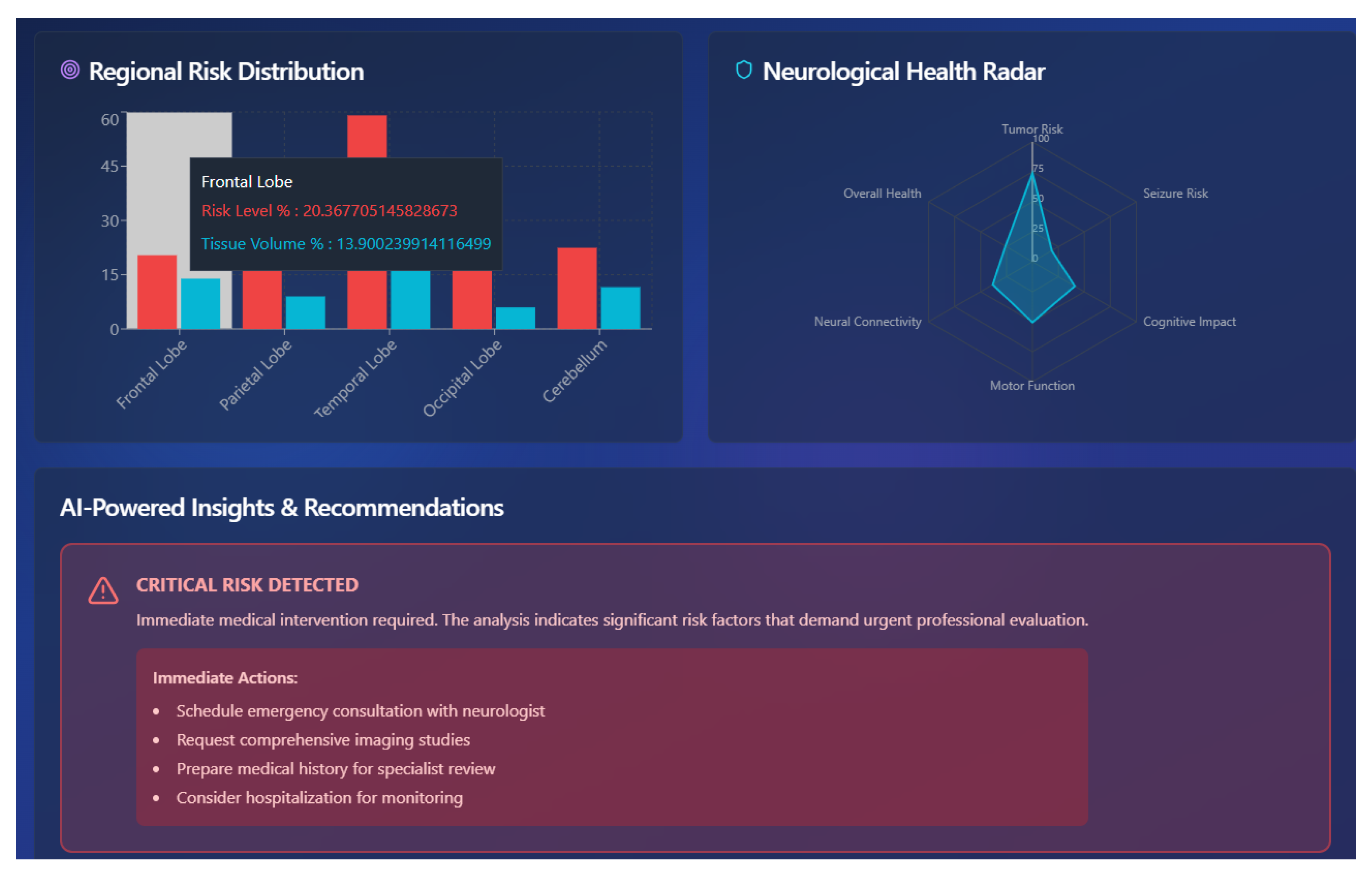
6.5. Tumor Localization Using GRAD-CAM
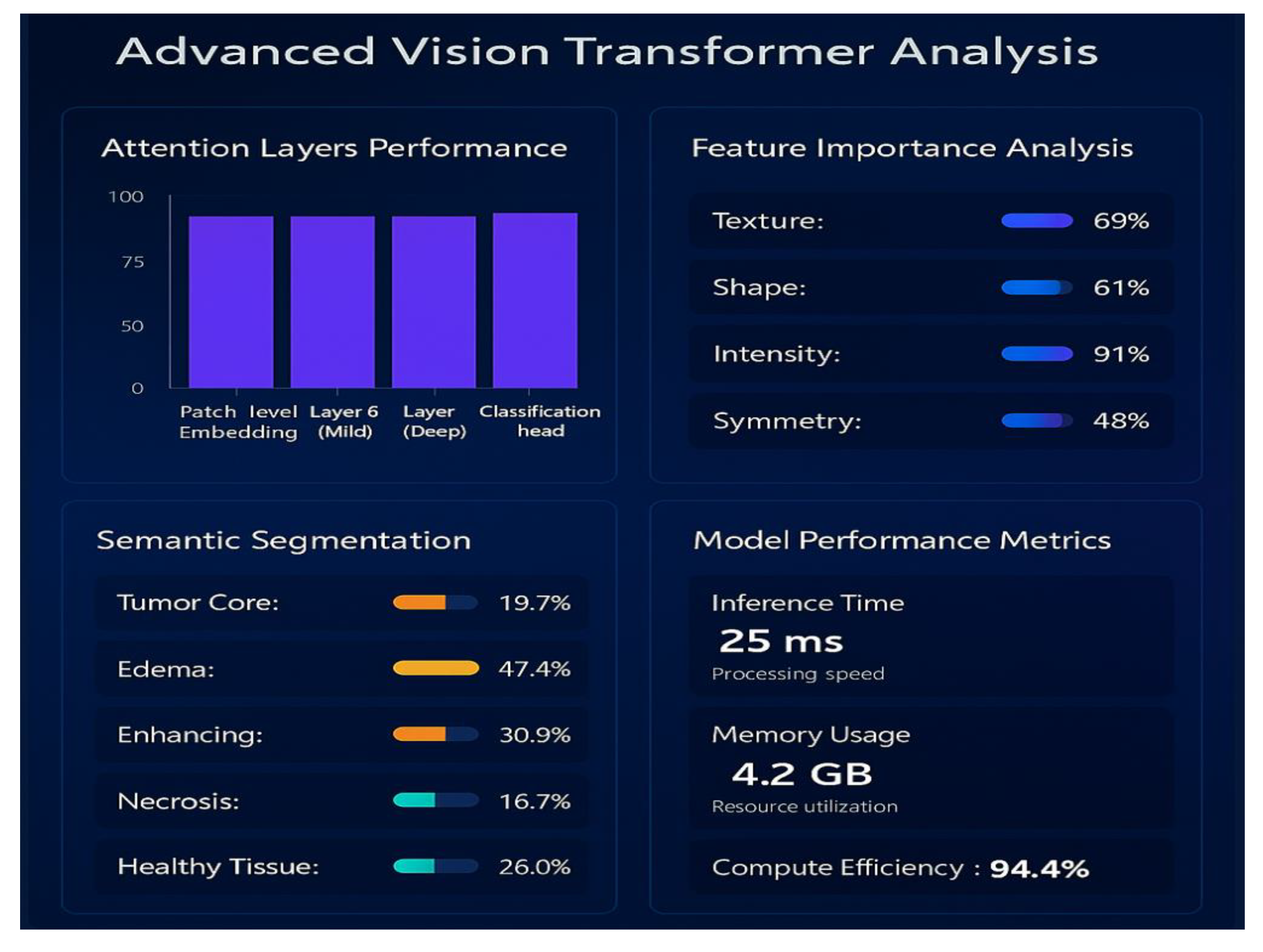
6.6. Predictive Insights Obtained from ViT++
6.7. Real-Time Neuro Analytics Captured Through EEG
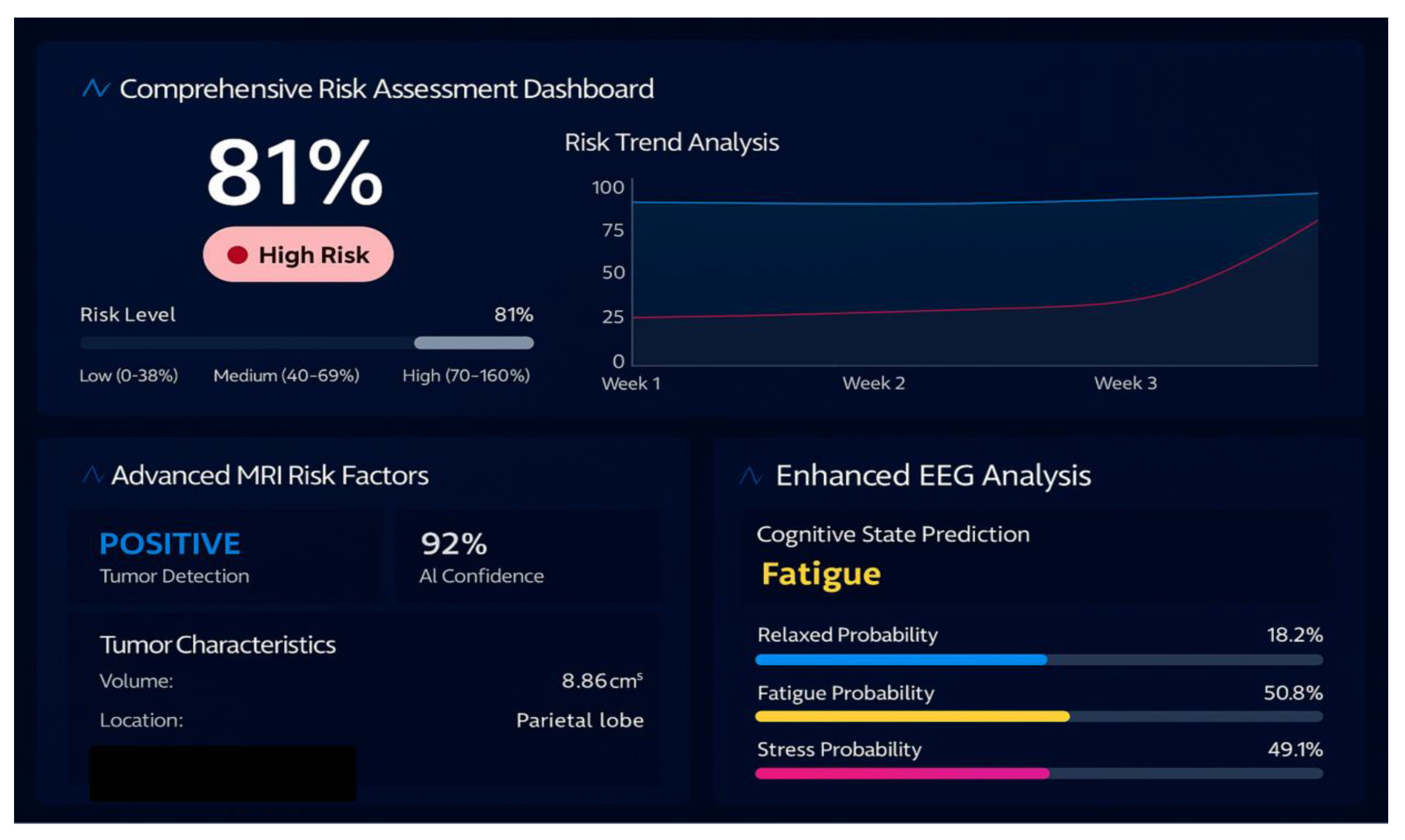
6.8. Unified Risk and Cognitive Brain State Prediction
6.9. AI Powered Insights
6.10. 3D Brain Interface
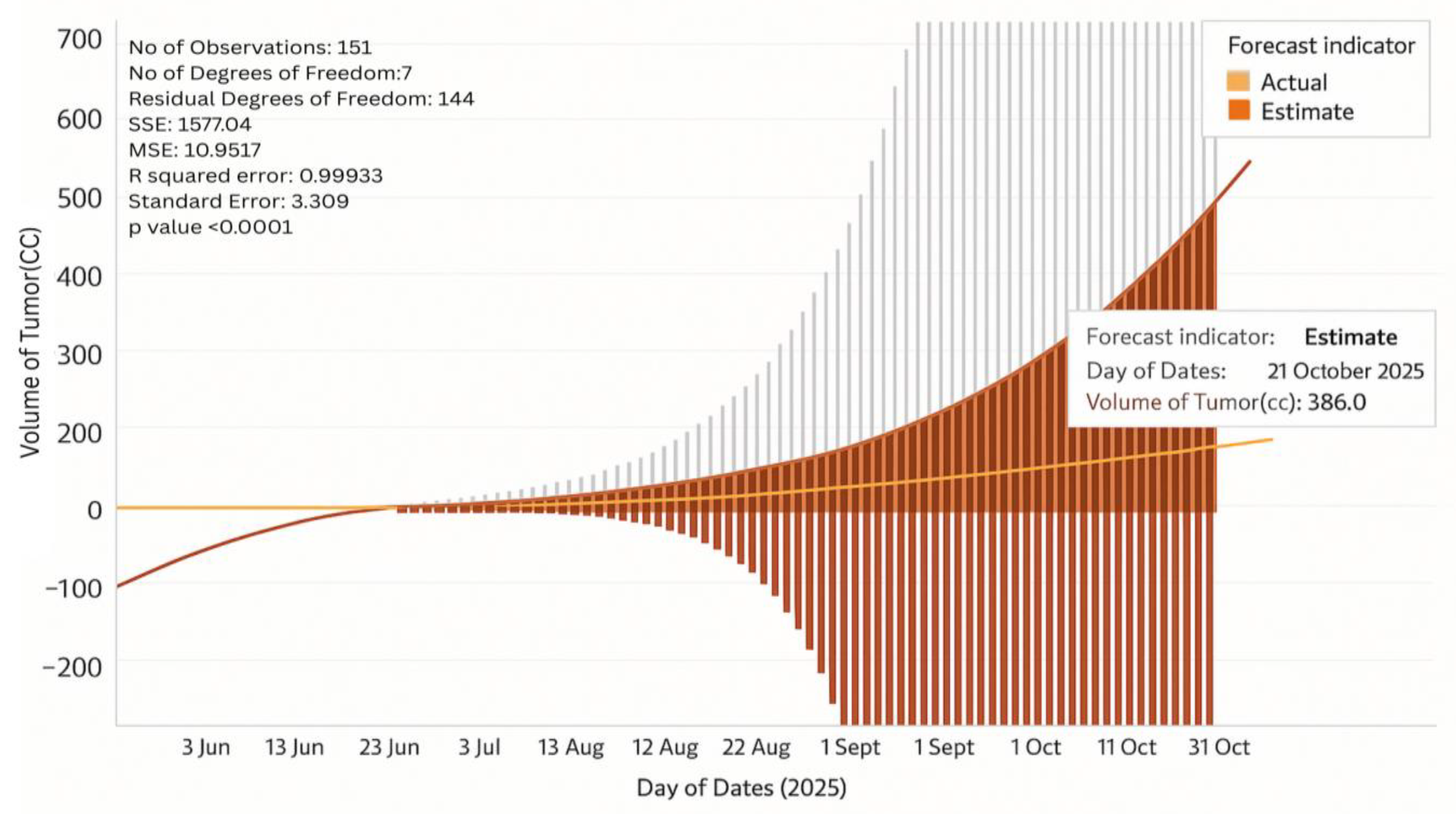
6.11. Tumor Growth Monitoring
- Stage I Glioblastoma
- Stage IV Glioblastoma
7. Conclusions
7.1. Future Works
8. Acknowledgement
References
- Hussain, A.; Malik, A.; Bangash, M.N.; Bashir, A.K.; Khan, M.; Kim, K. Deep transfer learning-based multi-modal digital twins for enhancement and diagnostic analysis of brain hemorrhage using attention ResNet and DCGAN. Frontiers in Neuroscience 2023, 17, 705323. [Google Scholar]
- Lv, Z.; Liu, J.; Wang, Y. DTBIA: An immersive visual analytics system for brain-inspired research. Scientific Reports 2023, 13, 14770. [Google Scholar] [CrossRef]
- Wang, J.; He, X.; Xiong, Y.; Chen, Y. Deep transfer learning framework for multimodal medical image fusion in digital twin-based healthcare. Journal of Cloud Computing: Advances, Systems and Applications 2024, 13, 22. [Google Scholar]
- Yao, Y.; Lv, Z.; Song, H. Digital Twin Brain-Immersive Analytics (DTBIA): A real-time VR-enabled visual analytics framework. Sensors 2023, 23, 1729. [Google Scholar]
- Khan, S.; Rathore, M.M.; Paul, A.; Hong, W.H. RF-based sensing and AI decision support for stroke patient monitoring: A digital twin approach. Journal of Biomedical Informatics: X 2023, 13, 100214. [Google Scholar] [CrossRef]
- Upadrista, R.S.; Subbiah, S.; Sudhakar, R. Blockchain-enabled digital twin framework for secure stroke prediction. In Neural Computing and Applications; 2023. [Google Scholar]
- Siyaev, A.; Jo, J. Neuro-symbolic reasoning framework for natural human interaction with digital twins. Frontiers in Neurorobotics 2023, 17, 1196203. [Google Scholar]
- Sultanpure, S.; Karthikeyan, R.; Kumari, V. Cloud-integrated digital twin system for brain tumor detection using particle swarm optimization and deep learning. Computers in Biology and Medicine 2023, 155, 106719. [Google Scholar] [CrossRef]
- Wan, J.; He, W.; Zhang, X. A hybrid AlexNet-S3VM digital twin model for multimodal brain image fusion and classification. Computers in Biology and Medicine 2023, 152, 106433. [Google Scholar]
- Cen, H.; Cai, Q.; Yang, Y.; Wang, Y. A digital twin approach for modeling the onset of thalamic atrophy in multiple sclerosis using MRI and mixed-effect spline regression. Computers in Biology and Medicine 2023, 152, 106419. [Google Scholar]
- Liu, X. BTSC-TNAS: A neural architecture search-based transformer for brain tumor segmentation and classification. Computerized Medical Imaging and Graphics 2023, 110, 102307. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. CKD-TransBTS: Clinical knowledge-driven hybrid transformer with modality-correlated cross-attention for brain tumor segmentation. IEEE Transactions on Medical Imaging 2023, 42, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.; Lunagaria, M.; Verma, D.K.; Vaghela, K.; Tejani, G.G.; Sharma, S.K.; Khan, A.R. PBVit: A patch-based vision transformer for enhanced brain tumor detection. IEEE Access 2024, 13, 13015–13029. [Google Scholar] [CrossRef]
- Massaro, A. Electronic Artificial Intelligence–Digital Twin Model for Optimizing Electroencephalogram Signal Detection. Electronics 2025, 14, 1122. [Google Scholar] [CrossRef]
- Kuang, Z. Epilepsy EEG Seizure Prediction Based on the Combination of Graph Convolutional Neural Network Combined with Long-and Short-Term Memory Cell Network. Applied Sciences 2024, 14, 11569. [Google Scholar] [CrossRef]


| Study/Approach | Data Modality | Dataset | Sensitivity (%) | Specificity (%) | AUC |
|---|---|---|---|---|---|
| Aftab Hussain et al. [1] | MRI (RSNA 2019) | 25,000 CT/MRI slices | 93.4 | 91.2 | 0.932 |
| Zhihan Lv et al. [2] | EEG only | ~100 subjects | 92.9 | 91.5 | 0.903 |
| Jinxia Wang et al. [3] | MRI + PET/SPECT | 120 patients | 89.3 | 90.8 | 0.917 |
| Yao et al. [4] | MRI (fMRI, DTI) | ~50 datasets | 88.6 | 89.7 | 0.905 |
| Sagheer Khan et al. [5] | RF signals (UWB) | 80 stroke patients | 93.4 | 92.3 | 0.911 |
| Upadrista et al. (2022) [6] | Clinical | 200 records | 92.8 | 93.1 | 0.928 |
| Siyaev et al. [7] | Voice + symbolic reasoning | 9000 queries | 91.2 | 90.4 | 0.914 |
| Sultanpure et al. [8] | MRI (IoT + Cloud) | 300 scans | 89.6 | 93.5 | 0.941 |
| Wan et al. [9] | MRI | 400 scans | 90.1 | 93.7 | 0.946 |
| Cen et al. [10] | MRI | HCP, ADNI | 85.7 | 90.2 | 0.902 |
| Liu et al. [11] | MRI (CNN + Transformer) | BraTS2019 + clinical | 91.0 | 92.0 | 0.941 |
| Lin et al. [12] | MRI (CKD-TransBTS) | BraTS2021 | 89.9 | 91.3 | 0.935 |
| Chauhan et al. [13] | MRI (ViT) | 2,327 MRIs | 93.2 | 95.3 | 0.958 |
| Massaro [14] | EEG (simulated + real) | Alcoholic EEG dataset | 90.4 | 89.6 | 0.921 |
| Kuang et al. [15] | EEG (multi-channel) | CHB-MIT | 99.12 | 95.72 | ≈1.0 |
| Proposed BrainTwin | MRI + EEG | 500 patients | 94.1 | 96.8 | 0.972 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.





