Submitted:
13 December 2025
Posted:
16 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Equipment and Reagents for Chemical Synthesis
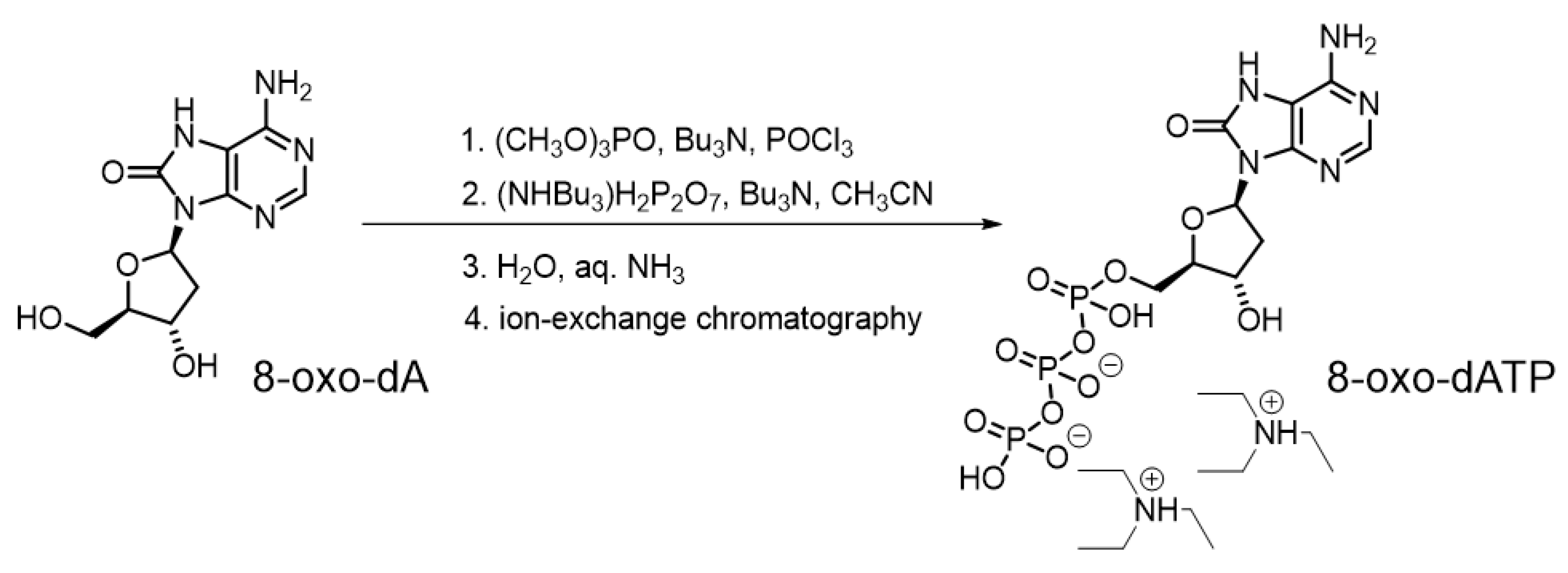
2.2. 8-oxo-dATP Synthesis
2.3. DNA Templates and Enzymes
2.4. DNA Polymerase Reactions for the Primer Extension Assay
3. Results
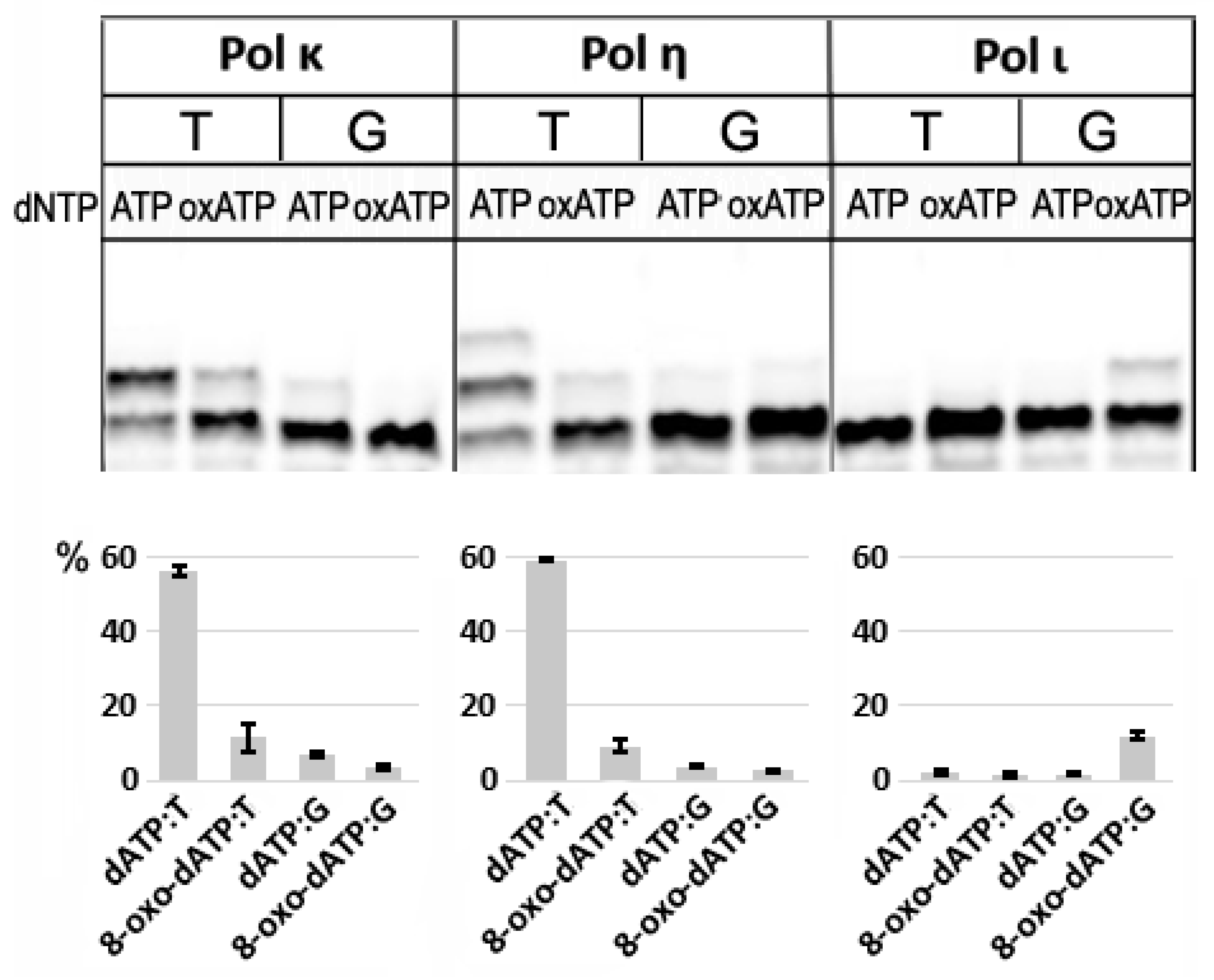
3.1. Negligible Incorporation of 8-oxo-dATP by Translesion DNA Polymerases of Family Y
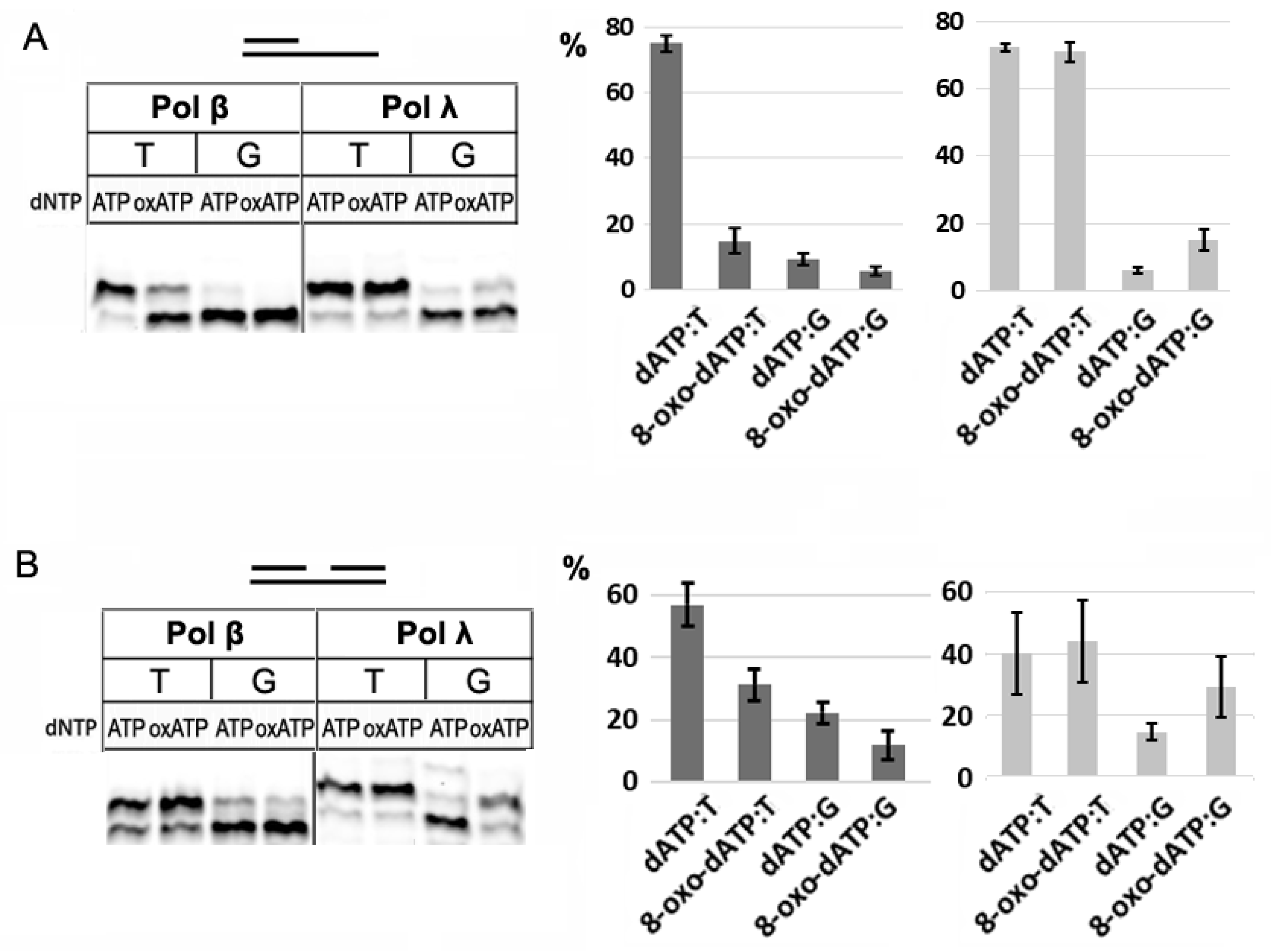
3.2. Incorporation of 8-oxo-dATP by Pol β and Pol λ of Family X
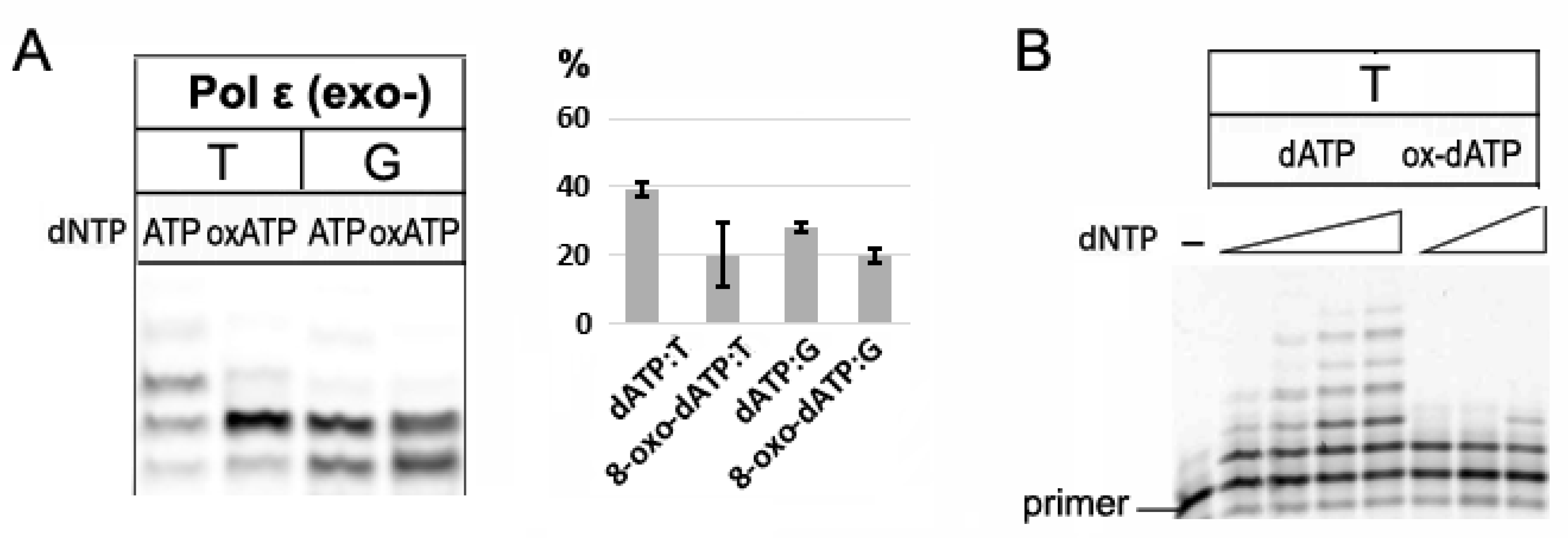
2.3. Incorporation of 8-oxo-dATP by Replicative B-family Pol ε
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Helbock, H.J.; Beckman, K.B.; Shigenaga, M.K.; Walter, P.B.; Woodall, A.A.; Yeo, H.C.; Ames, B.N. DNA Oxidation Matters: The HPLC–Electrochemical Detection Assay of 8-Oxo-Deoxyguanosine and 8-Oxo-Guanine. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 288–293. [Google Scholar] [CrossRef]
- Møller, P.; Cooke, M.S.; Collins, A.; Olinski, R.; Rozalski, R.; Loft, S. Harmonising Measurements of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine in Cellular DNA and Urine. Free Radical Research 2012, 46, 541–553. [Google Scholar] [CrossRef]
- Hayakawa, H.; Taketomi, A.; Sakumi, K.; Kuwano, M.; Sekiguchi, M. Generation and Elimination of 8-Oxo-7,8-Dihydro-2’-Deoxyguanosine 5’-Triphosphate, a Mutagenic Substrate for DNA Synthesis, in Human Cells. Biochemistry 1995, 34, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, Y.I.; Minnick, D.T.; Izuta, S.; Kunkel, T.A. DNA Replication Fidelity with 8-Oxodeoxyguanosine Triphosphate. Biochemistry 1994, 33, 4695–4701. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Ohta, E.; Abolhassani, N. MTH1 as a Nucleotide Pool Sanitizing Enzyme: Friend or Foe? Free Radical Biology and Medicine 2017, 107, 151–158. [Google Scholar] [CrossRef]
- Fuciarelli, A.F.; Wegher, B.J.; Gajewski, E.; Dizdaroglu, M.; Blakely, W.F. Quantitative Measurement of Radiation-Induced Base Products in DNA Using Gas Chromatography-Mass Spectrometry. Radiation Research 1989, 119, 219. [Google Scholar] [CrossRef]
- Tuo, J.; Jaruga, P.; Rodriguez, H.; Dizdaroglu, M.; Bohr, V.A. The Cockayne Syndrome Group B Gene Product Is Involved in Cellular Repair of 8-Hydroxyadenine in DNA. Journal of Biological Chemistry 2002, 277, 30832–30837. [Google Scholar] [CrossRef] [PubMed]
- Jałoszyński, P.; Jaruga, P.; Oliński, R.; Biczysko, W.; Szyfter, W.; Nagy, E.; Möller, L.; Szyfter, K. Oxidative DNA Base Modifications and Polycyclic Aromatic Hydrocarbon DNA Adducts in Squamous Cell Carcinoma of Larynx. Free Radical Research 2003, 37, 231–240. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Yu, D.; Sun, J.-F.; Zeng, L.; Wang, C.-Y.; Bai, L.-P.; Zhu, G.-Y.; Jiang, Z.-H.; Zhang, W. An Integrated Approach to Evaluate Acetamiprid-Induced Oxidative Damage to tRNA in Human Cells Based on Oxidized Nucleotide and tRNA Profiling. Environment International 2023, 178, 108038. [Google Scholar] [CrossRef]
- Fujikawa, K.; Kamiya, H.; Yakushiji, H.; Fujii, Y.; Nakabeppu, Y.; Kasai, H. The Oxidized Forms of dATP Are Substrates for the Human MutT Homologue, the hMTH1 Protein. Journal of Biological Chemistry 1999, 274, 18201–18205. [Google Scholar] [CrossRef]
- Petushkov, I.V.; Aralov, A.V.; Ivanov, I.A.; Baranov, M.S.; Zatsepin, T.S.; Kulbachinskiy, A.V. Effect of 8-Oxo-1,N6-Ethenoadenine Derivatives on the Activity of RNA Polymerases from SARS-CoV-2 and Escherichia Coli. Biochemistry Moscow 2024, 89, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Navacchia, M.L.; Postigo, A. A Facile One-Pot Synthesis of 8-Oxo-7,8-Dihydro-(2′-Deoxy)Adenosine in Water. Tetrahedron Letters 2006, 47, 711–714. [Google Scholar] [CrossRef]
- Kazachenko, K.Y.; Miropolskaya, N.A.; Gening, L.V.; Tarantul, V.Z.; Makarova, A.V. Alternative Splicing at Exon 2 Results in the Loss of the Catalytic Activity of Mouse DNA Polymerase Iota in Vitro. DNA Repair 2017, 50, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Shilkin, E.S.; Petrova, D.V.; Kruchinin, A.A.; Zharkov, D.O.; Makarova, A.V. The Effect of Methylation and Hydroxymethylation of Cytosine on Activity and Fidelity of Pol λ and Pol β. DNA Repair 2025, 148, 103815. [Google Scholar] [CrossRef] [PubMed]
- Boldinova, E.O.; Yudkina, A.V.; Shilkin, E.S.; Gagarinskaya, D.I.; Baranovskiy, A.G.; Tahirov, T.H.; Zharkov, D.O.; Makarova, A.V. Translesion Activity of PrimPol on DNA with Cisplatin and DNA–Protein Cross-Links. Sci Rep 2021, 11, 17588. [Google Scholar] [CrossRef]
- Boldinova, E.O.; Kruchinin, A.A.; Kamzeeva, P.N.; Aralov, A.V.; Makarova, A.V. Accurate DNA Synthesis Across 8-Oxoadenine by Human PrimPol. IJMS 2025, 26, 6796. [Google Scholar] [CrossRef]
- Bedaiwi, S.; Usmani, A.; Carty, M.P. Canonical and Non-Canonical Roles of Human DNA Polymerase η. Genes 2024, 15, 1271. [Google Scholar] [CrossRef]
- Grin, I.R.; Vasilyeva, S.V.; Dovgerd, A.P.; Silnikov, V.N.; Zharkov, D.O. Human and Bacterial DNA Polymerases Discriminate against 8-Oxo-2’-Deoxyadenosine- 5’-Triphosphate. Biopolym. Cell 2012, 28, 306–309. [Google Scholar] [CrossRef]
- Batra, V.K.; Beard, W.A.; Shock, D.D.; Pedersen, L.C.; Wilson, S.H. Structures of DNA Polymerase β with Active-Site Mismatches Suggest a Transient Abasic Site Intermediate during Misincorporation. Molecular Cell 2008, 30, 315–324. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Perera, L.; Shock, D.D.; Kim, T.; Schlick, T.; Wilson, S.H. Uncovering the Polymerase-Induced Cytotoxicity of an Oxidized Nucleotide. Nature 2015, 517, 635–639. [Google Scholar] [CrossRef]
- Zhao, Y.; Gregory, M.T.; Biertümpfel, C.; Hua, Y.-J.; Hanaoka, F.; Yang, W. Mechanism of Somatic Hypermutation at the WA Motif by Human DNA Polymerase η. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8146–8151. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Nair, D.T.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Replication across Template T/U by Human DNA Polymerase-ι. Structure 2009, 17, 974–980. [Google Scholar] [CrossRef]
- Yockey, O.P.; Jha, V.; Ghodke, P.P.; Xu, T.; Xu, W.; Ling, H.; Pradeepkumar, P.I.; Zhao, L. Mechanism of Error-Free DNA Replication Past Lucidin-Derived DNA Damage by Human DNA Polymerase κ. Chem. Res. Toxicol. 2017, 30, 2023–2032. [Google Scholar] [CrossRef]
- Roske, J.J.; Yeeles, J.T.P. Structural Basis for Processive Daughter-Strand Synthesis and Proofreading by the Human Leading-Strand DNA Polymerase Pol ε. Nat Struct Mol Biol 2024, 31, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- Gosavi, R.A.; Moon, A.F.; Kunkel, T.A.; Pedersen, L.C.; Bebenek, K. The Catalytic Cycle for Ribonucleotide Incorporation by Human DNA Pol λ. Nucleic Acids Research 2012, 40, 7518–7527. [Google Scholar] [CrossRef] [PubMed]
| Oligonucleotide | Sequence 5′-3′ |
| Сy5-Pr16 | Cy5-GTCACAGAGATACTAC |
| TemplateTA | GAGCAGTCGCACATGTAGTATCTCTGTGAC |
| TemplateGA | GAGCAGTCGCACAGGTAGTATCTCTGTGAC |
| Closing_oxodATP | /P/-TGTGCGACTGCTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





