1. Introduction
Redox processes are central to biological organization, energy transduction, and homeostasis. Life on Earth depends on a finely tuned interplay between electron donors, electron acceptors, and molecules capable of switching between these states (amphoteric species). This triad enables controlled metabolic reactions, cellular signaling, and the maintenance of oxidative balance [
1].
Understanding whether this redox architecture is a contingent feature of Earth’s biology or reflects a more universal chemical principle requires comparative analysis across different molecular populations. Interstellar molecular inventories represent the chemical environment that precedes and accompanies star and planet formation, including potential prebiotic precursors [
2]. Nutrient molecules, by contrast, represent a set shaped by billions of years of biological selection. PubChem, with tens of millions of heterogeneous compounds, reflects chemically possible structures without biological or cosmological selection pressures.
By comparing these three molecular spaces, it becomes possible to identify chemical signatures shared between prebiotic cosmic chemistry and biological functionality—and to separate them from signatures of random chemical diversity.
2. Materials and Methods
2.1. Molecular Descriptors: AQVN and EIIP
The electronic-structure descriptors AQVN (average quasivalence number) and EIIP (electron–ion interaction potential) are used to characterize the redox properties of organic molecules. These parameters originate from the “general model pseudopotential” [
3] framework and are mathematically defined as follows.
The EIIP value of a molecule is given by:
where Z* represents the AQVN, calculated according to
Here, Zi denotes the valence of the i-th element, ni is the number of atoms of that element, m is the number of distinct atomic species present, and N is the total atom count within the molecule. EIIP values derived from these equations are expressed in Rydbergs (Ry).
Molecular descriptors AQVN and EIIP have served as the foundation for studying diverse biological properties of proteins, DNA, and small molecules (with more than 400 peer-reviewed articles available at
https://electronicbiology.org/biomedical-articles/).
In earlier work, we analyzed 4,510,644 randomly selected molecules from the PubChem database [
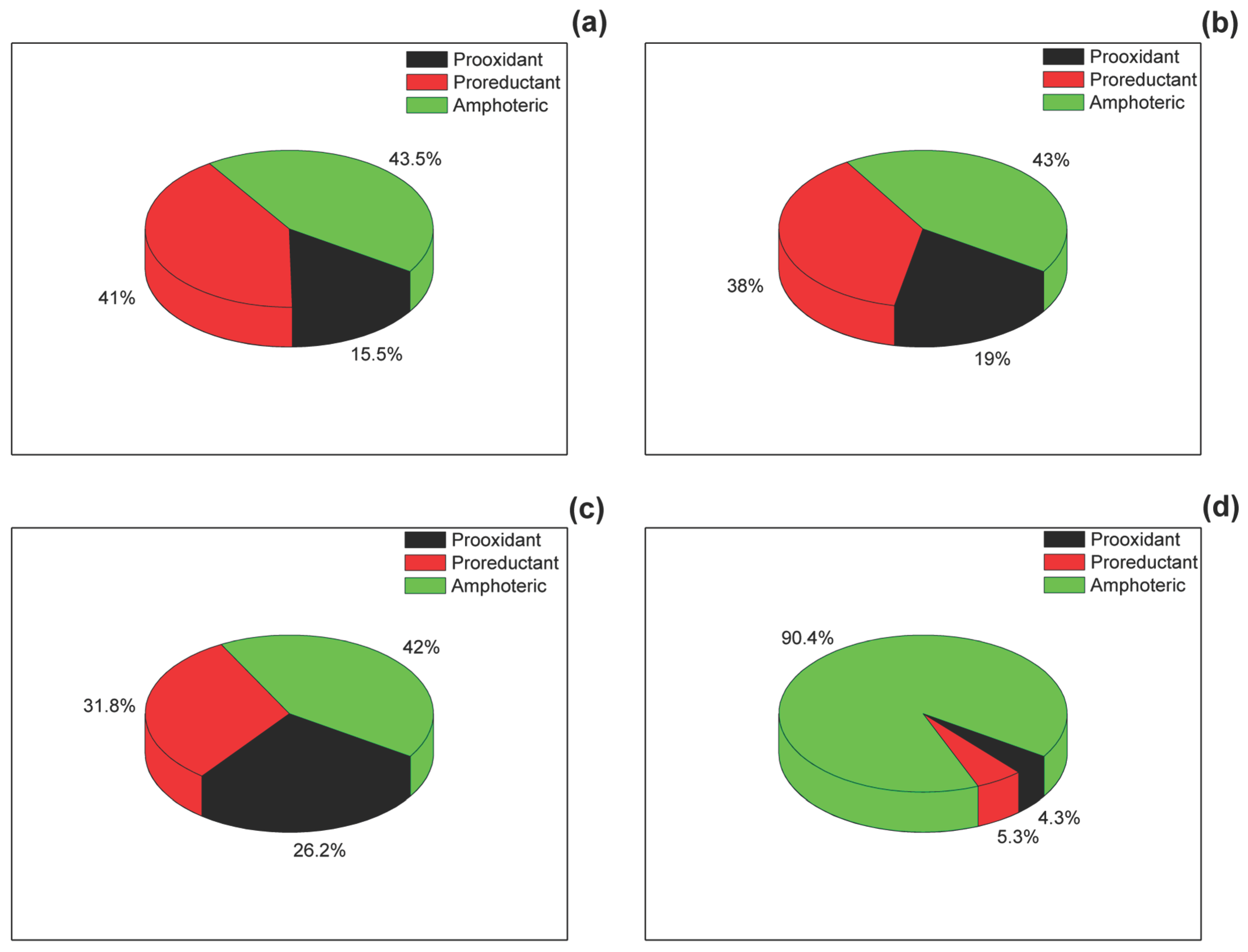
4]. Of these, 90.4% exhibited EIIP values between 0.00 and 0.10 Ry and AQVN values ranging from 2.4 to 3.2. This predominant region of the EIIP/AQVN space was designated the basic chemical space (BCS), as it contains the great majority of known chemical structures.
Molecules positioned to the left of the BCS show enhanced electron-donor capacity and correspond to pro-oxidant species. In contrast, those located to the right of the BCS act as pro-reductants with strong electron-accepting properties. These two groups represented 4.3% and 5.3% of the analyzed PubChem compounds, respectively. Molecules falling within the BCS display amphoteric behavior, with moderate redox characteristics.
2.2. Overview of Data and Classification Scheme
Interstellar compounds encompassing 380 molecules detected across astrophysical environments (Dataset 1 and Dataset 2) [
5,
6], 225 human nutrient molecules (Dataset 3) [
4], and >40 million compounds sourced from PubChem [
7] were classified into three redox categories:
- 1.
Prooxidants—predominant electron acceptors
- 2.
Proreductants—predominant electron donors
- 3.
Amphoteric molecules—able to act as either donors or acceptors depending on context.
While the PubChem dataset is vast and heterogeneous, its inclusion provides a baseline representing “chemistry at large,” free of biological or cosmic selection.
3. Results
In the Interstellar and nutritional molecules show nearly identical redox distributions in the AQVN/EIIP space. The two datasets exhibit a remarkably similar redox composition (
Figure 1):
Despite their completely different origins—cosmic molecular clouds vs. biologically selected metabolites—the distributions converge on a nearly identical tri-modal structure dominated by amfoteric molecules but with substantial fractions of specialized redox donors and acceptors.
The PubChem dataset stands in sharp contrast (
Figure 1):
This distribution reflects the unconstrained nature of synthetic and theoretical chemical diversity: most possible molecules do not adopt strongly polarized redox roles.
4. Discussion
4.1. Redox Diversity as a Universal Constraint on Functional Chemistry
The close alignment between interstellar chemistry and nutrient chemistry suggests that life did not emerge from a random subset of possible molecules. Instead, both cosmic prebiotic chemistry and biological evolution appear to favor a balanced redox diversity:
Enough prooxidants to support electron transfer chains
Enough proreductants to drive reductive synthesis
A large class of amphoteric molecules for buffering, stabilization, and dynamic switching
Such a distribution likely facilitates:
This implies that the precursors of life may have been drawn from a chemically privileged subset of the interstellar molecular inventory rather than from planet-specific contingencies.
4.2. Why PubChem Differs: The Distinction Between “Chemically Possible” and “Chemically Functional”
PubChem’s extreme skew toward amphoteric molecules highlights that most theoretically possible compounds do not support the balanced redox roles required for biological organization. The comparison suggests that redox specialization is not random but selected—cosmologically and biologically.
Thus, chemistry compatible with life appears to be a tiny, highly structured subset of all possible chemistry. PubChem serves as a control dataset, illustrating the extent to which life-relevant chemistry diverges from chemical arbitrariness.
4.3. Implications for Astrobiology: Redox Distribution as a Universal Biosignature
A central implication of this analysis is the following:
If the redox distribution observed in interstellar molecules is universal, then any life elsewhere in the universe—if it is based on chemical processes—would likely require a similar redox architecture. This does not imply identical biomolecules (e.g., DNA, proteins). Rather, it implies that the underlying chemical logic of life is constrained by redox balance:
Therefore, a balanced distribution of redox species within extraterrestrial molecular populations may serve as a functional biosignature—a chemical fingerprint indicating an environment capable of supporting life-like processes.
5. Conclusions
The comparative redox analysis of interstellar molecules, human nutrients, and the PubChem chemical universe reveals that:
Interstellar and biological molecules share a highly similar redox landscape, despite their radically different contexts.
The general chemical space is overwhelmingly amphoteric, lacking the redox diversity required for metabolic-like networks.
This pattern suggests a universal chemical constraint: life depends on balanced redox specialization, not on arbitrary molecular diversity.
If redox distributions in interstellar chemistry are universal, then life elsewhere in the galaxy would likely resemble Earth-life in its core chemical architecture, even if structurally distinct.
These findings open a new perspective on the chemical preconditions for life and suggest that redox diversity may be a powerful, quantifiable biosignature for astrobiological exploration.
Funding
This research received no external funding
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
A.1. Distributions in different libraries
Additional Chemical Libraries and Redox Distributions
We analyzed several well-established chemical libraries representing natural, metabolic, and pharmaceutical chemical space. Using the same EIIP/AQVN classification scheme applied to interstellar molecules and nutrients, we computed the distribution of n⁺, n⁻, and n⁰ species in each dataset.
Summary of distributions:
| Library |
n⁺ (%) |
n⁻ (%) |
n⁰ (%) |
N |
Predox = (n⁺+ n⁻)/N |
| COCONUT (natural products) |
6890 (10.5%) |
3942 (6.0%) |
54,699 (83.5%) |
65,531 |
0.165 |
| Plant metabolites |
478 (16.8%) |
406 (14.2%) |
1970 (69.0%) |
2,854 |
0.31 |
| KEGG metabolic compounds |
255 (3.4%) |
422 (5.7%) |
6,715 (90.9%) |
7,392 |
0.091 |
| DrugBank (approved) |
111 (7.6%) |
87 (6.0%) |
1,263 (86.4%) |
1,461 |
0.136 |
| DrugBank (experimental) |
286 (5.8%) |
676 (13.7%) |
3,969 (80.5%) |
4,931 |
0.195 |
| PubChem (random sample) |
~4.3% |
~5.3% |
~90% |
4,510,644 |
0.096 |
| Interstellar molecules |
— |
— |
— |
380 |
~0.57 |
| Human nutrients |
— |
— |
— |
225 |
~0.58 |
The central observation is that all additional control libraries fall in the 0.09–0.20 range for redox-specialized molecules, with plant metabolites (0.31) as the only partial exception. In contrast, interstellar molecules and nutrients cluster tightly around 0.57–0.58, which is far outside the range observed in any control dataset.
This confirms that PubChem was not an outlier: the low fraction of redox-specialized molecules is a general property of unconstrained chemical space.
A.2. Statistical Tests of Redox Distribution Differences
We performed two-proportion z-tests comparing interstellar molecules and nutrients against control libraries.
| Comparison |
z-value |
p-value |
| Interstellar vs PubChem |
~31.2 |
~10⁻²¹³ |
| Interstellar vs KEGG |
~28.5 |
~10⁻¹⁷⁸ |
| Interstellar vs COCONUT |
~21.0 |
~10⁻⁹⁷ |
| Interstellar vs DrugBank (approved) |
~17.9 |
~10⁻⁷¹ |
| Interstellar vs DrugBank (experimental) |
~16.8 |
~10⁻⁶³ |
| Interstellar vs plant metabolites |
~10.0 |
~10⁻²³ |
| Nutrients vs PubChem |
~24.6 |
~10⁻¹³³ |
| Nutrients vs KEGG |
~23.5 |
~10⁻¹²² |
| Nutrients vs COCONUT |
~16.7 |
~10⁻⁶² |
| Nutrients vs DrugBank (approved) |
~15.6 |
~10⁻⁵⁴ |
| Nutrients vs DrugBank (experimental) |
~13.8 |
~10⁻⁴³ |
| Nutrients vs plant metabolites |
~8.3 |
~10⁻¹⁶ |
Across all comparisons, p ≪ 10⁻¹⁰, confirming overwhelming statistical significance. The probability that interstellar or nutrient redox profiles arise from the same underlying distribution as any of the control libraries is effectively zero.
A.3. Conclusion of statistical testing
The similarity between interstellar molecules and nutrients is statistically robust.
The divergence between interstellar/nutrient chemistry and generic chemical libraries is highly significant and cannot be attributed to database size, sampling variation, or chance.
A.4. Discussion of Implications
Life-compatible redox chemistry appears as a distinct subset of cosmic chemistry
The joint redox profile of interstellar molecules and human nutrients—characterized by balanced proportions of electron donors, electron acceptors, and amphoteric species—is not observed in any of the large control libraries. This suggests that the chemical environment from which life emerged was not random, but drawn from a redox-structured subset of cosmic molecular space that already provided:
coupled redox pairs (n⁺–n⁻),
amphoteric intermediates enabling reversible reactions,
a distribution favorable to energy flow far from equilibrium.
This architecture is consistent with theoretical requirements for early metabolic networks and supports the idea that primordial chemistry contained pre-configured redox scaffolding.
Biological evolution preserved, rather than erased, the primordial redox pattern
It is striking that human nutrients—and, more broadly, molecules essential for cellular function—retain the same redox proportions as interstellar molecules. By contrast:
central metabolism (KEGG) is highly filtered toward amphoteric molecules (~91%),
pharmaceutical space deviates from natural redox balance (~10–20%),
generic chemical space is largely incompatible with life’s redox regime.
This implies a layered model of chemical selection:
Cosmic chemistry supplies a redox-balanced chemical landscape.
Early prebiotic chemistry / nutrient-level chemistry retains this balance.
Core metabolism evolves toward stability by filtering to neutral species.
Modern chemical libraries diverge further from life’s preferred redox distribution.
Astrobiology: a quantitative redox biosignature
Because the interstellar–nutrient redox profile is unique among all tested chemical spaces, the fraction of redox-specialized molecules (n⁺+ n⁻) may serve as a quantitative biosignature:
Thus, extraterrestrial environments with a high fraction of redox-specialized molecules may indicate chemical ecosystems conducive to life.
Implications for drug discovery and molecular design
The mismatch between life-like redox distributions and typical drug libraries suggests that much of pharmaceutical chemistry occupies electronically misaligned regions of chemical space. Redox-aware EIIP/AQVN filtering may therefore:
improve hit rates,
enrich scaffolds compatible with biological electron flow,
reduce attrition among candidates with non-functional electronic profiles.
A.5. Summary
The expanded analysis confirms that the shared redox profile of interstellar molecules and human nutrients is:
This strengthens the central conclusion of the preprint: life-compatible chemistry is rooted in a non-random, redox-structured subset of cosmic molecular diversity, preserved across planetary evolution and detectable through the EIIP/AQVN electronic parameters.
References
- Le Gal, K.; Schmidt, E.E.; Sayin, V.I. Cellular redox homeostasis. Antioxidants 2021, 10, 1377–1383. [Google Scholar] [CrossRef]
- Guelin, M.; Cernicharo, J. Organic molecules in interstellar space: Latest advances. Front. Astron. Space Sci. 2022, 9, 787567. [Google Scholar] [CrossRef]
- Veljković, V.; Slavić, I. Simple General-Model Pseudopotential. Phys. Rev. Lett. 1972, 29, 105–107. [Google Scholar] [CrossRef]
- Veljkovic, V.; Perovic, V.; Anderluh, M.; Paessler, S.; Veljkovic, M.; Glisic, S.; Nicolson, G. A simple method for calculation of basic molecular properties of nutrients and use as a criterion for healthy diet. F1000Res 2017, 6, 13–21. [Google Scholar] [CrossRef]
- List of Observed Interstellar Molecules. Available online: https://molecules-in.space/.
- List of interstellar and circumstellar compounds. Available online: https://en.wikipedia.org/wiki/List_of_interstellar_and_circumstellar_molecules.
- PubChem database. Available online: http://pubchem.ncbi.nlm.nih.gov.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).