Submitted:
01 December 2025
Posted:
03 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
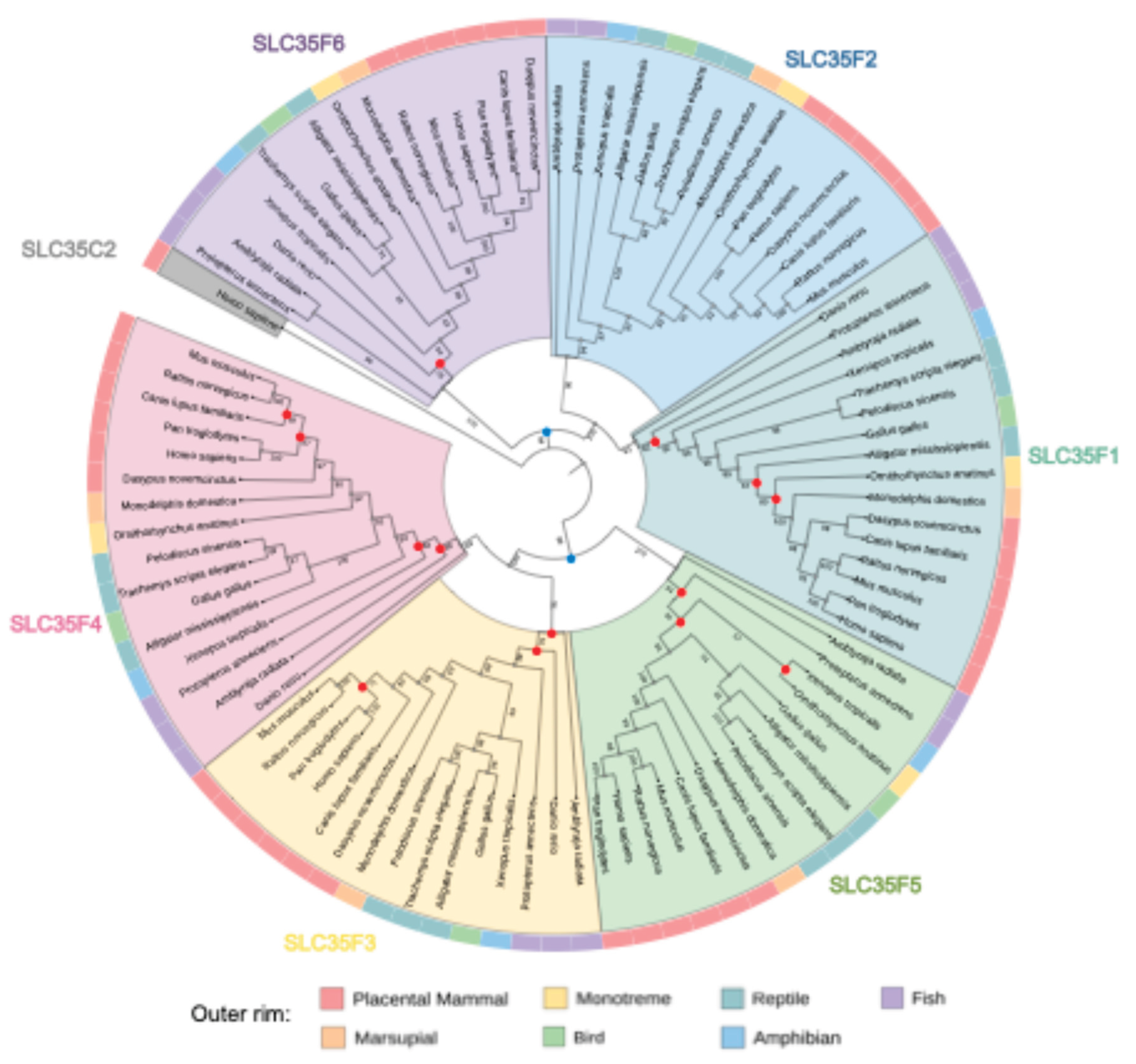
2.1. Phylogenetic Analysis of the SLC35F Family Reveals Two Divergent Evolutionary Lineages
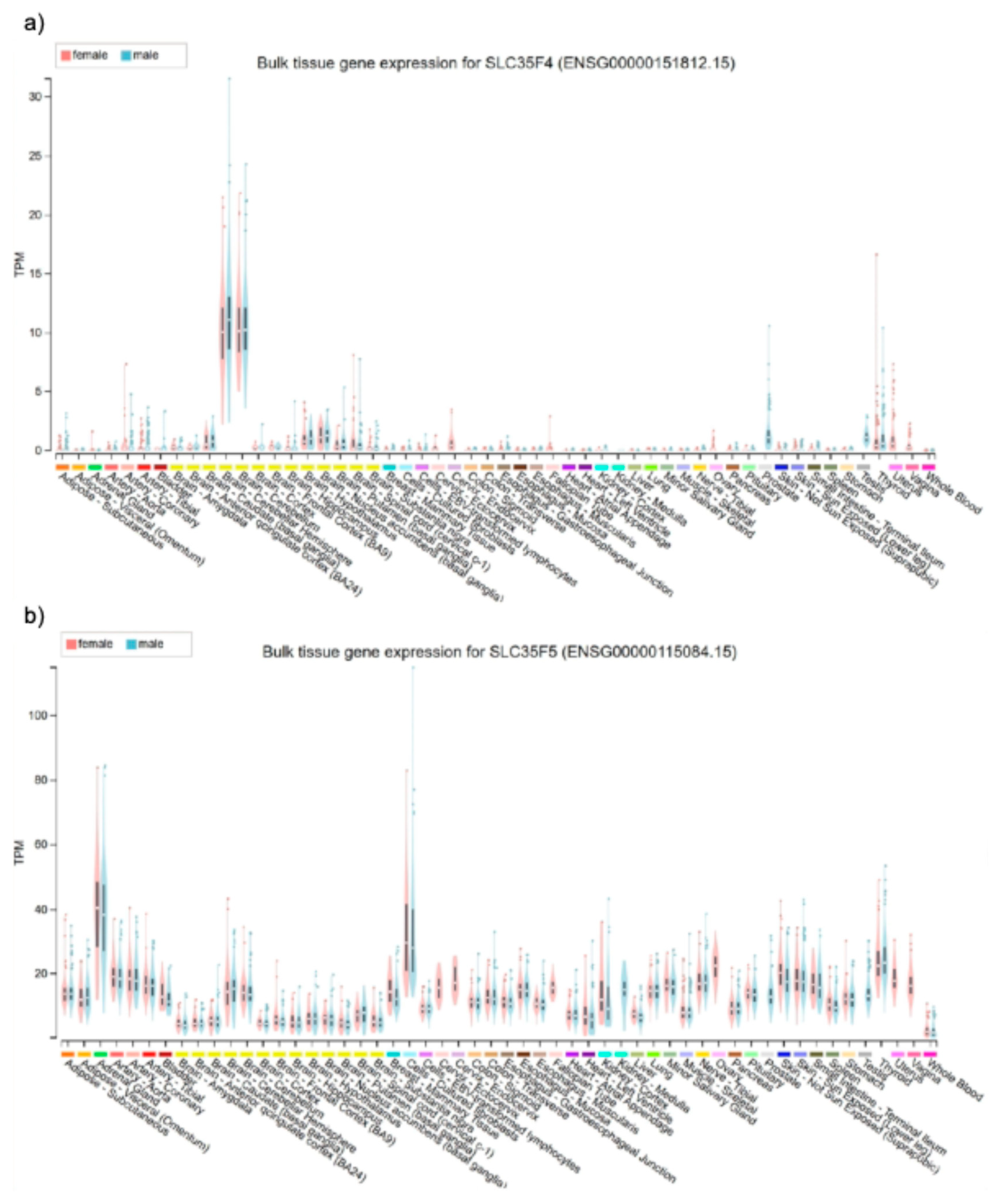
2.2. SLC35F4 and SLC35F5 Are Golgi-Localized Transmembrane Proteins with Distinct Tissue-Specific Expression Profiles
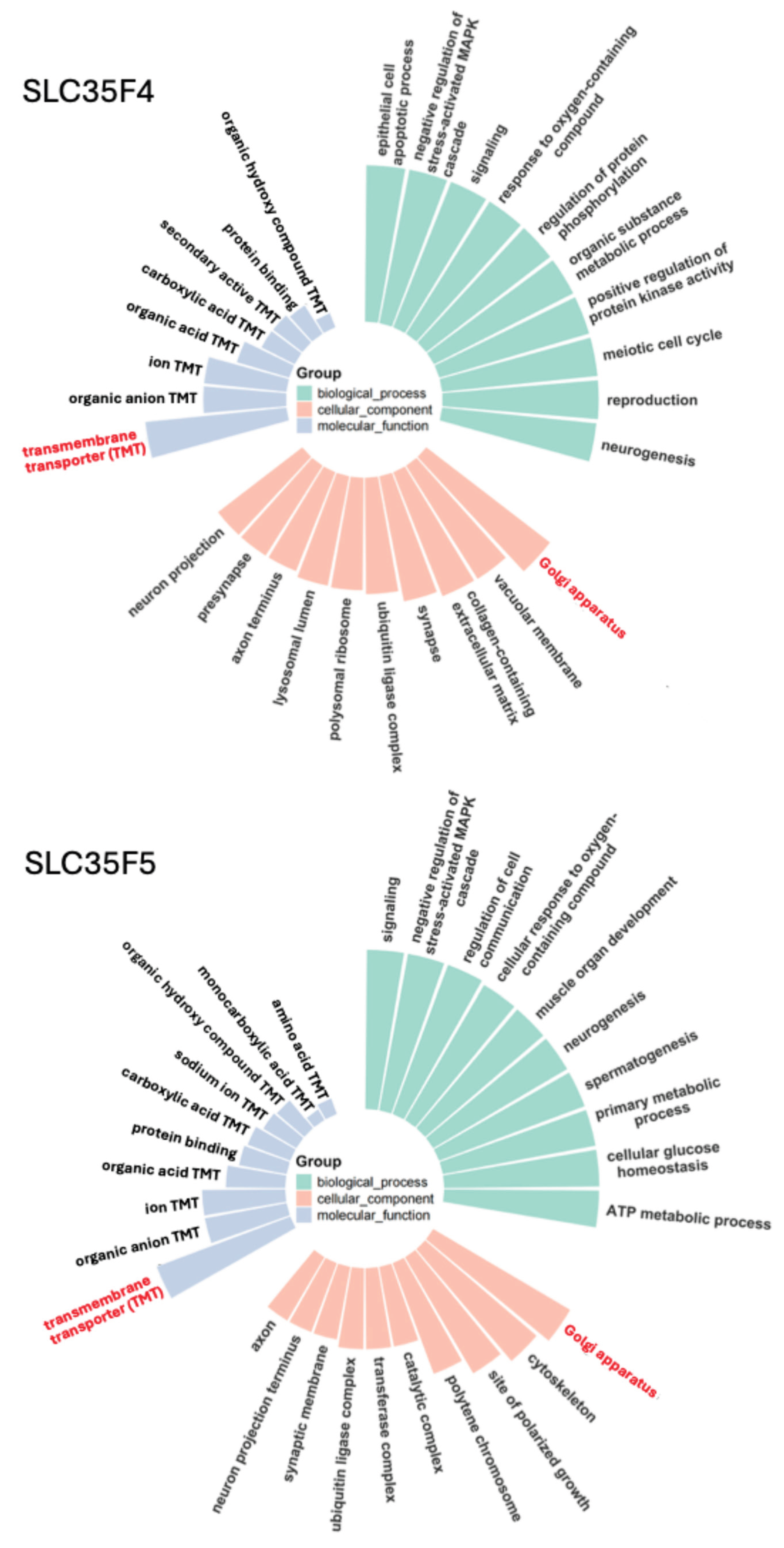
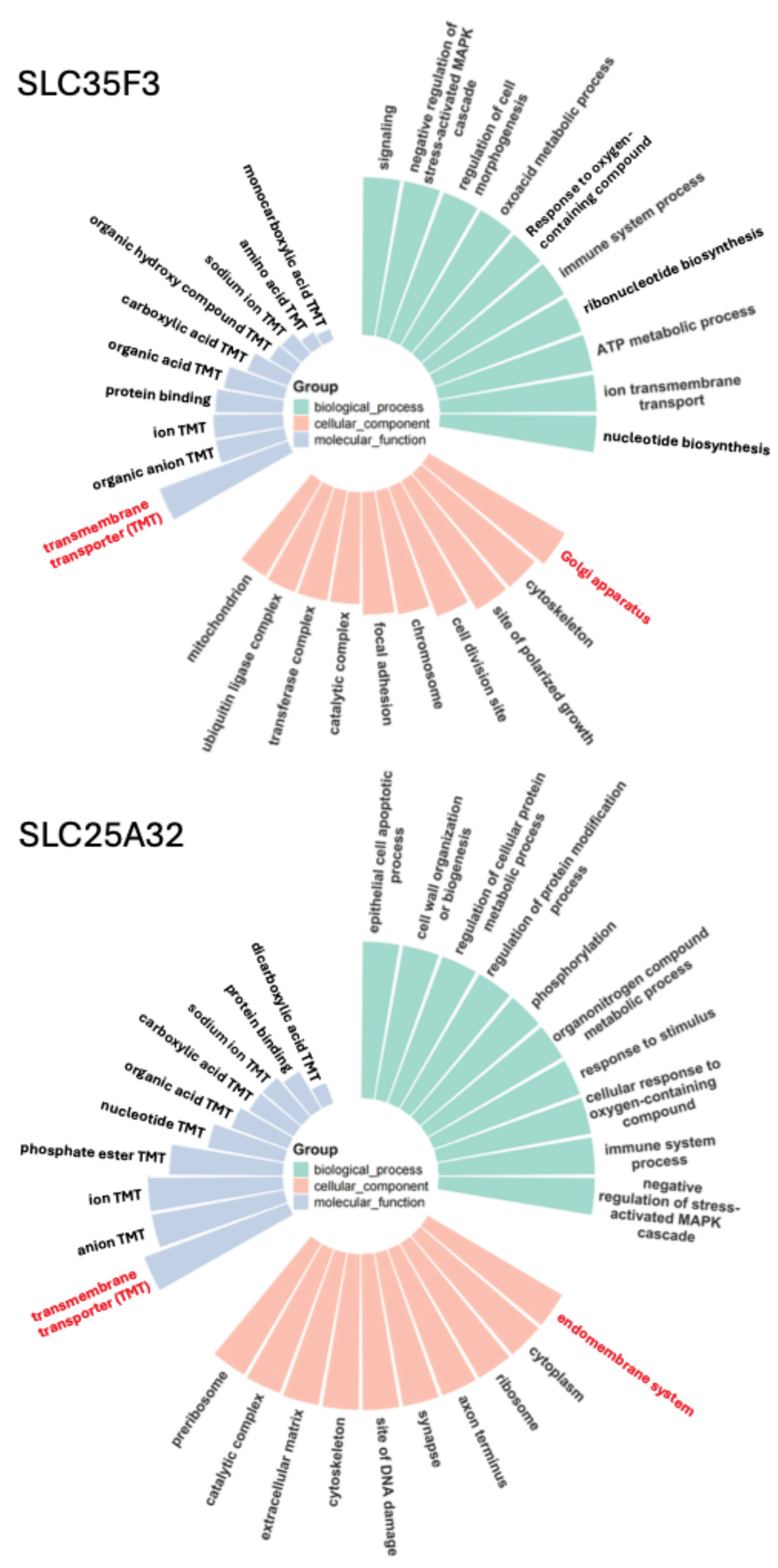
2.3. Gene Ontology Analysis Predicts Functions of SLC35F4 and SLC35F5 in Golgi-Localized Cofactor Transport Required for Post-Translational Modification
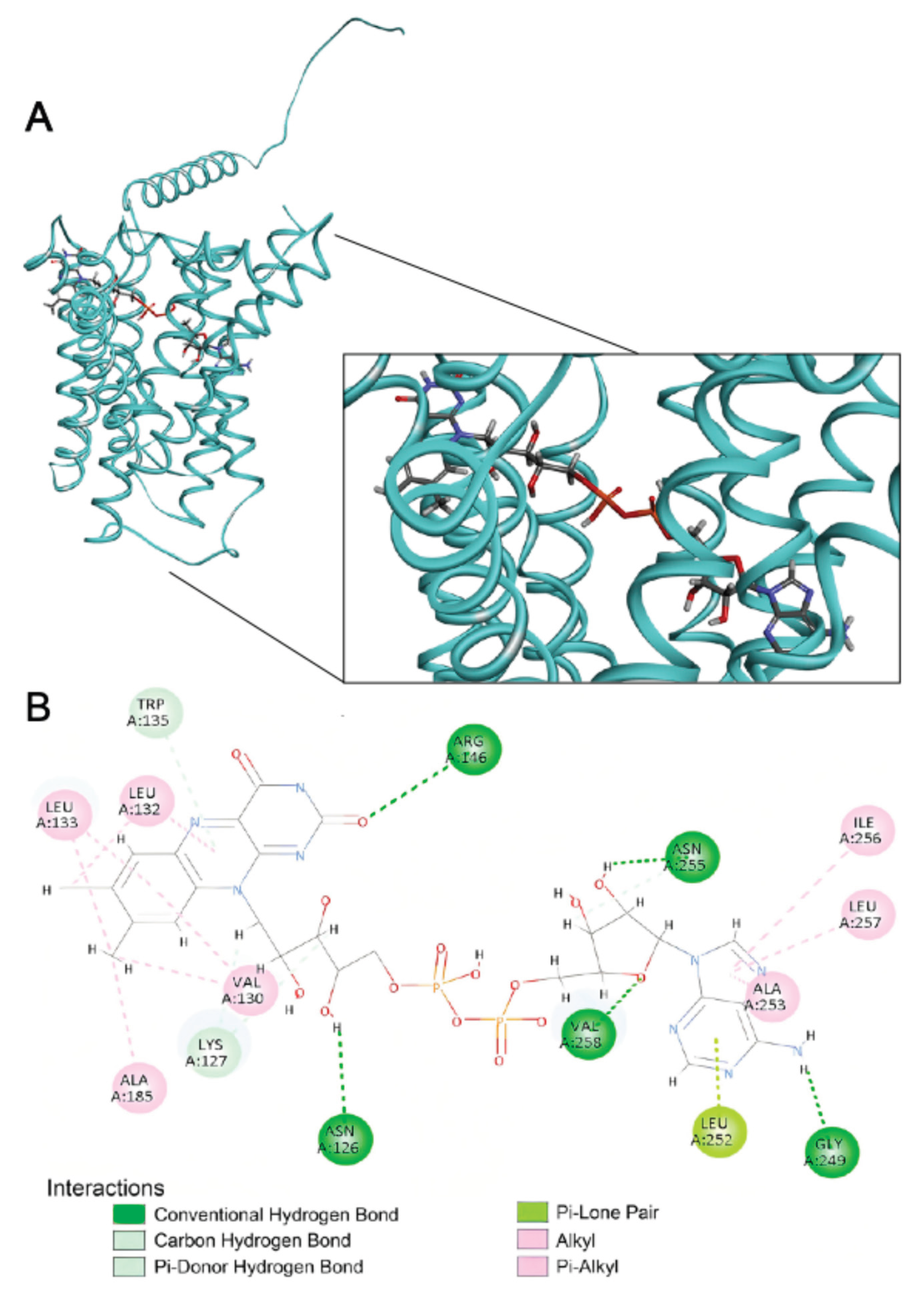
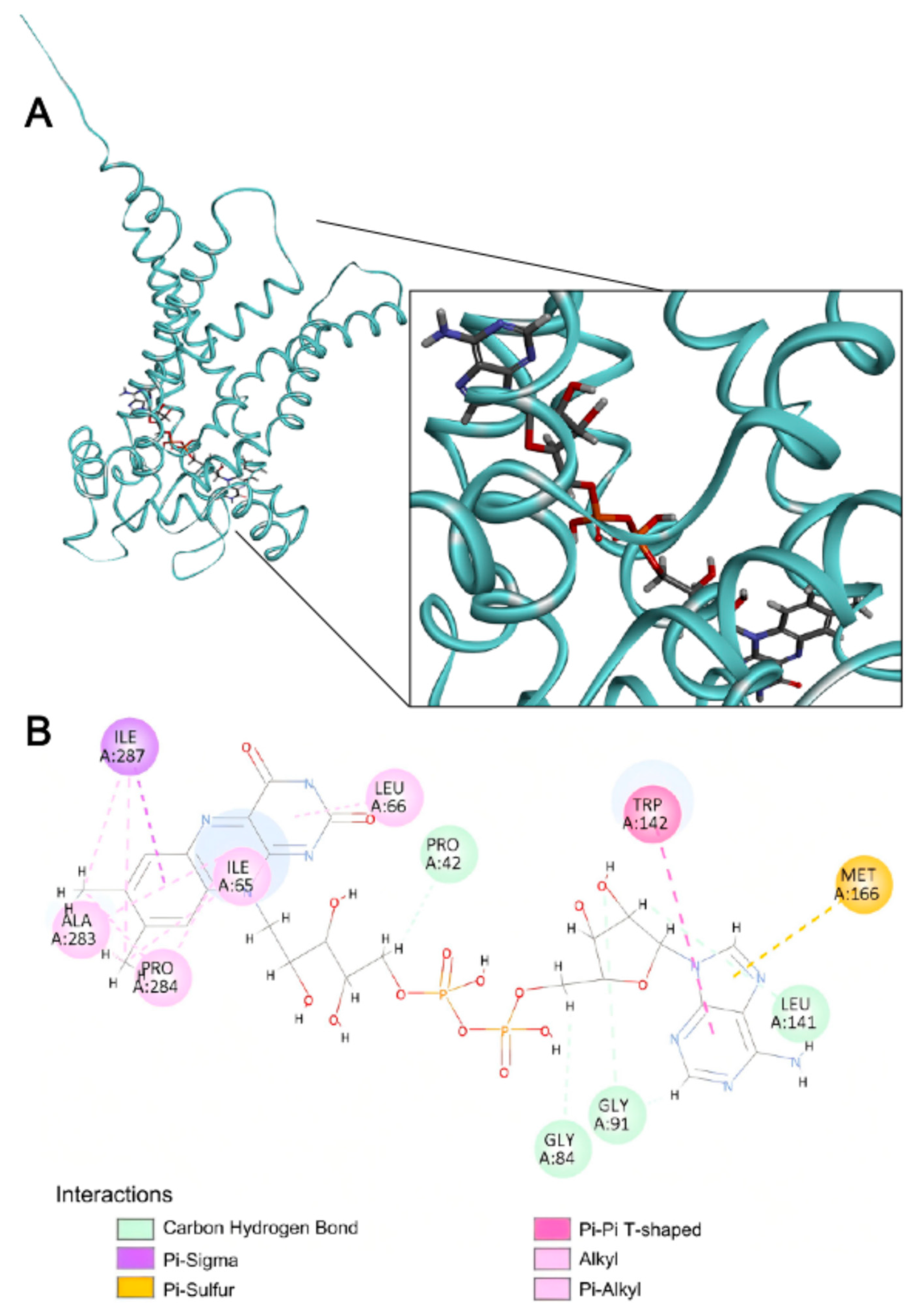
2.4. Structural Basis of FAD Binding by SLC35F4, SLC35F5, and the Canonical FAD Transporter SLC25A32
3. Discussion
4. Materials and Methods
4.1. Tissue-wide RNA Expression Analysis of SLC35F4 and SLC35F5
4.2. Transmembrane Topology and Subcellular Localization Prediction
4.3. Phylogenetic Analysis of the SLC35F Subfamily
4.4. Synteny Analysis
4.5. Gene Ontology (GO) Term Prediction
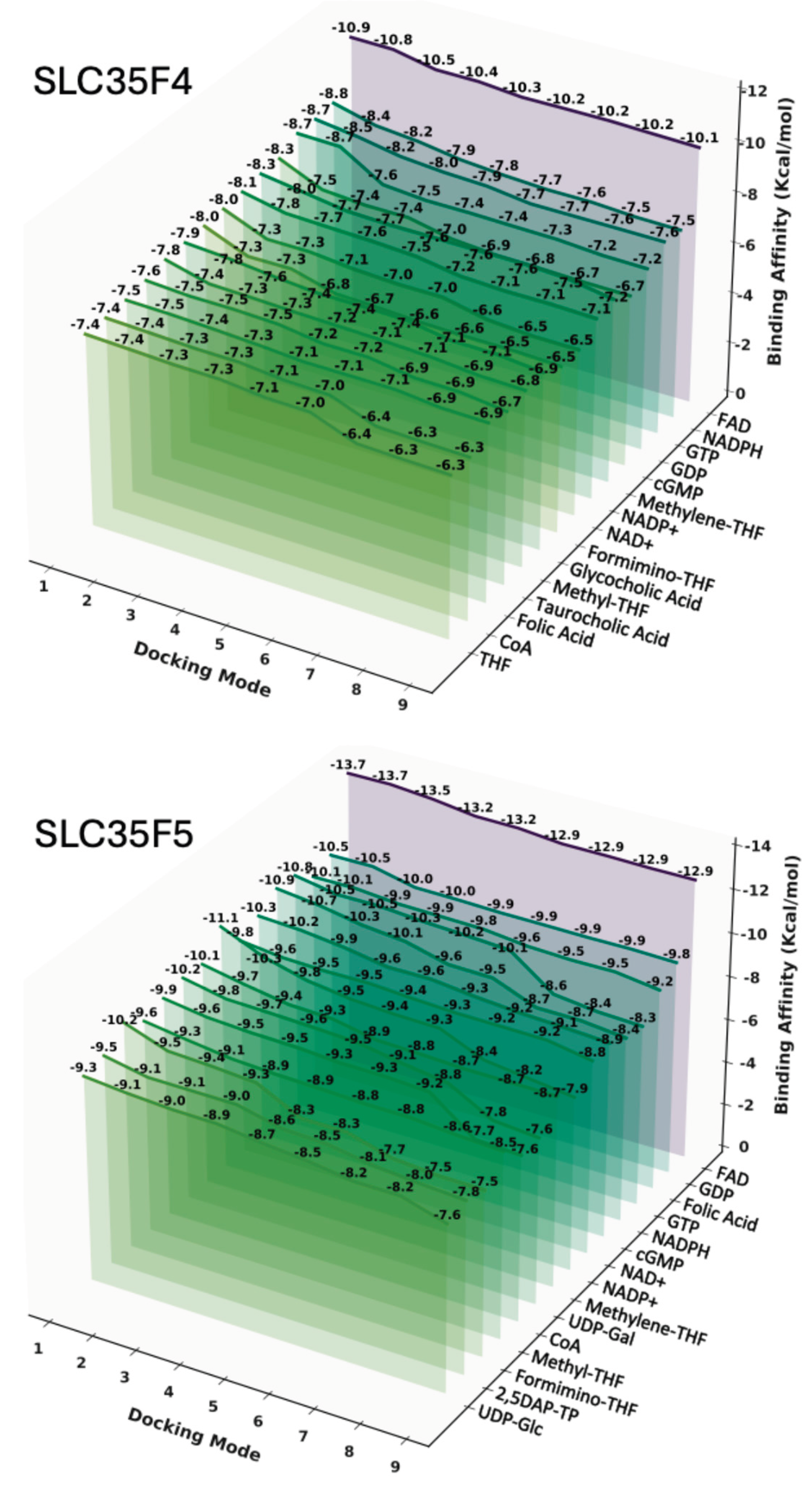
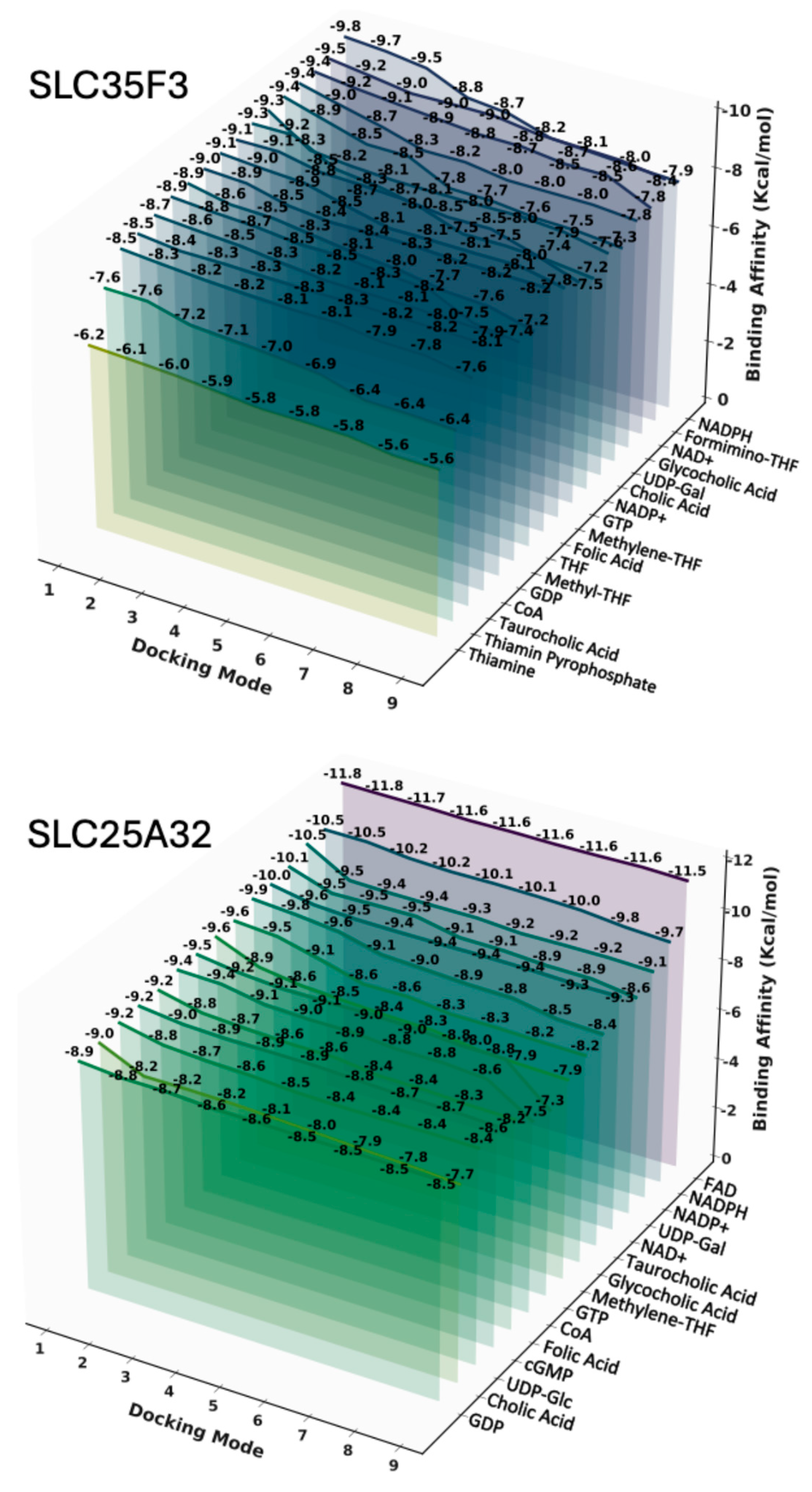
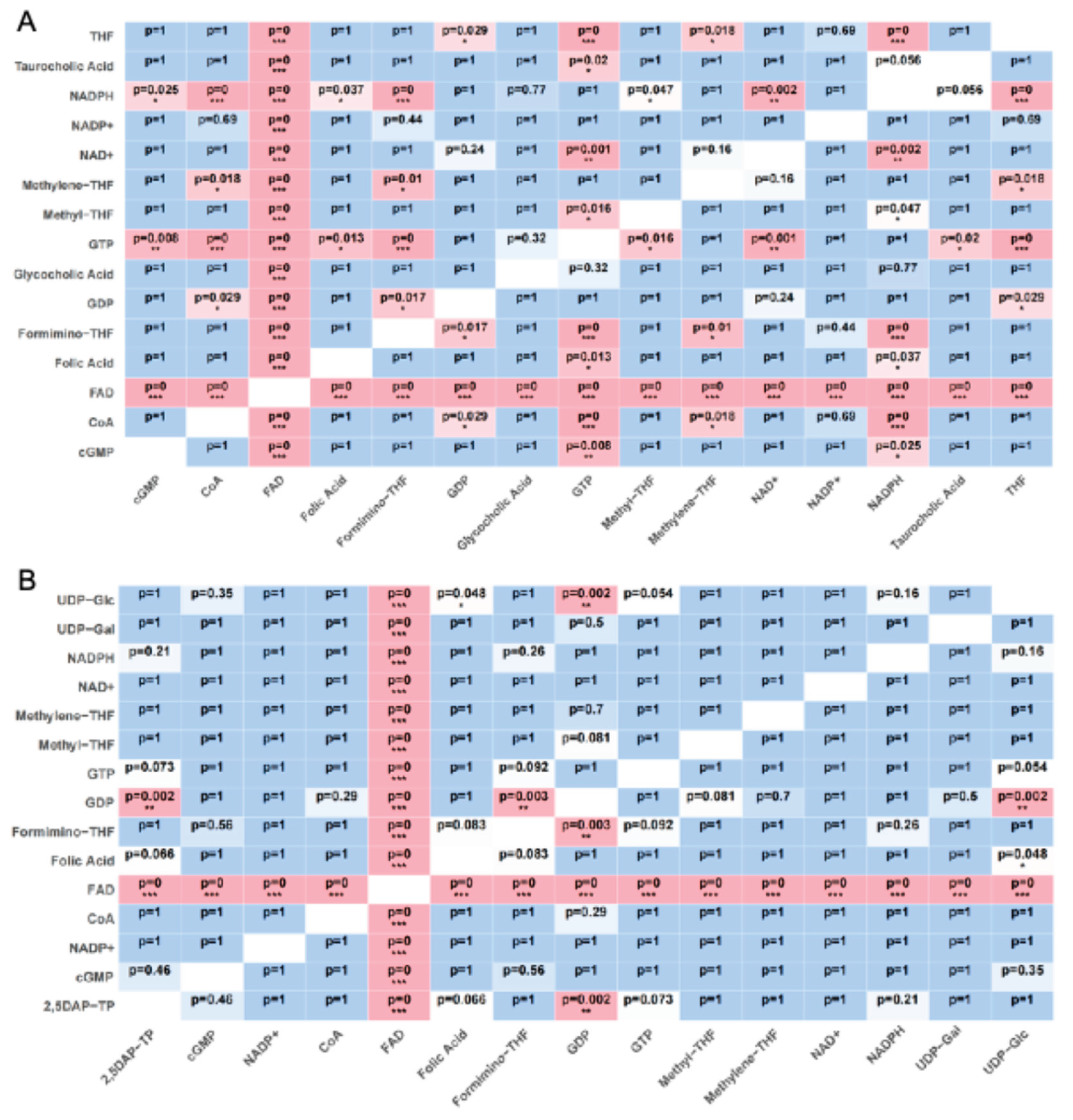
4.6. Ligand Docking and Pairwise Statistical Analysis
4.7. Docking Visualization and Interaction Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SLC | Solute Carrier |
| ER | Endoplasmic Reticulum |
| PAPS | Phosphoadenosine phosphosulfate |
| PTM | Post-translational Modifications |
| GO | Gene Ontology |
| TPP | Thiamine Pyrophosphate |
| FAD | Flavin Adenine Dinucleotide |
| THF | Tetrahydrofolate |
| UDP-Glc | Uridine Diphosphate Glucose |
| UDP-Gal | Uridine Diphosphate Galactose |
| 2,5 DAP-TP | 2,5-Diamino-6-(5’-triphosphoryl-3’,4’-trihydroxy-2’-oxopentyl)-amino-4-oxopyrimidine |
| Ero1 | ER Oxidase 1 |
| PDI | Protein Disulfide Isomerase |
References
- Pizzagalli, M. D.; Bensimon, A.; Superti-Furga, G., A guide to plasma membrane solute carrier proteins. Febs j 2021, 288, 2784-2835. [CrossRef]
- Liu, X., SLC Family Transporters. Adv Exp Med Biol 2019, 1141, 101-202.
- Huang, S.; Czech, M. P., The GLUT4 glucose transporter. Cell Metab 2007, 5, 237-52.
- Yiew, N. K. H.; Finck, B. N., The mitochondrial pyruvate carrier at the crossroads of intermediary metabolism. Am J Physiol Endocrinol Metab 2022, 323, E33-e52. [CrossRef]
- Martínez-Reyes, I.; Chandel, N. S., Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020, 11, 102. [CrossRef]
- Cimadamore-Werthein, C.; Jaiquel Baron, S.; King, M. S.; Springett, R.; Kunji, E. R., Human mitochondrial ADP/ATP carrier SLC25A4 operates with a ping-pong kinetic mechanism. EMBO Rep 2023, 24, e57127. [CrossRef]
- Zhang, Y.; Newstead, S.; Sarkies, P., Predicting substrates for orphan solute carrier proteins using multi-omics datasets. BMC Genomics 2025, 26, 130. [CrossRef]
- Hadley, B.; Litfin, T.; Day, C. J.; Haselhorst, T.; Zhou, Y.; Tiralongo, J., Nucleotide Sugar Transporter SLC35 Family Structure and Function. Comput Struct Biotechnol J 2019, 17, 1123-1134. [CrossRef]
- Ury, B.; Potelle, S.; Caligiore, F.; Whorton, M. R.; Bommer, G. T., The promiscuous binding pocket of SLC35A1 ensures redundant transport of CDP-ribitol to the Golgi. J Biol Chem 2021, 296, 100789. [CrossRef]
- Fang, R.; Jiang, Q.; Guan, Y.; Gao, P.; Zhang, R.; Zhao, Z.; Jiang, Z., Golgi apparatus-synthesized sulfated glycosaminoglycans mediate polymerization and activation of the cGAMP sensor STING. Immunity 2021, 54, 962-975.e8. [CrossRef]
- Van den Bossche, F.; Tevel, V.; Gilis, F.; Gaussin, J. F.; Boonen, M.; Jadot, M., Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway. Int J Mol Sci 2024, 25, 6718. [CrossRef]
- Kamiyama, S.; Sone, H., Solute Carrier Family 35 (SLC35)—An Overview and Recent Progress. In Biologics, 2024; Vol. 4, pp 242-279. [CrossRef]
- Zhang, K.; Huentelman, M. J.; Rao, F.; Sun, E. I.; Corneveaux, J. J.; Schork, A. J.; Wei, Z.; Waalen, J.; Miramontes-Gonzalez, J. P.; Hightower, C. M.; Maihofer, A. X.; Mahata, M.; Pastinen, T.; Ehret, G. B.; Schork, N. J.; Eskin, E.; Nievergelt, C. M.; Saier, M. H., Jr.; O’Connor, D. T., Genetic implication of a novel thiamine transporter in human hypertension. J Am Coll Cardiol 2014, 63, 1542-55. [CrossRef]
- Schiff, M.; Veauville-Merllié, A.; Su, C. H.; Tzagoloff, A.; Rak, M.; Ogier de Baulny, H.; Boutron, A.; Smedts-Walters, H.; Romero, N. B.; Rigal, O.; Rustin, P.; Vianey-Saban, C.; Acquaviva-Bourdain, C., SLC25A32 Mutations and Riboflavin-Responsive Exercise Intolerance. N Engl J Med 2016, 374, 795-7. [CrossRef]
- Peng, M. Z.; Shao, Y. X.; Li, X. Z.; Zhang, K. D.; Cai, Y. N.; Lin, Y. T.; Jiang, M. Y.; Liu, Z. C.; Su, X. Y.; Zhang, W.; Jiang, X. L.; Liu, L., Mitochondrial FAD shortage in SLC25A32 deficiency affects folate-mediated one-carbon metabolism. Cell Mol Life Sci 2022, 79, 375. [CrossRef]
- Zuckerkandl, E.; Pauling, L., Molecular Disease, Evolution, and Genic Heterogeneity. Academic Press 1962, 189-225.
- Zuckerkandl, E.; Pauling, L., Molecules as documents of evolutionary history. J Theor Biol 1965, 8, 357-66. [CrossRef]
- Eck, R. V.; Dayhoff, M. O., Evolution of the structure of ferredoxin based on living relics of primitive amino Acid sequences. Science 1966, 152, 363-6. [CrossRef]
- Dayhoff, M. O., Computer analysis of protein evolution. Sci Am 1969, 221, 86-95. [CrossRef]
- Hunt, L. T.; Dayhoff, M. O., The occurrence in proteins of the tripeptides Asn-X-Ser and Asn-X-Thr and of bound carbohydrate. Biochem Biophys Res Commun 1970, 39, 757-65. [CrossRef]
- Dayhoff, M. O.; Orcutt, B. C., Methods for identifying proteins by using partial sequences. Proc Natl Acad Sci U S A 1979, 76, 2170-4. [CrossRef]
- Buel, G. R.; Walters, K. J., Can AlphaFold2 predict the impact of missense mutations on structure? Nat Struct Mol Biol 2022, 29, (1-2. [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L. H.; Zielinski, M.; Sargeant, T.; Schneider, R. G.; Senior, A. W.; Jumper, J.; Hassabis, D.; Kohli, P.; Avsec, Ž., Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [CrossRef]
- Durairaj, J.; Waterhouse, A. M.; Mets, T.; Brodiazhenko, T.; Abdullah, M.; Studer, G.; Tauriello, G.; Akdel, M.; Andreeva, A.; Bateman, A.; Tenson, T.; Hauryliuk, V.; Schwede, T.; Pereira, J., Uncovering new families and folds in the natural protein universe. Nature 2023, 622, 646-653. [CrossRef]
- Barrio-Hernandez, I.; Yeo, J.; Jänes, J.; Mirdita, M.; Gilchrist, C. L. M.; Wein, T.; Varadi, M.; Velankar, S.; Beltrao, P.; Steinegger, M., Clustering predicted structures at the scale of the known protein universe. Nature 2023, 622, 637-645. [CrossRef]
- Hirschberg, C.B.; Robbins, P.W.; Abeijon, C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem 1998, 67, 49-69. [CrossRef]
- Parker, J.L.; Newstead, S. Gateway to the Golgi: molecular mechanisms of nucleotide sugar transporters. Curr Opin Struct Biol 2019, 57, 127-134. [CrossRef]
- Barile, M.; Giancaspero, T. A.; Leone, P.; Galluccio, M.; Indiveri, C., Riboflavin transport and metabolism in humans. J Inherit Metab Dis 2016, 39, 545-57. [CrossRef]
- Tu, B.P.; Ho-Schleyer, S.C.; Travers, K.J,; Weissman, J.S. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 2000, 290,1571-4. [CrossRef]
- Jessop, C. E.; Bulleid, N. J., Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem 2004, 279, 55341-7. [CrossRef]
- Joosten V.; van Berkel W.J. Flavoenzymes. Curr Opin Chem Biol 2007, 11, 195-202.
- Hudson, D.A.; Gannon, S.A.; Thorpe, C. Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic Biol Med 2015, 80,171-82. [CrossRef]
- Robinson PJ, Pringle MA, Fleming B, Bulleid NJ. Distinct role of ERp57 and ERdj5 as a disulfide isomerase and reductase during ER protein folding. J Cell Sci 2023, 136, jcs260656.
- Coppock, D. L.; Thorpe, C., Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal 2006, 8, 300-11. [CrossRef]
- Sevier, C. S., Erv2 and quiescin sulfhydryl oxidases: Erv-domain enzymes associated with the secretory pathway. Antioxid Redox Signal 2012, 16, 800-8. [CrossRef]
- Reznik, N.; Fass, D., Disulfide bond formation and redox regulation in the Golgi apparatus. FEBS Lett 2022, 596, 2859-2872. [CrossRef]
- Yang, Y.; Peng, H.; Meng, D.; Fa, Z.; Yao, C.; Lin, X.; Schick, J.; Jin, X. Stress Management: How the Endoplasmic Reticulum Mitigates Protein Misfolding and Oxidative Stress by the Dual Role of Glutathione Peroxidase 8. Biomolecules 2025, 15, 847. [CrossRef]
- Lake, D. F.; Faigel, D. O., The emerging role of QSOX1 in cancer. Antioxid Redox Signal 2014, 21, 485-96. [CrossRef]
- Knutsvik, G.; Collett, K.; Arnes, J.; Akslen, L. A.; Stefansson, I. M., QSOX1 expression is associated with aggressive tumor features and reduced survival in breast carcinomas. Mod Pathol 2016, 29, 1485-1491. [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z., GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017, 45, W98-w102. [CrossRef]
- Nishimura, M.; Suzuki, S.; Satoh, T.; Naito, S. Tissue-specific mRNA expression profiles of human solute carrier 35 transporters. Drug Metab Pharmacokinet 2009, 24, 91-9. [CrossRef]
- Lackey, E. P.; Heck, D. H.; Sillitoe, R. V., Recent advances in understanding the mechanisms of cerebellar granule cell development and function and their contribution to behavior. F1000Res 2018, 7, 1142. [CrossRef]
- Lopera, F.; Marino, C.; Chandrahas, A. S.; O’Hare, M.; Villalba-Moreno, N. D.; Aguillon, D.; Baena, A.; Sanchez, J. S.; Vila-Castelar, C.; Ramirez Gomez, L.; Chmielewska, N.; Oliveira, G. M.; Littau, J. L.; Hartmann, K.; Park, K.; Krasemann, S.; Glatzel, M.; Schoemaker, D.; Gonzalez-Buendia, L.; Delgado-Tirado, S.; Arevalo-Alquichire, S.; Saez-Torres, K. L.; Amarnani, D.; Kim, L. A.; Mazzarino, R. C.; Gordon, H.; Bocanegra, Y.; Villegas, A.; Gai, X.; Bootwalla, M.; Ji, J.; Shen, L.; Kosik, K. S.; Su, Y.; Chen, Y.; Schultz, A.; Sperling, R. A.; Johnson, K.; Reiman, E. M.; Sepulveda-Falla, D.; Arboleda-Velasquez, J. F.; Quiroz, Y. T., Resilience to autosomal dominant Alzheimer’s disease in a Reelin-COLBOS heterozygous man. Nat Med 2023, 29, 1243-1252. [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M., The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015, 19, A68-77. [CrossRef]
- Weibel, E. R.; Palade, G. E., New cytoplasmic components in arterial endothelia. J Cell Biol 1964, 23, 101-12. [CrossRef]
- Helle, K. B.; Metz-Boutigue, M. H.; Cerra, M. C.; Angelone, T., Chromogranins: from discovery to current times. Pflugers Arch 2018, 470, 143-154. [CrossRef]
- Huang, T. T., Jr.; Hardy, D.; Yanagimachi, H.; Teuscher, C.; Tung, K.; Wild, G.; Yanagimachi, R., pH and protease control of acrosomal content stasis and release during the guinea pig sperm acrosome reaction. Biol Reprod 1985, 32, 451-62. [CrossRef]
- Hardy, D. M.; Oda, M. N.; Friend, D. S.; Huang, T. T., Jr., A mechanism for differential release of acrosomal enzymes during the acrosome reaction. Biochem J 1991, 275, 759-66. [CrossRef]
- Baba, T.; Hoff, H. B., 3rd; Nemoto, H.; Lee, H.; Orth, J.; Arai, Y.; Gerton, G. L., Acrogranin, an acrosomal cysteine-rich glycoprotein, is the precursor of the growth-modulating peptides, granulins, and epithelins, and is expressed in somatic as well as male germ cells. Mol Reprod Dev 1993, 34, 233-43.
- Bierring, F., Electron microscopic observations on the mucus production in human and rat intestinal goblet cells. Acta Pathol Microbiol Scand 1962, 54, 241-52. [CrossRef]
- Afzelius, B. A., The ultrastructure of the cortical granules and their products in the sea urchin egg as studied with the electron microscope. Exp Cell Res 1956, 10, 257-85. [CrossRef]
- Valentijn, K. M.; Sadler, J. E.; Valentijn, J. A.; Voorberg, J.; Eikenboom, J., Functional architecture of Weibel-Palade bodies. Blood 2011, 117, 5033-43. [CrossRef]
- Barbosa, J. A.; Gill, B. M.; Takiyyuddin, M. A.; O’Connor, D. T., Chromogranin A: posttranslational modifications in secretory granules. Endocrinology 1991, 128, 174-90. [CrossRef]
- Bi, M.; Hickox, J. R.; Winfrey, V. P.; Olson, G. E.; Hardy, D. M., Processing, localization and binding activity of zonadhesin suggest a function in sperm adhesion to the zona pellucida during exocytosis of the acrosome. Biochem J 2003, 375, 477-88. [CrossRef]
- Olson, G. E.; Winfrey, V. P.; Bi, M.; Hardy, D. M.; NagDas, S. K., Zonadhesin assembly into the hamster sperm acrosomal matrix occurs by distinct targeting strategies during spermiogenesis and maturation in the epididymis. Biol Reprod 2004, 71, 1128-34. [CrossRef]
- Tardif, S.; Wilson, M. D.; Wagner, R.; Hunt, P.; Gertsenstein, M.; Nagy, A.; Lobe, C.; Koop, B. F.; Hardy, D. M., Zonadhesin is essential for species specificity of sperm adhesion to the egg zona pellucida. J Biol Chem 2010, 285, 24863-70. [CrossRef]
- Gum, J. R., Jr.; Hicks, J. W.; Toribara, N. W.; Siddiki, B.; Kim, Y. S., Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem 1994, 269, 2440-6. [CrossRef]
- DeSilva, U.; D’Arcangelo, G.; Braden, V. V.; Chen, J.; Miao, G. G.; Curran, T.; Green, E. D., The human reelin gene: isolation, sequencing, and mapping on chromosome 7. Genome Res 1997, 7, 157-64. [CrossRef]
- Yasui, N.; Kitago, Y.; Beppu, A.; Kohno, T.; Morishita, S.; Gomi, H.; Nagae, M.; Hattori, M.; Takagi, J., Functional importance of covalent homodimer of reelin protein linked via its central region. J Biol Chem 2011, 286, 35247-56. [CrossRef]
- Frappaolo, A.; Karimpour-Ghahnavieh, A.; Sechi, S.; Giansanti, M. G., The Close Relationship between the Golgi Trafficking Machinery and Protein Glycosylation. Cells 2020, 9, 2652. [CrossRef]
- Chen, M.; Xu, L.; Wu, Y.; Soba, P.; Hu, C., The organization and function of the Golgi apparatus in dendrite development and neurological disorders. Genes Dis 2023, 10, 2425-2442. [CrossRef]
- Huang, S.; Wang, Y., Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res 2017, 6, 2050. [CrossRef]
- Berninsone, P. M.; Hirschberg, C. B., Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol 2000, 10, 542-7. [CrossRef]
- Song, Z., Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol Aspects Med 2013, 34, 590-600. [CrossRef]
- Ilani, T.; Reznik, N.; Yeshaya, N.; Feldman, T.; Vilela, P.; Lansky, Z.; Javitt, G.; Shemesh, M.; Brenner, O.; Elkis, Y.; Varsano, N.; Jaramillo, A. M.; Evans, C. M.; Fass, D., The disulfide catalyst QSOX1 maintains the colon mucosal barrier by regulating Golgi glycosyltransferases. Embo j 2023, 42, e111869.
- Kang, C.; Wu, H.L.; Xu, M.L.; Yan, X.F.; Liu, Y.J.; Yu, R.Q. Simultaneously quantifying intracellular FAD and FMN using a novel strategy of intrinsic fluorescence four-way calibration. Talanta 2019, 197, 105-112.
- Tzur, A.; Kafri, R.; LeBleu, V.S.; Lahav, G.; Kirschner, M.W. Cell growth and size homeostasis in proliferating animal cells. Science 2009, 325, 167-71.
- Zhao, L.; Kroenke, C.D.; Song, J.; Piwnica-Worms, D.; Ackerman, J.J.; Neil, J.J. Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. NMR Biomed 2008, 21,159-64.
- Hühner, J.; Ingles-Prieto, Á.; Neusüß, C.; Lämmerhofer, M.; Janovjak, H. Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection. Electrophoresis 2015, 36, 518-25. [CrossRef]
- Redeker, K.M.; Brockmöller, J. Several orphan solute carriers functionally identified as organic cation transporters: Substrates specificity compared with known cation transporters. J Biol Chem 2024, 300, 107629. [CrossRef]
- Burtnyak, L.; Yuan, Y.; Stojek, E.; Pan, X.; Gunaratne, L.; Silveira d’Almeida, G.; Fergus, C.; Martinelli, M.; C, J. R.; Fernandez, J.; Patel, B. I.; Marquez, I.; Ehrenhofer-Murray, A. E.; Swairjo, M. A.; Alfonzo, J. D.; Green, B. D.; Kelly, V. P.; de Crécy-Lagard, V., The oncogene SLC35F2 is a high-specificity transporter for the micronutrients queuine and queuosine. Proc Natl Acad Sci U S A 2025, 122, e2425364122.
- Ferrada, E.; Superti-Furga, G., A structure and evolutionary-based classification of solute carriers. iScience 2022, 25, 105096.
- Roberts, E. K.; Tardif, S.; Wright, E. A.; Platt, R. N., 2nd; Bradley, R. D.; Hardy, D. M., Rapid divergence of a gamete recognition gene promoted macroevolution of Eutheria. Genome Biol 2022, 23, 155.
- Roberts, E. K.; Wright, E. A.; Worsham, A. E.; Hardy, D. M.; Bradley, R. D., Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels. Diversity 2023, 15, 1145. [CrossRef]
- The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318-1330.
- Bernsel, A.; Viklund, H.; Hennerdal, A.; Elofsson, A., TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 2009, 37, (Web Server issue), W465-8.
- Tsirigos, K. D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A., The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 2015, 43, W401-7. [CrossRef]
- Thumuluri, V.; Almagro Armenteros, J. J.; Johansen, A. R.; Nielsen, H.; Winther, O., DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res 2022, 50, W228-W234.
- Edgar, R. C., MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004, 32, 1792-7. [CrossRef]
- Minh, B. Q.; Schmidt, H. A.; Chernomor, O.; Schrempf, D.; Woodhams, M. D.; von Haeseler, A.; Lanfear, R., IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020, 37, 1530-1534.
- Letunic, I.; Bork, P., Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res 2024, 52, W78-W82. [CrossRef]
- Nguyen, N. T. T.; Vincens, P.; Dufayard, J. F.; Roest Crollius, H.; Louis, A., Genomicus in 2022: comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res 2022, 50, D1025-D1031.
- Boadu, F.; Cao, H.; Cheng, J., Combining protein sequences and structures with transformers and equivariant graph neural networks to predict protein function. Bioinformatics 2023, 39, i318-i325. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S. A. A.; Ballard, A. J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A. W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D., Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583-589.
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A. J.; Bambrick, J.; Bodenstein, S. W.; Evans, D. A.; Hung, C. C.; O’Neill, M.; Reiman, D.; Tunyasuvunakool, K.; Wu, Z.; Žemgulytė, A.; Arvaniti, E.; Beattie, C.; Bertolli, O.; Bridgland, A.; Cherepanov, A.; Congreve, M.; Cowen-Rivers, A. I.; Cowie, A.; Figurnov, M.; Fuchs, F. B.; Gladman, H.; Jain, R.; Khan, Y. A.; Low, C. M. R.; Perlin, K.; Potapenko, A.; Savy, P.; Singh, S.; Stecula, A.; Thillaisundaram, A.; Tong, C.; Yakneen, S.; Zhong, E. D.; Zielinski, M.; Žídek, A.; Bapst, V.; Kohli, P.; Jaderberg, M.; Hassabis, D.; Jumper, J. M., Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493-500.
- Trott, O.; Olson, A. J., AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010, 31, 455-61. [CrossRef]
- Forli, S.; Huey, R.; Pique, M. E.; Sanner, M. F.; Goodsell, D. S.; Olson, A. J., Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 2016, 11, 905-19.

| Transporter | Subcellular Compartment |
Predicted Probability |
Threshold |
|---|---|---|---|
| SLC35F4 | Golgi apparatus | 0.7088 | 0.6494 |
| Cell membrane | 0.4742 | 0.5646 | |
| Lysosome/Vacuole | 0.4302 | 0.5848 | |
| Endoplasmic reticulum | 0.3601 | 0.6090 | |
| Peroxisome | 0.1715 | 0.7364 | |
| Cytoplasm | 0.1148 | 0.4761 | |
| Nucleus | 0.1114 | 0.5014 | |
| Mitochondrion | 0.0865 | 0.6220 | |
| Extracellular | 0.0799 | 0.6173 | |
| Plastid | 0.0278 | 0.6395 | |
| SLC35F5 | Golgi apparatus | 0.7691 | 0.6494 |
| Cell membrane | 0.4655 | 0.5646 | |
| Lysosome/Vacuole | 0.3943 | 0.5848 | |
| Endoplasmic reticulum | 0.3426 | 0.6090 | |
| Peroxisome | 0.1690 | 0.7364 | |
| Cytoplasm | 0.1174 | 0.4761 | |
| Nucleus | 0.0930 | 0.5014 | |
| Mitochondrion | 0.0858 | 0.6220 | |
| Extracellular | 0.0654 | 0.6173 | |
| Plastid | 0.0223 | 0.6395 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





