1. Introduction
GRAPHS are mathematical structures that stem from pairwise interactions between entities, which are widely used in a diverse range of domains. Due to their expressive power in modeling relational data, graph generative models (GGMs), with the goal of learning the underlying statistical patterns of observed graphs and generating new samples, have attracted significant attention [
1]. This capability has enabled impactful applications across various domains, including scientific research, social network analysis, and protein design [
2].
In general, we focus on two broad categories of graphs [
3] widely adopted in GGMs: (1) Geometric graphs, typically consisting of a large set of small graphs (e.g., chemical compounds or protein graphs) [
2]. (2) Scale-free graphs, which are typically large graphs (possibly with several components) where node degrees follow a power-law distribution. These graphs are often generated by the temporal evolution of nodes, as seen in social networks or user-item bipartite graphs [
4].
Naturally, the algorithms used for these two types of graphs differ significantly due to their differing structural formation mechanisms [
5]. To achieve unbiased coverage of the entire set of relevant graphs, GGMs can be accordingly categorized into two probabilistic modeling methods: (1) For scale-free graphs, early works focused primarily on statistical-based simulation models, which encode hypothesized rules for network formation [
6,
7]. These models aim to replicate key topological properties observed in real-world networks, such as power-law degree distribution. More recently, LLM-based simulators have revisited these classical mechanisms by directly simulating interactions between nodes through language-guided roles and rules [
8,
9]. Unlike traditional statistical-based simulation, which relies on predefined rules for graph generation, LLM-based role-play agents simulate human-like interactions, where the evolution of the graph is governed by dynamic and context-sensitive interactions. However, the need for tailored models to capture different properties of scale-free graphs complicates the integration of these methods into a unified framework. (2) For geometric graphs, given their smaller scale and rich supervisory information, deep learning-based generative models have become the dominant approach. With the rise of deep learning, techniques such as auto-regressive (AR) models, variational autoencoders (VAEs), generative adversarial networks (GANs), normalizing flows, and denoising diffusion models have become popular for learning expressive graph distributions and generating high-quality samples [
2].
In recent years, substantial progress has been made in GGMs, leading to a comprehensive and diverse range of methods in the literature. However, despite the fundamental differences in graph categories and generation mechanisms, existing surveys often overlook these distinctions [
10,
11]. To address this gap, we present a comprehensive survey that systematically categorizes GGMs based on graph categories (geometric vs. scale-free) and probabilistic modeling methods (simulation-based vs. deep learning-based). As shown in the
Awesome-Graph-Generation resource
1, we present a comprehensive review of the field to help the community track its progress. The rest of the survey is organized as follows:
In
Section 2 and
Section 3, we introduce a three-axis taxonomy of GGMs, encompassing their target graph categories, graph attribute modality and probabilistic modeling methods. In
Section 4, we further delve into the specific model architectures employed within each framework, including neural network architectures and modeling strategies. In
Section 5, we summarize and categorize the evaluation metrics used to assess GGMs. In
Section 6, we present the various applications of GGMs. Finally, in
Section 7, we discuss open challenges and promising research directions.
Figure 1.
The organization of this survey. Our survey systematically examines GGMs across five key aspects: graph types categorization, probabilistic modeling methods, model architectures, evaluation metrics and applications. For each aspect, we discuss both geometric graph generators and scale-free graph generators, analyzing their unique characteristics and challenges.
Figure 1.
The organization of this survey. Our survey systematically examines GGMs across five key aspects: graph types categorization, probabilistic modeling methods, model architectures, evaluation metrics and applications. For each aspect, we discuss both geometric graph generators and scale-free graph generators, analyzing their unique characteristics and challenges.
1.1. Related Works
Existing surveys relevant to our topic fall into three strands.
Statistical-based GGMs
The first strand synthesizes graph generation from classical graph theory [
3]. These surveys emphasize random graph models and analytical properties, but they predate the modern wave of deep generative neural networks for graphs.
Deep learning-based GGMs
The second strand reviews neural generative models for graphs, offering complementary emphases and taxonomies. For example, [
10,
12] primarily categorize methods by backbone paradigms: auto-regressive, autoencoder-based, RL-based and etc. While [
1] analyzes techniques for both unconditional and conditional generation. [
13] focuses on diffusion-based graph generators.
LLM-based social simulation
This strand surveys LLM-based social simulation, focusing on comparisons to traditional agent-based modeling [
8,
14]. These works examine how interactions and social influence propagate over network structures (with nodes as individuals and edges as relations). But they rarely focus on the formation and topology of the social networks.
Despite their breadth, the above strands share two blind spots. First, existing surveys rarely classify the graph dataset themselves. In practice, graph categories imply different inductive biases that necessitate distinct GGMs, yet current surveys treat these regimes uniformly. Second, existing surveys neglect LLM-based GGMs. Growing interest in LLM-based agent simulation has elevated network evolution to a central role. Yet to our knowledge, no survey unifies this emerging research [
15]. To address these gaps, we propose: (a) a graph-category-centric taxonomy that distinguishes between geometric, scale-free, and general graphs; (b) a systematic organization of the probabilistic modeling methods, neural network architectures, and modeling strategies in GGMs; and (c) the first structured survey of LLM-based GGMs, analyzing their unique capabilities compared to conventional GGMs while identifying future directions.
Figure 2.
Comparison of two graph categories: (1) Geometric Graph, (2) Scale-Free Graph. Each graph category is illustrated with representative types of graphs, highlighting their distinct structural formation mechanisms.
Figure 2.
Comparison of two graph categories: (1) Geometric Graph, (2) Scale-Free Graph. Each graph category is illustrated with representative types of graphs, highlighting their distinct structural formation mechanisms.
2. Preliminaries
Formally, for one given graph dataset
, a Graph Generative Model (GGM) can be abstractly represented as
where
p denotes the probabilistic modeling method that defines the data distribution
, and
A denotes the model architecture that parameterizes and learns this distribution. In the following sections, we systematically describe the categorization and construction of
,
p, and
A.
2.1. Graph Category
This survey focuses on the two most representative categories of graph datasets commonly used in GGMs: geometric graphs and scale-free graphs. Other graph structures, such as those derived from text clustering or semantic relations, knowledge graphs, abstract syntax trees, and workflow DAGs, are excluded due to their task-specific nature and distinct structural characteristics.
Geometric Graph
A geometric graph is a graph whose vertices correspond to points in a geometric space (typically
, often
or 3), and whose edges are determined by geometric or spatial relationships between these points [
16]. In practical applications, geometric graphs arise in domains such as molecular modeling (e.g., atoms as 3D points connected by bonds), protein structure analysis, and transportation networks. For geometric graph datasets, we typically observe a set of
M graphs, denoted by
. GGMs seek to learn the underlying data distribution
and synthesize new samples
.
Scale-Free Graph
Compared to geometric graphs, scale-free graphs are rare in practice [
17]. A scale-free graph is characterized by a power-law degree distribution, where a few nodes act as high-degree hubs while most nodes have small degrees [
18]. Such networks commonly arise in social and technological domains. For scale-free graph datasets, typically only one large graph (
) is given. Therefore, the goal of the generation model is to learn the distribution of the single large graph
and generate a new graph
.
In both cases, GGMs that sample from the learned distribution must preserve essential topological patterns (e.g., connectivity, motifs) and attribute correlations (e.g., node/edge features). The distinct formation mechanisms of graph categories require different designs for GGMs.
2.2. Graph Representation
One graph G is defined as a quadruple , where is the node set with N nodes, each node is associated with an a-dimensional attribute vector . These encodings are organized in a matrix . is the edge set, representing pairwise connections between nodes, each edge is associated with b-dimensional attribute vector . Similarly, a matrix groups the one-hot encoding of each edge. We use to denote the adjacency matrix. In graph generation models, the graph representation can mainly be categorized into two types: sequential representation and matrix representation.
Sequential Representation
In sequential representation,
G is represented by a sequence of components
, where each
is a generation unit. The distribution
is the joint (or conditional) probability over these components. The components are hierarchically defined: (1) Node-level:
represents a single node, capturing atomic building blocks for graph construction [
19]. (2) Edge-level:
corresponds to an edge, encoding pairwise connections between nodes [
20,
21]. (3) Motif-level:
encodes a higher-order subgraph pattern (e.g., triangles, cliques, or star motifs) as a
k-tuple of nodes
, enabling the generation of complex structural motifs [
22]. This hierarchical decomposition enables flexible control over the generation process, allowing for progressive refinement from coarse-grained structures to fine-grained details. This kind of graph representation is more widely used in auto-regressive generation models [
19,
23], where the graph is constructed sequentially by conditioning on previously generated components. A critical aspect of sequential representation is the ordering of components
S. The choice of ordering strategy (e.g., breadth-first traversal [
24], depth-first traversal [
25] or random walk [
26]) directly impacts the model’s ability to generate structurally valid graphs.
Matrix Representation
The matrix representation encodes the graph
G using its adjacency matrix
, where
if
and 0 otherwise. Matrix representation is widely used in one-shot generation models [
27,
28,
29], where the entire graph is generated in a single step. These approaches leverage the adjacency matrix
A and attribute matrices (e.g.,
for node features) as inputs. Despite their efficiency in encoding global structure, matrix representations face scalability challenges for large
N, as their memory complexity scales quadratically
due to the adjacency matrix. Consequently, matrix-based models are often restricted to small-scale graphs, such as molecular or protein structures, where
N remains modest.
3. Probabilistic Modeling
To generate these three types of graphs, two core probabilistic modeling methods have emerged. Firstly,
deep learning-based models, which use neural architectures to learn and sample complex graph distributions automatically [
3]. They are particularly effective for multi-graph settings (e.g., geometric graphs) where rich supervision enables expressive modeling. Some models treat both geometric and scale-free graphs similarly by learning from sampled subgraphs of scale-free graphs, making them applicable to general graphs. Secondly,
simulation-based models, which instantiate predefined mechanisms (e.g., preferential attachment) to reproduce target network characteristics [
30]. Recent LLM-based simulation models develop stable interaction patterns for graph generation through LLM-based agents [
31]. These models are especially suitable for single graph settings (e.g., scale-free graphs), where scalability and controllability are crucial.
Building upon this foundation, we propose a three-axis taxonomy for graph generative models as illustrated in
Figure 1. Firstly, GGMs are classified by the types of graphs they generate: geometric, scale-free, or general graphs (including both geometric and scale-free). Secondly, they are categorized based on the attributes of the generated graphs, including non-attribute, categorical/numeric attributes, and textual attributes, reflecting the increasing complexity and richness of graph information. Finally, they are grouped by their underlying probabilistic modeling methods, namely deep learning-based and simulation-based approaches. All models are organized chronologically to highlight methodological advancements.
3.1. Deep Learning-Based Probabilistic Modeling
Deep learning-based probabilistic modeling can be broadly divided into two paradigms:
explicit and
implicit probabilistic modeling [
32]. Explicit models define explicit density forms and allow exact likelihood inference of graph data. This family includes auto-regressive models, Variational Autoencoders (VAE), and flow-matching models.
In contrast, implicit models directly learn a transformation from the prior to the data distribution [
33]. Classical approaches in this category include Generative Adversarial Networks (GANs) and diffusion models.
Auto-regressive Models
Given an input sequential representation
S of a graph
G, at step
i, auto-regressive models [
34] learn the conditional probability distribution
over graph components using the chain rule of probability, where
represents the subsequence of graph components prior to step
i.
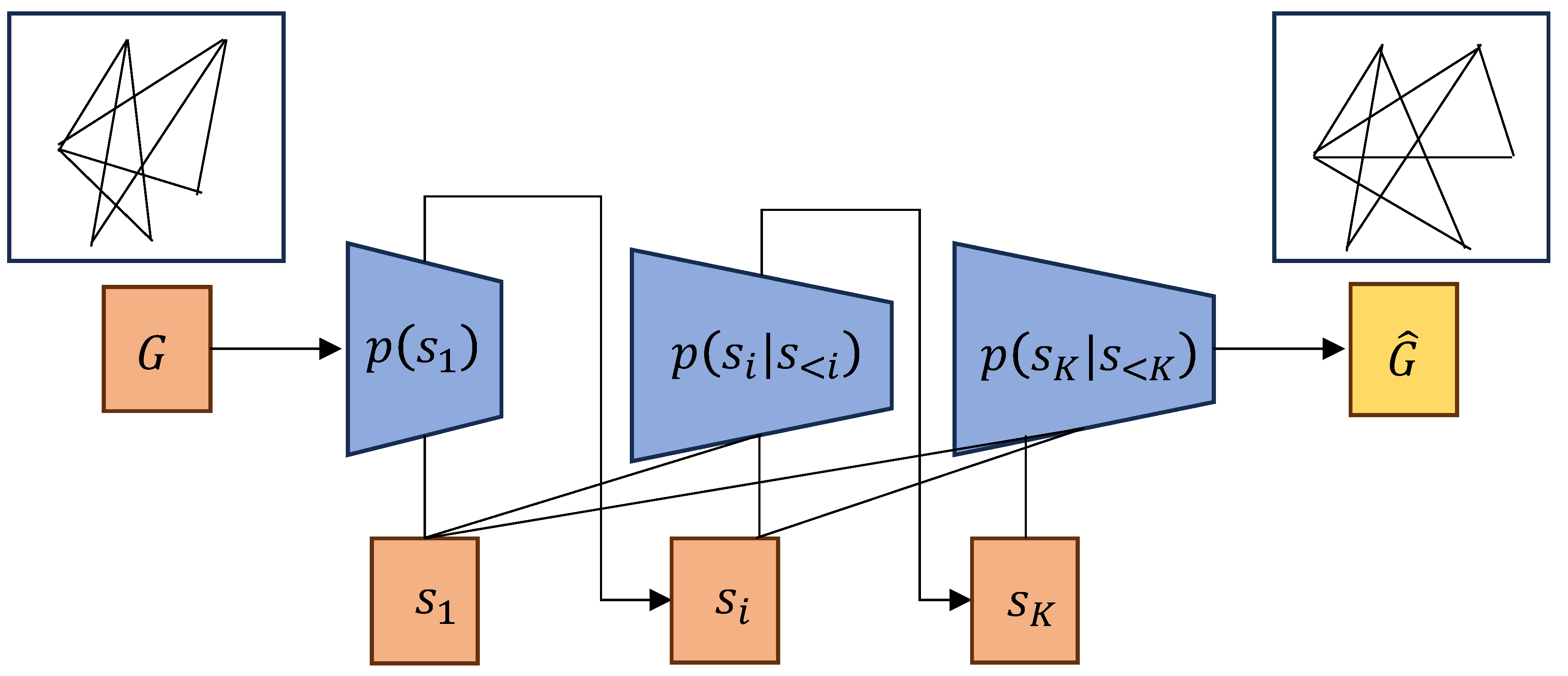
Figure 3.
Auto-regressive models for graph generation. The model generates the next component based on the previously generated components .
Figure 3.
Auto-regressive models for graph generation. The model generates the next component based on the previously generated components .
Auto-regressive models are commonly employed for sequential graph representation [
19,
23], where the input sequence consists of nodes, edges, or motifs, and the model predicts the next node or edge based on the previously generated sequence. Specifically, this model factorizes the generation process into sequential steps, each determining the next action based on the current subgraph. The general formulation of auto-regressive models is expressed as:
This approach enables the flexible generation of graphs with varying structures and sizes, typically through the sequential sampling of nodes, edges or motifs. Representative works, such as GraphRNN [
19] and NetGAN [
26], utilize an node-list sequential representation, which add new nodes one at a time before connecting these nodes with edges to the previously generated components. In contrast, models like BiGG [
20] utilize an edge-list sequential representation and employ a tree-structured auto-regressive approach for generating the edges associated with each node. GraphGPT(2) [
35] employs a motif-list based representation, which translates graphs into sequences of tokens representing nodes, edges, and attributes in a reversible manner using Eulerian paths.
VAE Models
Figure 4.
VAE models for graph generation.
Figure 4.
VAE models for graph generation.
Variational Autoencoders (VAEs) [
36] provide a robust framework for data representation by learning a probabilistic mapping between the graph domain and a latent space. Specifically, a graph encoder
maps input graphs
G to a low-dimensional continuous latent representation
, while a graph decoder
reconstructs the graph from sampled latent variables [
27]. This formulation approximates the intractable posterior distribution
through a variational approximation
, which is optimized via the Evidence Lower Bound (ELBO):
where the first term (reconstruction loss) measures the fidelity of graph reconstruction, and the second term (KL divergence) enforces the latent distribution to align with a prior
, typically a standard Gaussian. Graph-structured VAEs (GraphVAEs) typically parameterize the encoder and decoder using graph neural networks (GNNs), such as GCNs [
37] or GATs [
38], to capture relational inductive biases. Notable variants include: NeVAE [
39], which incorporates masking strategies to ensure chemical validity in molecular graph generation; Graphite [
40], which integrates spectral graph convolutions into both encoder and decoder, leveraging permutation invariance and locality in node representations; MiCaM [
41], which uses an iterative subgraph merging algorithm based on frequency to identify frequent molecular fragments (motifs) and their inter-connections.
While these methods excel in static graph generation, where node/edge structures remain fixed. However, real-world graphs are inherently dynamic: their topologies and features evolve over time [
11]. To address this, recent works like VRDAG [
42] introduce temporal modeling capabilities, which employs a bidirectional message-passing mechanism to encode both structural and attribute information, combined with a recurrence state updater to capture temporal dependencies in graph generation.
Figure 5.
Flow matching models for graph generation.
Figure 5.
Flow matching models for graph generation.
Flow Matching Models
Normalizing flows estimate the density of graph data
by establishing an invertible and deterministic mapping between latent variables and graph structures via the change of variables theorem [
43]. These models employ a sequence of invertible functions
to transform a simple prior distribution
(e.g., a standard Gaussian) into a complex data distribution
. The inverse function
enables graph generation by transforming latent samples back into the data space. For a graph
, the forward transformation is defined as:
where
is sampled from the prior, and
represents the latent variable corresponding to the graph
G. During training, the log-likelihoods of observed graph samples are maximized to update the parameters of the forward transformations
:
The determinant of the Jacobian (det) in normalizing flows quantifies how a learnable transformation alters the volume of the data space. MoFlow [
44] is an early work that adopts a two-step generation process, which first generates molecular bonds (edges) using a Glow-based flow and then predicts atom types (nodes) through a graph-conditional flow conditioned on the bond structure. A subsequent post-processing correction ensures chemical validity by resolving structural issues such as invalid valencies. Building on this, GraphDF [
45] leverages discrete normalizing flows based on invertible modulo shift transforms to map latent variables to graph structures, achieving exact invertibility and mitigating the limitations inherent in continuous relaxation approaches. Most recently, GGFlow [
46] introduces discrete flow matching with optimal transport, and incorporates an edge-augmented graph transformer to directly model interactions among chemical bonds, thereby further improving molecular graph generation.
GAN Models
Generative Adversarial Networks (GANs) are a class of implicit generative models that have achieved significant success in the computer vision domain and have since been extended to graph data [
47]. A typical GAN framework consists of a generator, which learns to produce synthetic graphs from random noise, and a discriminator, which aims to distinguish generated graphs from real ones. The generator
and discriminator
are trained in an adversarial min-max objective functions:
To address the unique challenges of graph data, including structural dependencies, permutation invariance, and conditional graph generation, various GAN-based models have been proposed. For example, GraphGAN [
24] models vertex connectivity through adversarial learning, while NetGAN [
26] generates graphs by learning the distribution of biased random walks with a recurrent neural-network-based generator and a discriminator, optimized using the Wasserstein GAN objective. CONDGEN [
48] further advances this direction by integrating variational methods with adversarial training, enabling conditional graph generation based on semantic contexts and achieving permutation-invariant output. Despite these advances, GAN-based models for graphs still face several limitations such as training instability, convergence difficulties, and mode collapse, motivating ongoing research for more robust and scalable solutions [
49].
Figure 6.
GAN models for graph generation.
Figure 6.
GAN models for graph generation.
Diffusion Models
Diffusion models generate data by defining a forward process that gradually adds Gaussian noise to the input data
over
T steps, producing a sequence of noisy samples
, with the noise at each step controlled by a schedule
(commonly Gaussian). The forward process is defined as:
where
represents the conditional distribution of the noisy data
given the previous state
, and
controls the amount of noise added at each step. As
, the final state
approaches a standard Gaussian distribution. To sample from the data distribution, this process is reversed by learning an approximate reverse transition
, since the true reverse kernel
is generally intractable. A neural network is trained to parameterize the reverse generative process as
where
represents the learned denoising transition used to iteratively reconstruct clean graphs from noise. Training optimizes a variational lower bound (VLB) on the data log-likelihood, expressed as
Three major paradigms of diffusion models have emerged, each addressing probabilistic modeling through distinct methodologies [
13]. Score Matching with Langevin Dynamics (SMLD), represented by DeepRank-GNN [
50], trains a score function to estimate the gradient of the log data density and employs Langevin dynamics for sampling, prioritizing efficiency in high-dimensional spaces. Denoising Diffusion Probabilistic Models (DDPM), represented by Digress [
51], formalize the reverse process probabilistically, drawing on principles from nonequilibrium thermodynamics to iteratively denoise data. Score-based Generative Models (SGM), represented by GraphGDP [
52] and GDSS [
28], generalize discrete-time diffusion to continuous-time formulations via stochastic differential equations, offering enhanced flexibility for modeling complex temporal and spatial dependencies. Although these diffusion-based models have demonstrated strong sample quality and stable training, they remain computationally intensive due to the large number of reverse diffusion steps required for generation, typically making them slower than GANs and VAEs [
53].
Figure 7.
Diffusion models for graph generation.
Figure 7.
Diffusion models for graph generation.
Deep learning-based generative models employ diverse training objectives as summarized in
Table 1. VAEs approximate data log-likelihood through the ELBO in Equation (
2), representing an approximate maximum likelihood estimation (MLE) approach. Flow Matching and AR models directly maximize exact likelihood via sequential probability decomposition, explicitly modeling graphs in their respective formulations in Equations (
1) and (
3). GANs adopt adversarial training between generator and discriminator networks, following the objective in Equation (
4). Diffusion models optimize either the VLB for reverse diffusion processes in Equation (
5) or employ score matching techniques to estimate gradients of data distributions [
28], with recent advancements enhancing stability and efficiency [
54].
3.2. Simulation Based Probabilistic Modeling
Deep learning-based graph generators rely on high-quality reference data to train robust GGMs. However, obtaining real-world graph data remains challenging due to confidentiality constraints, legal restrictions, and the high cost of data collection. Furthermore, these models often face scalability issues for large-scale graph generation [
10].
To overcome this limitation, simulation-based random graph generators offer a scalable alternative. Random graph models are essential for analyzing complex networks, aiding in understanding, controlling, and predicting various phenomena [
30]. Historically, such approaches can be divided into two categories:
statistical-based simulation and
LLM-based simulation. Early models design predefined statistical rules to generate graphs with target properties. Recent advances in LLM-based agent simulations have demonstrated remarkable capabilities in autonomous decision-making, enabling the dynamic simulation of interaction processes within complex networks [
8,
55]. These models are increasingly recognized as a novel paradigm for text-attributed graph generation.
Statistical-based Simulation
Early graph generators assume real-world networks adhere to predefined structural rules (e.g., degree distributions, clustering patterns) and employ probabilistic sampling to replicate target properties. Three foundational properties have emerged as pivotal in scale-free graph generation: (1) The first key property is the small-world phenomenon. Watts and Strogatz [
56] showed that many real-world networks exhibit high clustering: two nodes are more likely to connect if they share neighbors. They introduced a tunable small world model that rewires a regular lattice to interpolate between order and randomness. (2) The second key property of scale-free graphs emphasized in recent work is shrinking diameter, where network diameter decreases to a constant value over time [
57]. The Forest Fire model [
57] explains this behavior through a modified preferential attachment process known as community-guided attachment. Subsequently, [
58] proposed a social affiliation network model which extended this idea by modeling network evolution via bipartite representations of agents and communities, naturally reproducing heavy-tailed degree distributions, community structure, and shrinking diameter. (3) The third key property of scale-free graphs is their heavy-tailed degree distribution, first highlighted by Albert and Barabási [
7]. The Barabási-Albert (BA) model [
7] formalizes this through preferential attachment, where new nodes connect to existing nodes with probability proportional to their degree (
), resulting in a "rich-get-richer" dynamic that produces power-law degree distributions
(where
). Edge copying models [
59] extend this principle by simulating local growth mechanisms, where new nodes select a prototype node uniformly at random, then probabilistically copy a fraction of its edges. The prototype selection probability remains proportional to degree (
), implicitly enforcing power-law scaling while introducing local clustering.
Later advancements, such as the affiliation network model [
58], unify preferential attachment and edge copying to jointly reproduce power-law degrees and community structure. Moreover, exponential random graph models (ERGMs), also known as
models [
60], constitute a foundational class of statistical frameworks for analyzing and generating social networks. ERGMs define the probability distribution over the space of graphs
with
N nodes by weighting graph statistics
(e.g., edge counts, triangle densities, degree distributions) via parameters
. Each graph
is assigned probability
where
is the normalizing constant ensuring probabilities sum to 1.
Figure 8.
Scalable graph generators based on statistical-based simulation. The left figure shows the graph growth mechanism based on preferential attachment, where edges of new node are added to the graph based on the degree of existing nodes. The right figure shows the graph growth mechanism based on edge coping, where edges of new node are added coping edges of chosen node prototype.
Figure 8.
Scalable graph generators based on statistical-based simulation. The left figure shows the graph growth mechanism based on preferential attachment, where edges of new node are added to the graph based on the degree of existing nodes. The right figure shows the graph growth mechanism based on edge coping, where edges of new node are added coping edges of chosen node prototype.
LLM-based Simulation
With LLMs demonstrating advanced capabilities in human-like responses and autonomous planning [
14], they have emerged as a new paradigm for simulation across diverse domains, including education [
61], social dynamics [
62], and economics [
63]. Particularly, LLM-based multi-agent systems excel in simulating complex interactions, motivating the development of LLM-driven graph generators for dynamic, text-attributed graphs where temporal node and edge formation emerges organically from agent behaviors [
8,
31]. Early efforts in LLM-driven graph generation focused on capturing fundamental properties like degree power-law distributions, small-world phenomenon and community structure [
64,
65], with frameworks such as LLM4GraphGen [
55] formalizing graph synthesis as a textual prompting task. Subsequent works, including IGDA [
66], introduced iterative reasoning mechanisms to model causal relationships in graph evolution, while [
67] explored additional social graph characteristics in political domain. However, these approaches often lack realism due to oversimplified interaction pattern and scalability, typically limited to fewer than 100 agents. Recently, GAG [
68] adopts LLM agents to generate dynamic social graphs attributed to text by large-scale multi-agent simulations, which scales to graphs with 100,000 nodes or 10 million edges. Building on GAG, GraphMaster [
69] extends graph generation beyond social networks, proposing a multi-agent framework with specialized agents that collaboratively optimize graph synthesis process.
4. Model Architecture
Probabilistic modeling defines the underlying generation mathematical formulation, while the model architecture serves as the backbone for learning graph representations and capturing complex dependencies. The latter comprises two components: neural network architectures, which parameterize the model; and modeling strategies, which determine how graphs are represented and generated.
4.1. Neural Network Architecture
Neural network architectures play a crucial role in graph generation models by providing the necessary representational capacity to capture complex dependencies and structures inherent in graph data. Different architectures are suited to various types of probabilistic modeling methods, as summarized in
Table 2. Below, we discuss several prominent neural network architectures commonly employed in graph generation tasks.
Feedforward Neural Networks
Feedforward Neural Networks (FFN), also referred to as multi-layer perceptrons (MLPs), represent the most basic form of neural networks. They are particularly effective for input data that can be represented as fixed-size vectors, such as node or edge attributes. Models like NeVAE [
39] and GraphVAE [
27] employ FNNs to learn latent representations of graphs, which are sampled to generate new graphs. Specifically, NeVAE uses an MLP-based probabilistic encoder that aggregates information from a variable number of hops for each node into an embedding vector. Coupled with a decoder, this approach can generate molecular structures by predicting the spatial coordinates of atoms. Furthermore, FNNs are utilized in human motion generation tasks, as in MoGlow [
114] and MotionDiffuse [
127], where they generate motion sequences based on learned representations of temporal human motion graphs.
Graph Neural Network
Graph Neural Networks (GNNs) are specialized architectures designed to operate on graph-structured data, enabling the modeling of complex relational dependencies among nodes and edges. GNNs leverage message-passing mechanisms to aggregate information from neighboring nodes, allowing them to capture both local and global graph structures. Initial works like GRAN [
23], Graphite [
40], MiCaM [
41] integrated GNNs as decoders within AR, VAE, or flow matching models for molecular graph generation. MolFlow [
44] innovatively replaced traditional affine transformations with graph convolution layers [
37] for node representations, and adopt Glow [
163] for edge generation, enabling topology-aware invertible mappings for molecular graphs. For GAN-based GGMs, GCPN [
123] utilized GCNs to model subgraph topology and scaffold interactions in the generator, with a discriminator validated actions (e.g., node connections, edge types, termination signals) to ensure structural feasibility. GDSS [
28] employs message-passing GNNs as link predictors within a score-based diffusion framework, enhancing generation stability and fidelity by iteratively refining noisy graph structures through learned gradient scores.
Recurrent Neural Network
Recurrent neural network (RNN), due to their ability to process sequential data through hidden state dynamics, are particularly suited for graph generation tasks with sequential representations. GraphRNN [
19] exemplifies this paradigm through a two-tiered architecture: a graph-level RNN maintains the state of the growing graph and generates new nodes, while an edge-level RNN generates the edges for newly generated node. Building upon this foundation, the MolecularRNN [
72] extends GraphRNN for generating graphs with node and edge types. Tigger [
21] adapts the auto-regressive paradigm for dynamic graph generation by decomposing temporal interaction graphs into temporal random walks. Tigger employs RNNs to learn sequential patterns in temporal random walks, with synthetic walk sequences later reconstructed into dynamic graphs through post-processing.
Long Short-Term Memory Network
Long Short-Term Memory (LSTM) networks, a specialized variant of RNNs, further advance graph generation by addressing long-term dependency challenges inherent in sequential modeling. Like standard RNNs, LSTMs process graphs as stepwise sequences but excel in scalability for large-scale graphs due to their memory-cell design. For edge-sequential representation, GEEL [
74] employs LSTM to model the generation of edges in a graph, which reduces vocabulary complexity via gap encoding and bandwidth constraints. It further extends to attributed graphs through grammar-based adaptations, ensuring scalable and structured generation aligned with edge counts. For node-sequential representation, OLR [
73] employs LSTM to learn node-wise dependencies, introducing a regularization term that enforces invariance of hidden states to node ordering permutations. Moreover, LSTM has been applied to dynamic graph generation tasks, such as TG-GAN [
119], which uses LSTM to help GAN-based GGMs for continuous-time temporal graph generation, by modeling the deep generative process for truncated temporal random walks and their compositions. It models truncated temporal random walks by jointly capturing edge sequences, timestamp, and node attributes through LSTM modules. A complementary discriminator combines recurrent architectures with time-node encodings to distinguish synthetic sequences from real-world temporal patterns.
Other Architectures
Beyond the mainstream architectures discussed earlier, several non-typical frameworks have been proposed to address specialized challenges in graph generation. For statistical-based simulation, classical algorithms such as the Barabási-Albert (BA) model [
7] and Erdős-Rényi (ER) model [
6] define explicit generative rules that produce graphs with characteristic structural properties, including scale-free degree distributions and random connectivity patterns. For deep learning-based generative models, D2G2 [
88] integrates factorized bayesian models of dynamic graphs to generate temporally coherent graph snapshots, enabling the modeling of evolving network structures over time. MolGrow [
115] adopts a flow-based generative model using the RealNVP architecture [
166], which ensures efficient sampling and exact likelihood estimation through invertible transformations. MoGlow [
114] adopts the Glow architecture and extends it to model controllable motion sequences. Grammar-VAE [
100] incorporates probabilistic context-free grammars (PCFGs) to enforce chemical validity in molecular graph generation. By representing discrete graph structures as parse trees derived from context-free grammar rules, it guarantees syntactically valid chemical configurations. PCFG assigns probabilities to each production rule in the grammar, and thus defines a probability distribution over parse trees and, consequently, over valid molecular graphs.
4.2. Modeling Strategy
After learning the probability distributions of observed graphs’ latent representations, GGMs sample synthetic graphs from
. Specifically, the post-processing process includes two dimensions:
generation strategies and
sampling strategies, as illustrated in
Figure 9.
Generation Strategy
Due to the discrete, high dimensional, and unordered nature of graph data representations, GGMs typically employ two main strategies: one-shot generation and sequential generation [
10].
One-shot generation, typically paired with matrix-based representations, generate the entire graph in a single step, offering computational efficiency by avoiding sequential dependencies on node ordering. This approach is particularly widely adopted in VAE, Flow, Diffusion-based models for geometric graph generation. For example, Digress [
51] generate the entire graph structure in one step by predicting the adjacency matrix and node features simultaneously. Similarly, flow-based models like MoFlow [
44] and ChemFlow [
101] generate molecular graphs in a single pass.
While one-shot graph generation offer computational efficiency, they face critical limitations in flexibility compared to sequential representations, particularly in generating large-scale graphs (e.g., social networks). Firstly, output space complexity. Generating a graph with N nodes requires the model to output values to fully define its adjacency matrix. Second, non-unique representations. In general graph generation tasks, a graph with N nodes admits up to equivalent adjacency matrices due to arbitrary node orderings.
Sequential generation has proven particularly effective for scale-free graphs, where iterative node or edge additions align naturally with edge-list or node-list sequential representations [
19,
20,
77]. Pioneering work like GraphRNN [
19] formalized this approach, using recurrent networks to generate nodes and edges sequentially while capturing complex structural dependencies. Extensions such as MTM [
167] further refine temporal dynamics by incorporating motif-list sequential representations to model evolving graph patterns. Recent advancements have also adapted edge-list sequential representations to dynamic graph generation, demonstrating their utility in downstream tasks like link prediction [
76].
Sampling Strategy
In graph generation, sampling plays a crucial role in determining how synthetic graphs are produced. Since generation tasks often require control over specific graph properties or contextual conditions, two primary sampling strategies are commonly employed: random sampling and conditional sampling [
10]. Random sampling involves drawing latent codes from a learned distribution
, producing graphs without explicit control over their structural or properties. In contrast, conditional sampling aims to sample the latent code to generate new graphs with desired properties [
48]. As shown in
Figure 9, such constraints can encompass semantic objectives, structural patterns, graph-level property or node/edge-level attributes.
Semantic conditions involve natural language prompts that guide generating graphs with desired textual description. Early work focused on extracting graph-structured representations from text [
169] or generating graphs based on specified semantic description [
127,
170]. More recently, pre-trained language models have been integrated into conditional generation frameworks; GraphGPT(1) [
77] and LGGM [
143] encode textual prompts and align them with graph embeddings to enforce semantic consistency during generation.
Structural conditions refer to the constraints on the graph structure, such as the number of nodes, edges, or specific connectivity patterns. Random graph models like FastSGG [
151] explicitly enforce degree distribution constraints, while ROLL-Tree [
150] ensures scale-free property of generated graphs. Darwini [
171] further incorporates degree-dependent clustering coefficient distributions, enabling scalable synthesis of graphs that mirror real-world networks while varying size. These approaches are critical for domains requiring strict adherence to domain-specific structural norms.
Property conditions extend structural constraints by incorporating categorical or quantitative attributes of graphs(e.g., hydrophobicity, protein validity). Deep learning models often embed these properties into training objectives via loss functions [
94,
98]. For example, GGDiff [
172] frames conditional diffusion as a stochastic control problem, dynamically adjusting sampling process to generate graphs of target properties.
Edge/Node attributes enforce constraints on discrete or continuous node/edge features (e.g., labels, embeddings). EDGE [
29] generates graphs by first sampling node types and then constructing graphs conditioned on these attributes, while GraphMaker [
130] generalizes this to large attributed graphs by decoupling attribute and structure generation. However, most frameworks struggle with high-dimensional attributes (e.g., text embeddings), often truncating dimensions to fit model limits [
76]. Since real-world graphs often contain high-dimensional attributes [
173], such as node- or edge-level textual features [
173], generating such graphs has attracted increasing attention. LLM-based GGMs [
68,
69] simulate text-driven interactions through agent-based modeling, enabling the organic emergence of complex graph structures.
5. Evaluation
The evaluation of GGMs encompasses three main paradigms: statistic-based metrics, neural-based metrics, and downstream task performance. Statistic-based metrics quantify generation quality by comparing distributions of structural features and domain-specific attributes between generated and real graphs. To overcome their inability to capture high-level semantics, neural-based metrics employ deep models like graph neural networks to extract unified graph representations, enabling comprehensive evaluation of attributed graphs. Finally, downstream evaluation tests practical utility in scenarios including data augmentation, pre-training, and conditional generation, ensuring functional value beyond statistical similarity. These three approaches form a complementary, multi-faceted evaluation framework.
Table 3.
Summary of Statistic-based Metrics for GGMs Across Tasks. Each dataset is cited with its original proposing paper.
Table 3.
Summary of Statistic-based Metrics for GGMs Across Tasks. Each dataset is cited with its original proposing paper.
| Graph Type |
Domain |
Evaluation Metrics |
Datasets |
| Molecule Graph |
Chemistry |
Atom Stability, Molecule Stability,
Validity, Uniqueness, Novelty, Diversity,
FCD
Coverage,
AMR,
PlogP,
Quantitative Estimate of Drug-likeness,
Synthetic Accessibility,
Dipole Moment, Polarizability,
HOMO Energy, LUMO Energy,
Orbital Energy Gap, Heat Capacity |
QM9 [174]
ZINC [175],
MOSES [176],
ChEMBL [177],
CEPDB [178],
PCBA [179],
GEOM-Drugs [180] |
| Protein Graph |
Biology |
Contact Accuracy, Perplexity,
Fitness |
Enzymes [180,181],
Lobster [182],
Protein [183] |
| Geo-Spatial or Spatial Graph |
Engineering |
RMSE, NRMSE of OD Matrix,
JSD of OD Flow Volumes,
CPC |
LODES [184],
METR-LA [185] |
| Human Motion Graph |
Computer Vision |
MSE, NPSS,
NDMS, PCK,
FID, IS
|
PiGraphs [186],
Human3.6M [187] |
| Social Graph |
Social Science |
Power-law Exponent Gap, Claw Count,
Wedge Count, KOL Distribution |
TwitterNet [188] |
| User-item Bipartite Graph |
Recommendation |
MMR, Hit Rate,
Recall@k
|
MovieLens [189] |
| Synthetic Graph |
N/A |
Degree, Clustering Coefficient,
Spectral Eigenvalues,
Orbit Counts
|
ER [6],
BA [7],
|
5.1. Statistic-Based Metric
For generative models aiming to capture the distribution of real-world graph data, evaluation primarily focuses on comparing statistical property distributions between generated and reference graphs. Common graph statistic-based metrics include the degree distribution (the probability distribution of per-node connection counts), motif counts (subgraphs recurring within or across networks), clustering coefficient (the extent of local node aggregation), orbit counts and etc.
For distributional comparison of statistic-based metrics, researchers often evaluate the graph generation performance by the standard Maximum Mean Discrepancy (MMD) between generated and reference graphs
[
190],
where
is a general kernel function, usually use RBF kernel following [
19]:
where
is pairwise distance, specifically, GraphRNN [
19] employs the Earth Mover’s Distance (EMD) over the set of graph statistics. In addition to MMD, other works use root-mean-square deviation (RMSD) [
191] or mean absolute error (MAE) [
192] as alternative distance metrics. Since graphs encompass distinct structural or attribute priors, statistic-based metrics are inherently domain-specific. We summarize evaluation metrics and datasets for seven graph types in
Table 3, with dataset details in
Table 4. Detailed descriptions of the metrics and datasets are provided in Appendix.
5.2. Neural-Based Metric
Purely statistic-based metrics have three key limitations [
190]: (1) They produce multiple disjoint scores, complicating model selection. (2) They focus on topology, while under-representing node/edge attributes and missing semantic information. (3) Higher-order statistics (e.g., subgraph counts, spectral distances) are computationally expensive and inefficient. To address these issues, recent work proposes neural-based metrics for evaluating the quality of generative models. [
193,
194] propose untrained neural networks for static graph feature extraction. Building on this, subsequent studies [
190,
195] demonstrate that random graph neural networks can serve as feature extractors for synthetic and real graphs, and then compute a scalar score by applying distances (e.g., MMD, cosine similarity) to the embeddings. Empirical results demonstrate that these neural metrics effectively capture both topology and attribute.
Dynamic graph generation has recently garnered increasing attention [
76]. Early evaluation protocols apply static graph metrics by discretize dynamic graphs into static snapshots. This approach has several drawbacks: (1) it treats temporally dependent events as independent, ignoring temporal evolution; (2) it lacks a unified score sensitive to both attributes and topology; (3) it requires multiple, incomparable metrics; (4) it materializes each snapshot, leading to high runtime and poor scalability. To address these limitations, the JL-metric [
196] is proposed as a dynamic graph evaluation method. It uses dynamic GNNs as temporal encoders to capture the joint evolution of node states and topology, enabling end-to-end assessment without snapshot decomposition. Recently, the Graph Embedding Metric [
197] further improves JL-metric, incorporating textual information from node attributes alongside dynamic GNNs. This method jointly models the evolution of both textual attributes and graph structure, providing a unified evaluation of text-attributed dynamic graph generation.
5.3. Downstream Task
Beyond distributional similarity to reference graphs, a complementary line of work evaluates generated graphs by their utility on downstream tasks. Rather than directly matching structures, these protocols assess the practical usefulness of generated graphs in standard graph learning scenarios. Under this evaluation framework, the generated graphs are primarily assessed across five types of downstream tasks: (1) Machine learning tasks involving graph generation evaluate whether synthetic graphs can substitute reference graphs in training discriminative models, with performance ratios near 1 indicating comparability in topology and attributes [
130]. (2) Data augmentation tasks address data sparsity, particularly for long-tail categories [
198]. (3) Robust graph generation tasks enhance graph resilience against noise, such as in relation extraction, with frameworks like LLM-CG [
199] improving robustness and detecting anomalous structures. (4) Generative pretraining tasks leverage self-supervised graph generation to boost downstream performance, including link prediction, node classification and etc [
91,
143]. (5) Conditional graph generation focuses on synthesizing graphs that meet specific conditions, crucial in applications like drug discovery, with methods such as ChemFlow [
101] and Next-Mol [
145] generating molecular structures based on target properties or protein interactions for drug design.
6. Application
Building upon
Section 3, we categorize and discuss real-world applications of GGMs in alignment with their probabilistic modeling methods. Deep learning-based GGMs automatically learn complex graph data distributions, excelling in high-fidelity applications like molecule design, protein engineering, and transportation network modeling. In contrast, simulation-based GGMs capture graph evolution through predefined rules or agent behaviors, making them ideal for social network analysis and recommender systems, where interpretability and dynamic modeling are key. These paradigms complement each other, advancing graph generation in scientific and social science research applications.
6.1. Deep-Learning Based Graph Generator
Deep learning-based probabilistic modeling for graph-structured data have emerged as a transformative force across diverse domains, especially in geometric graph generation. This section explores six key applications of graph generation: (1) molecule generation, (2) protein design, (3) transportation network modeling, (4) human motion prediction, (5) dynamic graph modeling, and (6) general graph modeling. These advancements underscore the critical role of GGMs in addressing high-dimensional combinatorial challenges under domain-specific constraints.
Molecule Generation
In molecular graph generation, molecules are graphs: atoms as nodes, bonds as edges [
200]. The goal is chemically valid structures with desirable properties, spanning 2D and 3D molecule generation. 2D molecule generation focuses on constructing a standard molecular graph. Most existing methods fall into several categories based on their generative paradigms. Auto-regressive models, such as GraphAF [
82], MolecularRNN [
72], and Lingo3DMol [
84], generate atoms and bonds sequentially, capturing the step-by-step formation of molecules. VAE-based methods, such as JT-VAE [
93], learn a latent space to encode and decode molecular graphs while enforcing chemical validity. Diffusion-based approaches, like Digress [
51], model the generation process as a denoising trajectory from noise to structured molecules. Flow-based models, such as GraphCNF [
104], GraphNVP [
95], and GGFlow [
46], use invertible transformations to map between molecular graphs and latent distributions, enabling efficient and tractable generation. While 3D molecule generation can be regarded as the construction of a special type of graph, where each node represents an atom characterized by atom type and 3D coordinates. Edges are bonding relationships and inferred based on pairwise distances between atoms. Most models use diffusion- or flow-based generative methods. For example, EDM [
131] was the first to apply diffusion models for 3D molecule generation, followed by models such as GeoLDM [
128] and CDGS [
133], which build upon this paradigm. Alternatively, methods like ENF [
102], EquiFM [
103], GeoBFN [
105], and GOAT [
109] utilize flow-based models to generate 3D molecular graphs. Recently, Uni-3DAR [
85] introduced an auto-regressive framework for 3D molecule generation and also achieved competitive results.
Protein Design
Protein generation tasks have also become an essential branch of graph generation research. The 3D structures of proteins are formed by the folding of amino acid sequences and can be naturally represented as graphs, where each node typically corresponds to an amino acid, and edges are defined between residues that are either spatially close in 3D space or sequentially adjacent [
201]. AlphaFold 3 [
202] introduced a diffusion-based generative framework capable of predicting full-atom coordinates of biomolecular complexes. Due to limitations in computational scale and complexity, more studies have primarily focused on generating partial protein structures or lower-level structural representations. For example, in antibody design tasks, current methods [
203] aim to generate only the complementarity-determining regions (CDRs), which are then combined with external structure prediction models to obtain complete antibody structures. Other works, such as [
203,
204], focus on generating peptides, which are smaller in scale and structurally simpler. At even lower levels of structural representation, recent methods [
205,
206] generate 2D contact maps, which are subsequently decoded into 3D conformations. In addition, works like [
207,
208] utilize tertiary or secondary structural information as guidance for protein sequence generation.
Table 4.
Summary of commonly used datasets in deep-learning-based, statistical simulation-based and LLM simulation-based graph generation models (GGMs). Columns: #Graphs (number of graphs), #Nodes (average node count per graph; ranges [a, b], alternates (b) denoting commonly observed preprocessed sizes, or for bipartite graphs a/b for user/item counts respectively), #Edges (average; N/S = not specific), whether nodes/edges have attributes ( = text-attributed), source, and representative models. Numerical values are normalized to thousands (k) or millions (M).
Table 4.
Summary of commonly used datasets in deep-learning-based, statistical simulation-based and LLM simulation-based graph generation models (GGMs). Columns: #Graphs (number of graphs), #Nodes (average node count per graph; ranges [a, b], alternates (b) denoting commonly observed preprocessed sizes, or for bipartite graphs a/b for user/item counts respectively), #Edges (average; N/S = not specific), whether nodes/edges have attributes ( = text-attributed), source, and representative models. Numerical values are normalized to thousands (k) or millions (M).
| Graph Type |
Probabilistic Modeling |
Name |
# Graphs |
# Nodes |
#Edge |
Node Attr. |
Edge Attr. |
Source |
Model Used |
| Geometric |
| molecular |
deep-learning based |
QM9 |
133.9K |
18 |
19 |
✓ |
✓ |
[174] |
[27,29,82,87,110,116,143,209,210,211] |
| molecular |
deep-learning based |
ZINC250k |
250K |
23 |
25 |
✓ |
✓ |
[175] |
[25,72,94,96,110,111,143,209,211,212,213] |
| protein |
deep-learning based |
Lobster |
100 |
53 |
52 |
|
|
[182] |
[20,25,28,51,74,214] |
| protein |
deep-learning based |
Enzymes |
600 (587) |
15 |
149 |
|
|
[180,181] |
[23,25,51,74,143,214] |
| protein |
deep-learning based |
Protein |
1.1K (918) |
1.6k |
646 |
|
|
[183] |
[20,25,28,51,74,143,214,215] |
| geo-spatial |
deep-learning based |
California |
1 |
7.9k |
2.6M |
|
✓ |
[216] |
[83] |
| geo-spatial |
deep-learning based |
Massachusetts |
1 |
2.4k |
58.2k |
|
✓ |
[216] |
[83] |
| geo-spatial |
deep-learning based |
Texas |
1 |
7.9k |
756.2k |
|
✓ |
[216] |
[83] |
| synthetic, spatial |
deep-learning based |
Planar |
200 |
64 |
N/S |
|
|
[217] |
[28,74,214] |
| synthetic, spatial |
deep-learning based |
Grid |
100 |
[100, 400] |
N/S |
|
|
[19] |
[20,51,215] |
| synthetic, spatial |
deep-learning based |
DRG Graph |
1 |
N/S |
N/S |
|
|
[218] |
[219] |
| synthetic, spatial |
deep-learning based |
DWR Graph |
1 |
N/S |
N/S |
|
|
[218] |
[219] |
| Scale-free |
| synthetic, social |
deep-learning based |
SBM |
200 |
[20, 40] |
N/S |
|
|
[217] |
[20,25,28,74,214,215] |
| synthetic, social |
deep-learning based |
Ego |
500 |
[60, 160] |
N/S |
|
|
[19] |
[19,29,40,82,215,220,221,222,223] |
| synthetic, social |
deep-learning based |
Community |
757 |
[50, 399] |
N/S |
|
|
[19] |
[19,29,82,215,220,221] |
| social |
deep-learning based |
Polblogs |
1 |
1.2K |
16.7K |
|
|
[224] |
[29,215,225] |
| social |
deep-learning based |
DBLP |
1 |
17.8K |
51.1K |
|
|
[226] |
[124,126,143,227] |
| social |
deep-learning based |
WIKI |
1 |
9.2K |
157.4K |
|
|
[228] |
[21,76,124] |
| social |
deep-learning based |
Facebook |
1 |
1.0K |
53.5K |
|
|
[229] |
[168] |
| social |
deep-learning based |
Citeseer |
1 |
3.3K |
4.7K |
✓ |
|
[230] |
[25,40,91,129,168,215] |
| social |
deep-learning based |
Cora |
1 |
2.7K |
5.4K |
✓ |
|
[231] |
[25,26,29,40,91,129,168,215,232] |
| |
statistical-based simulation |
|
|
|
|
✓ |
|
|
[153] |
| social |
deep-learning based |
Enron Emails |
1 |
785 |
5.8K |
|
|
[233,234] |
[235] |
| |
statistical-based simulation |
|
|
|
|
|
|
|
[22,236] |
| social |
statistical-based simulation |
ego-Twitter |
1 |
81,306 |
1.8M |
✓ |
|
[229] |
[151] |
| social |
LLM-based simulation |
WARRIORS |
1 |
100.0K |
285.0K |
|
|
[158] |
[158] |
| social |
LLM-based simulation |
IMDb-text |
1 |
125.7K |
1.5M |
|
|
[197] |
[197] |
| social |
LLM-based simulation |
Cora-text |
1 |
48.8K |
110.8K |
|
|
[231] |
[68,197] |
| social |
LLM-based simulation |
WeiboTech |
1 |
20.7K |
109.3K |
|
|
[197] |
[197,237] |
| social |
LLM-based simulation |
WeiboDaily |
1 |
66.5K |
354.1K |
|
|
[197] |
[197,237] |
| social |
LLM-based simulation |
Metoo |
1 |
1.0K |
31.9K |
|
|
[238] |
[239,240] |
| social |
LLM-based simulation |
Roe |
1 |
1.0K |
121.5K |
|
|
[241] |
[239,240] |
| social |
LLM-based simulation |
Twitter |
1 |
1.0M |
30.2M |
|
|
[242] |
[242] |
| user-item bipartite |
LLM-based simulation |
Movielens-1M |
1 |
6.0K/3.9K |
1.0M |
✓ |
✓ |
[189] |
[154,243] |
| user-item bipartite |
LLM-based simulation |
Amazon review |
1 |
54.4M/48.2M |
571.5M |
✓ |
|
[244] |
[155,243,245] |
| user-item bipartite |
LLM-based simulation |
Steam |
1 |
2.6M/15.4K |
7.8M |
✓ |
|
[246] |
[243,247] |
| user-item bipartite |
LLM-based simulation |
Lastfm |
1 |
1.9K/17.6K |
92.8K |
|
|
[248] |
[249,250,251] |
Transportation Network Modeling
Spatial graphs, or specifically geo-spatial graphs, model dynamic interactions in transportation systems, captured through an OD matrix that quantifies trip volumes and encodes connectivity [
252,
253]. GGMs jointly capture the temporal and spatial distributions of spatial graphs, enabling their application to multivariate time series forecasting in transportation networks. For example, STGEN [
125] learns the multi-modal distribution of spatial graphs by modeling the distribution of spatio-temporal walks using a novel heterogeneous probabilistic sequential model. This allows it to generate GPS coordinates for spatial graphs that align with real-world geographic maps.
Human Motion Prediction
In human motion graph generation, models need to reconstruct occluded or incomplete skeletal markers in historical data. MGCU [
254] extracts hierarchical spatial-temporal features using a multi-scale graph computational unit that models interactions across joint hierarchies. CSGN [
255] employs a graph-based framework to holistically generate entire motion sequences rather than auto-regressive sequential prediction, transforming latent vectors sampled from a Gaussian process into skeletal trajectories. MoGlow [
114] and MotionDiffuse [
127] leverage auto-regressive normalizing flows with FFN-based architectures to model motion sequences. Building upon MoGlow [
114], ST-GCN-2 [
256] integrates spatial graph convolutional networks to explicitly encode human skeletal topology within normalizing flows, uses a spatial graph convolutional network to extract features from past motion sequences.
Dynamic Graph Modeling
Most existing methods focus on static graphs, while many real-world graphs are dynamic, with nodes, edges, and attributes evolving over time. Recent advancements in dynamic graph learning have primarily targeted discrete-time dynamic graphs (DTDGs) and continuous-time dynamic graphs (CTDGs), with significant progress made through dynamic graph neural networks (DGNNs) [
257,
258]. These models excel in discriminative tasks such as future link prediction and node classification. In contrast, generative dynamic graph learning, which aims to synthesize realistic dynamic graph structures, remains in its nascent stage. Early works primarily focus on DTDGs, generating graph snapshots through spatiotemporal embedding learning akin to static graph generation [
23,
26]. Recent advances have shifted toward CTDGs, enabling fine-grained temporal modeling that better aligns with real-world application requirements [
21,
124]. Recent work is beginning to explore attribute-aware generation. VRDAG [
42] implements node feature generation through graph-based VAEs, while DG-Gen [
76] models edge attributes via joint conditional probability distributions. However, the field currently lacks standardized benchmarks, as most works directly adopt datasets designed for discriminative tasks [
21,
124].
General Graph Modeling
Recent years have witnessed unprecedented success achieved by Large Generative Models (LGMs) [
259]. The key to their success lies in their usage of the world knowledge inherited from the pre-training stage. Compared with the recent LGMs in NLP and computer vision, exploration into LGMs for graphs remains limited. Notably, LLGM [
143] proposes a large-scale training paradigm that uses a large corpus of graphs (over 5000 graphs) from 13 domains, offers superior zero-shot generative capability to existing graph generative models. Similarly, G2PT [
260] processes an auto-regressive transformer architecture that learns graph structures through edge-list based sequential representation on graphs in different graph domains.
6.2. Simulation-Based Graph Generator
In contrast to deep learning-based GGMs, simulation-based GGMs prioritize capturing the dynamic evolution of scale-free graphs, such as social graphs and user-item bipartite graphs. This section explores three prominent real-world applications of scalable GGMs, including social network analysis and recommender systems.
Social Network Analysis
Social network analysis (SNA) is foundational in computational social science, with simulation-based graph models emerging as powerful tools for real-world network dynamics. We highlight three key SNA applications: (1) Network pattern understanding. Statistical probabilistic modeling enables the discovery of emergent topological patterns, offering insights into their formation mechanisms. The Barabási-Albert (BA) model [
7] employs preferential attachment to generate scale-free networks. Similarly, edge-copying models [
59] simulate processes like gene duplication in biological networks, reflecting structural conservation through node neighborhood replication. (2) Network evolution modeling. Graph evolution simulations are instrumental in studying dynamic processes. Recent advances integrate LLM-based agents to simulate human-like behaviors for large-scale social network evolution, particularly in online social environments. Studies leverage either real data [
159,
239], or synthetic user profiles [
68] to initialize networks. Hybrid approaches, such as OASIS [
160], combine limited real-world relationships with synthetic data, while others employ homophily-driven assumptions to connect similar users [
162]. (3) Influence maximization. Identifying key opinion leaders (KOLs) within social networks is essential for effective information dissemination and marketing strategies. Simulation-based graph generators can model the influence diffusion process through networks, enabling researchers to pinpoint nodes with the greatest potential for widespread information propagation. Early graph generation studies often relied on IC/LT models to simulate influence propagation and identify key nodes [
261]. More recent work has shifted toward using LLM-based agents to simulate user behaviors and interactions within social networks. These models simulate influence propagation to study information diffusion and dynamic attitude shifts in response to events, such as opinion leadership dynamics [
9] and rumor spread [
157]. (4) Policy analysis. Simulation-based graph generators provide a controlled environment to evaluate the impact of various policies on social networks. In economic domains, frameworks like SRAP-Agent [
156] simulate decision-making under resource allocation policies, enabling policy evaluation through synthetic network interactions.
Recommender System
In recommender systems, predicting user-item interactions fits a bipartite graph: users and items as separate node sets, edges for ratings, purchases, or clicks [
262]. LLM-driven simulations build controllable user-item interaction environments, enabling analysis of recommender challenges that resist traditional approaches. (1) Addressing sparsity and cold-start challenges. Traditional collaborative filtering methods struggle with sparse data, especially for new users or items. Simulation-based approaches alleviate this by generating realistic interactions for cold-start scenarios. For example, the Lusifer environment [
263] uses LLM agents to update user states dynamically and generate feedback for new items, directly targeting the cold-start problem. Similarly, LLM-powered user simulators [
165] address sparse training data by combining logical preference reasoning with data-driven statistics to simulate reliable behaviors for underrepresented users and items. (2) Modeling preference evolution. Static bipartite graphs cannot model evolving user preferences and item popularity. Simulation frameworks address this by embedding temporal patterns into graph generation. SUBER [
264] employs LLM-based agents to simulate user behavior trajectories, generating dynamic bipartite graphs that capture preference changes over time. In contrast to modeling individual users, RecAgent [
154] incorporates social interactions where users influence others’ preferences. RecUserSim [
265] extends this approach by incorporating memory and behavioral modules in conversational settings to model evolution across dialogue interactions. Agent4Rec [
243] shows how generative agents with personalized profiles and memory can simulate diverse long-term user behaviors.
7. Future Opportunity
We explore several promising future opportunities, including improving scalability of deep learning models, enhancing controllability with target properties, advancing multimodal graph generation using diverse data sources, and establishing more robust and generalizable evaluation metrics.
Scalability
Deep learning-based GGMs capture complex high-order dependencies but suffer from super-linear time complexity, limiting them to small graphs. Only a handful of approaches achieve linear time complexity (O(M)) [
20,
29]. Simulation-based models scale linearly to millions of nodes but rely on oversimplified assumptions, causing unrealistic topologies. As illustrated in
Figure 1, simulation-based models dominate large-scale graph generation due to their efficiency, while deep learning-based excel in capturing complex, real-world graph statistics.Bridging this gap requires hybrid frameworks that combine scalability and expressiveness.
Controllability
Existing GGMs provide limited control beyond basic graph-level statistics. While domains such as molecule and protein design require constraint satisfaction [
266,
267,
268,
269], and social networks demand behavior-aligned structures [
9,
55]. Control interfaces that encode prior knowledge are crucial. Moreover, current methods still lack semantic and functional control of node and edge attributes.
Multimodality
Real-world graphs include rich multimodal information (e.g., text, attributes, temporal signals). As illustrated in
Figure 1, recent research has progressively advanced the attribute modalities of generated graphs, evolving from non-attributed structures to categorical/numeric attributes, and ultimately to textual or multimodal representations [
68,
270]. Challenges persist in effectively capturing attributed graphs, such as textual descriptions, OD flows, and molecule types, which are crucial for generating results that are both structurally sound and semantically meaningful [
1].
Evaluation
Like generative models for image and language, graph generation lacks unique ground truth, making evaluation inherently difficult. Neural-based metrics capture structure but do not scale; statistic-based metrics miss important properties [
190]. Many domains also require costly or domain-specific validation [
271]. More robust, efficient, and standardized evaluation protocols are needed.
8. Conclusion
This survey provides a systematic overview of graph generative models (GGMs). First, we propose a novel taxonomy that categorizes GGMs by target graph categories (geometric vs. scale-free), generated attribute modality (non-attribute, categorical/numeric, or textual), and probabilistic modeling methods (deep learning-based vs. simulation-based). For geometric graphs, we analyze how deep learning-based GGMs leverage advanced architectures to capture structural dependencies; For scale-free graphs, we show how simulation-based approaches effectively model network growth mechanisms. Additionally, we examine model architectures and evaluation metrics across different graph categories, analyzing their strengths and limitations in various application scenarios. Finally, we identify four key research directions: improving scalability, enhancing controllability, advancing multimodal graph generation, and developing robust evaluation metrics. This survey aims to provide a foundational reference for researchers in graph generation by consolidating recent advances and offering structured insights into this rapidly evolving field.
References
- Guo, X.; Zhao, L. A Systematic Survey on Deep Generative Models for Graph Generation. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 5370–5390. [CrossRef]
- Han, J.; Cen, J.; Wu, L.; Li, Z.; Kong, X.; Jiao, R.; Yu, Z.; Xu, T.; Wu, F.; Wang, Z.; et al. A survey of geometric graph neural networks: Data structures, models and applications. CoRR arXiv:2403.00485 2024.
- Bonifati, A.; Holubová, I.; Prat-Pérez, A.; Sakr, S. Graph Generators: State of the Art and Open Challenges. ACM Comput. Surv., CSUR 2021, 53, 36:1–36:30. [CrossRef]
- Tang, H.; Wu, S.; Xu, G.; Li, Q. Dynamic graph evolution learning for recommendation. In Proceedings of the Proc. Int. ACM SIGIR Conf. Res. Dev. Inf. Retr., SIGIR, 2023, pp. 1589–1598.
- Sakr, S.; Pardede, E., Eds. Graph Data Management: Techniques and Applications; IGI Global, 2011. [CrossRef]
- Erdos, P.; Rényi, A.; et al. On the evolution of random graphs. Publ. math. inst. hung. acad. sci 1960, 5, 17–60.
- Barabási, A.L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512.
- Mou, X.; Ding, X.; He, Q.; Wang, L.; Liang, J.; Zhang, X.; Sun, L.; Lin, J.; Zhou, J.; Huang, X.; et al. From Individual to Society: A Survey on Social Simulation Driven by Large Language Model-based Agents. CoRR arXiv:2412.03563 2024, [2412.03563]. [CrossRef]
- Zhang, X.; Chen, X.; Liu, Y.; Wang, J.; Hu, Z.; Yan, R. A Large-scale Time-aware Agents Simulation for Influencer Selection in Digital Advertising Campaigns. CoRR arXiv:2411.01143 2024, [2411.01143]. [CrossRef]
- Zhu, Y.; Du, Y.; Wang, Y.; Xu, Y.; Zhang, J.; Liu, Q.; Wu, S. A Survey on Deep Graph Generation: Methods and Applications. In Proceedings of the Learning on Graphs Conf., LoG, 2022, Vol. 198, p. 47.
- Zhang, Z.; Cui, P.; Zhu, W. Deep Learning on Graphs: A Survey. IEEE Trans. Knowl. Data Eng. 2022, 34, 249–270. [CrossRef]
- Faez, F.; Ommi, Y.; Baghshah, M.S.; Rabiee, H.R. Deep Graph Generators: A Survey. IEEE Access 2021, 9, 106675–106702. [CrossRef]
- Liu, C.; Fan, W.; Liu, Y.; Li, J.; Li, H.; Liu, H.; Tang, J.; Li, Q. Generative Diffusion Models on Graphs: Methods and Applications. In Proceedings of the Proc. Int. Joint Conf. Artif. Intell., IJCAI, 2023, pp. 6702–6711. [CrossRef]
- Gao, C.; Lan, X.; Li, N.; Yuan, Y.; Ding, J.; Zhou, Z.; Xu, F.; Li, Y. Large language models empowered agent-based modeling and simulation: A survey and perspectives. Humanities and Social Sciences Communications 2024, 11, 1–24.
- Hao, G.; Wu, J.; Pan, Q.; Morello, R. Quantifying the uncertainty of LLM hallucination spreading in complex adaptive social networks. Sci. Rep. 2024, 14, 16375.
- Pach, J. Geometric graph theory. London Mathematical Society Lecture Note Series 1999, pp. 167–200.
- Broido, A.D.; Clauset, A. Scale-free networks are rare. Nat. Commun. 2019, 10, 1017.
- Holme, P. Rare and everywhere: Perspectives on scale-free networks. Nat. Commun. 2019, 10, 1016.
- You, J.; Ying, R.; Ren, X.; Hamilton, W.L.; Leskovec, J. GraphRNN: Generating Realistic Graphs with Deep Auto-regressive Models. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2018, Vol. 80, pp. 5694–5703.
- Dai, H.; Nazi, A.; Li, Y.; Dai, B.; Schuurmans, D. Scalable Deep Generative Modeling for Sparse Graphs. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2020, Vol. 119, pp. 2302–2312.
- Gupta, S.; Manchanda, S.; Bedathur, S.; Ranu, S. TIGGER: Scalable Generative Modelling for Temporal Interaction Graphs. Proc. AAAI Conf. Artif. Intell., AAAI 2022, 36, 6819–6828. [CrossRef]
- Zeno, G.; La Fond, T.; Neville, J. DYMOND: DYnamic MOtif-NoDes Network Generative Model. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW, New York, NY, USA, 2021; pp. 718–729. [CrossRef]
- Liao, R.; Li, Y.; Song, Y.; Wang, S.; Hamilton, W.L.; Duvenaud, D.; Urtasun, R.; Zemel, R.S. Efficient Graph Generation with Graph Recurrent Attention Networks. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2019, pp. 4257–4267.
- Wang, H.; Wang, J.; Wang, J.; Zhao, M.; Zhang, W.; Zhang, F.; Xie, X.; Guo, M. GraphGAN: Graph Representation Learning With Generative Adversarial Nets. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2018, pp. 2508–2515. [CrossRef]
- Goyal, N.; Jain, H.V.; Ranu, S. GraphGen: A Scalable Approach to Domain-agnostic Labeled Graph Generation. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW, 2020, pp. 1253–1263. [CrossRef]
- Bojchevski, A.; Shchur, O.; Zügner, D.; Günnemann, S. NetGAN: Generating Graphs via Random Walks. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2018, Vol. 80, pp. 609–618.
- Simonovsky, M.; Komodakis, N. GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders. In Proceedings of the Artificial Neural Networks and Machine Learning, ICANN, 2018, Vol. 11139, pp. 412–422. [CrossRef]
- Jo, J.; Lee, S.; Hwang, S.J. Score-based Generative Modeling of Graphs via the System of Stochastic Differential Equations. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2022, Vol. 162, pp. 10362–10383.
- Chen, X.; He, J.; Han, X.; Liu, L. Efficient and Degree-Guided Graph Generation via Discrete Diffusion Modeling. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2023, Vol. 202, pp. 4585–4610.
- Drobyshevskiy, M.; Turdakov, D. Random graph modeling: A survey of the concepts. ACM Comput. Surv., CSUR 2019, 52, 1–36.
- Schneider, P.J.; TIAN, L.; Rizoiu, M.A. Learning to Make Friends: Coaching LLM Agents toward Emergent Social Ties. In Proceedings of the Workshop on Scaling Environments for Agents, 2025.
- Eigenschink, P.; Reutterer, T.; Vamosi, S.; Vamosi, R.; Sun, C.; Kalcher, K. Deep Generative Models for Synthetic Data: A Survey. IEEE Access 2023, 11, 47304–47320. [CrossRef]
- dos Santos, C.N.; Mroueh, Y.; Padhi, I.; Dognin, P.L. Learning Implicit Generative Models by Matching Perceptual Features. In Proceedings of the Proc. IEEE Int. Conf. Comput. Vis., ICCV, 2019, pp. 4460–4469. [CrossRef]
- Bengio, Y.; Ducharme, R.; Vincent, P. A Neural Probabilistic Language Model. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2000, pp. 932–938.
- Zhao, Q.; Ren, W.; Li, T.; Xu, X.; Liu, H. GraphGPT: Graph Learning with Generative Pre-trained Transformers. CoRR arXiv:2401.00529 2024, [2401.00529]. [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2014.
- Zhang, S.; Tong, H.; Xu, J.; Maciejewski, R. Graph Convolutional Networks: Algorithms, Applications and Open Challenges. In Proceedings of the Inte. Conf. Computational Social Networks, CSoNet, 2018, Vol. 11280, pp. 79–91. [CrossRef]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2018.
- Samanta, B.; De, A.; Jana, G.; Chattaraj, P.K.; Ganguly, N.; Rodriguez, M.G. NeVAE: A Deep Generative Model for Molecular Graphs. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2019, pp. 1110–1117. [CrossRef]
- Grover, A.; Zweig, A.; Ermon, S. Graphite: Iterative Generative Modeling of Graphs. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2019, Vol. 97, pp. 2434–2444.
- Geng, Z.; Xie, S.; Xia, Y.; Wu, L.; Qin, T.; Wang, J.; Zhang, Y.; Wu, F.; Liu, T. De Novo Molecular Generation via Connection-aware Motif Mining. CoRR arXiv:2302.01129 2023, [2302.01129]. [CrossRef]
- Li, F.; Wang, X.; Cheng, D.; Chen, C.; Zhang, Y.; Lin, X. Efficient Dynamic Attributed Graph Generation, 2024, [arXiv:cs.DB/2412.08810].
- Rezende, D.J.; Mohamed, S. Variational Inference with Normalizing Flows. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2015, Vol. 37, pp. 1530–1538.
- Zang, C.; Wang, F. MoFlow: An Invertible Flow Model for Generating Molecular Graphs. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, 2020, pp. 617–626. [CrossRef]
- Luo, Y.; Yan, K.; Ji, S. GraphDF: A Discrete Flow Model for Molecular Graph Generation. CoRR arXiv:2102.01189 2021, [2102.01189].
- Hou, X.; Zhu, T.; Ren, M.; Bu, D.; Gao, X.; Zhang, C.; Sun, S. Improving Molecular Graph Generation with Flow Matching and Optimal Transport. CoRR arXiv:2411.05676 2024, [2411.05676]. [CrossRef]
- Pan, Z.; Yu, W.; Yi, X.; Khan, A.; Yuan, F.; Zheng, Y. Recent progress on generative adversarial networks (GANs): A survey. IEEE access 2019, 7, 36322–36333.
- Yang, C.; Zhuang, P.; Shi, W.; Luu, A.; Li, P. Conditional Structure Generation through Graph Variational Generative Adversarial Nets. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2019, pp. 1338–1349.
- Wiatrak, M.; Albrecht, S.V.; Nystrom, A. Stabilizing generative adversarial networks: A survey. CoRR arXiv:1910.00927 2019.
- Réau, M.; Renaud, N.; Xue, L.C.; Bonvin, A.M.J.J. DeepRank-GNN: a graph neural network framework to learn patterns in protein-protein interfaces. Bioinform. 2023, 39. [CrossRef]
- Vignac, C.; Krawczuk, I.; Siraudin, A.; Wang, B.; Cevher, V.; Frossard, P. Digress: Discrete Denoising diffusion for graph generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2023.
- Huang, H.; Sun, L.; Du, B.; Fu, Y.; Lv, W. GraphGDP: Generative Diffusion Processes for Permutation Invariant Graph Generation. In Proceedings of the Proc. IEEE Int. Conf. Data Min. ICDM, 2022, pp. 201–210. [CrossRef]
- Cao, H.; Tan, C.; Gao, Z.; Xu, Y.; Chen, G.; Heng, P.; Li, S.Z. A Survey on Generative Diffusion Models. IEEE Trans. Knowl. Data Eng. 2024, 36, 2814–2830. [CrossRef]
- Ma, Y.; Zhan, K. Self-Contrastive Graph Diffusion Network. In Proceedings of the Proc. ACM Int. Conf. Multimed., MM, 2023, pp. 3857–3865. [CrossRef]
- Yao, Y.; Wang, X.; Zhang, Z.; Qin, Y.; Zhang, Z.; Chu, X.; Yang, Y.; Zhu, W.; Mei, H. Exploring the Potential of Large Language Models in Graph Generation. CoRR arXiv:2403.14358 2024, [2403.14358]. [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ’small-world’ networks. Nature 1998, 393, 440–442.
- Leskovec, J.; Kleinberg, J.; Faloutsos, C. Graphs over time: densification laws, shrinking diameters and possible explanations. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, 2005, pp. 177–187.
- Lattanzi, S.; Sivakumar, D. Affiliation networks. In Proceedings of the Proc. Annu. ACM Symp. Theory Comput., STOC, 2009, pp. 427–434. [CrossRef]
- Kleinberg, J.M.; Kumar, R.; Raghavan, P.; Rajagopalan, S.; Tomkins, A.S. The web as a graph: Measurements, models, and methods. In Proceedings of the Lect. Notes Comput. Sci., COCOON. Springer, 1999, pp. 1–17.
- Robins, G.; Pattison, P.; Kalish, Y.; Lusher, D. An introduction to exponential random graph (p*) models for social networks. Social networks 2007, 29, 173–191.
- Chen, W.; Su, Y.; Zuo, J.; Yang, C.; Yuan, C.; Chan, C.; Yu, H.; Lu, Y.; Hung, Y.; Qian, C.; et al. AgentVerse: Facilitating Multi-Agent Collaboration and Exploring Emergent Behaviors. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2024.
- Park, J.S.; O’Brien, J.C.; Cai, C.J.; Morris, M.R.; Liang, P.; Bernstein, M.S. Generative Agents: Interactive Simulacra of Human Behavior. In Proceedings of the Proc. Annu. ACM Symp. User Interface Softw. Technol., UIST, 2023, pp. 2:1–2:22. [CrossRef]
- Li, N.; Gao, C.; Li, M.; Li, Y.; Liao, Q. EconAgent: Large Language Model-Empowered Agents for Simulating Macroeconomic Activities. In Proceedings of the Proc. Annu. Meet. Assoc. Comput Linguist., ACL, 2024, pp. 15523–15536. [CrossRef]
- Marzo, G.D.; Pietronero, L.; Garcia, D. Emergence of Scale-Free Networks in Social Interactions among Large Language Models. CoRR arXiv:2312.06619 2023, [2312.06619]. [CrossRef]
- Papachristou, M.; Yuan, Y. Network Formation and Dynamics Among Multi-LLMs. CoRR arXiv:2402.10659 2024.
- Havrilla, A.; Alvarez-Melis, D.; Fusi, N. IGDA: Interactive Graph Discovery through Large Language Model Agents. CoRR arXiv:2502.17189 2025, [2502.17189]. [CrossRef]
- Chang, S.; Chaszczewicz, A.; Wang, E.; Josifovska, M.; Pierson, E.; Leskovec, J. LLMs Generate Structurally Realistic Social Networks but Overestimate Political Homophily. In Proceedings of the Proc. Int. AAAI Conf. on Web and Social Media, ICWSM, 2025, pp. 341–371. [CrossRef]
- Ji, J.; Lei, R.; Bi, J.; Wei, Z.; Chen, X.; Lin, Y.; Pan, X.; Li, Y.; Ding, B. LLM-Based Multi-Agent Systems are Scalable Graph Generative Models, 2025, [arXiv:cs.CL/2410.09824].
- Du, E.; Li, X.; Jin, T.; Zhang, Z.; Li, R.; Wang, G. GraphMaster: Automated Graph Synthesis via LLM Agents in Data-Limited Environments. CoRR arXiv:2504.00711 2025, [2504.00711]. [CrossRef]
- Grisoni, F.; Moret, M.; Lingwood, R.; Schneider, G. Bidirectional Molecule Generation with Recurrent Neural Networks. J. Chem. Inf. Model. 2020, 60, 1175–1183. [CrossRef]
- Lai, X.; Yang, P.; Wang, K.; Yang, Q.; Yu, D. MGRNN: Structure generation of molecules based on graph recurrent neural networks. Mol. Inf. 2021, 40, 2100091.
- Popova, M.; Shvets, M.; Oliva, J.; Isayev, O. MolecularRNN: Generating realistic molecular graphs with optimized properties. CoRR arXiv:1905.13372 2019, [1905.13372].
- Cohen-Karlik, E.; Rozenberg, E.; Freedman, D. Overcoming Order in Autoregressive Graph Generation for Molecule Generation. Trans. Mach. Learn. Res. 2024, 2024.
- Jang, Y.; Lee, S.; Ahn, S. A Simple and Scalable Representation for Graph Generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2024.
- Bacciu, D.; Podda, M. Graphgen-redux: a Fast and Lightweight Recurrent Model for labeled Graph Generation. In Proceedings of the Int. Joint Conf. on Neural Networks, IJCNN, 2021, pp. 1–8. [CrossRef]
- Hosseini, R.; Simini, F.; Vishwanath, V.; Hoffmann, H. A Deep Probabilistic Framework for Continuous Time Dynamic Graph Generation. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2025, pp. 17249–17257. [CrossRef]
- Lu, H.; Wei, Z.; Wang, X.; Zhang, K.; Liu, H. Graphgpt: A graph enhanced generative pretrained transformer for conditioned molecular generation. Int. J. Mol. Sci. 2023, 24, 16761.
- Jensen, J.H. A graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space. Chem. Sci. 2019, 10, 3567–3572.
- Bongini, P.; Bianchini, M.; Scarselli, F. Molecular generative Graph Neural Networks for Drug Discovery. Neurocomputing 2021, 450, 242–252. [CrossRef]
- Mahmood, O.; Mansimov, E.; Bonneau, R.; Cho, K. Masked graph modeling for molecule generation. Nat. Commun. 2021, 12, 3156.
- Alhamoud, K.; Ghunaim, Y.; Alshehri, A.S.; Li, G.; Ghanem, B.; You, F. Leveraging 2D molecular graph pretraining for improved 3D conformer generation with graph neural networks. Comput. Chem. Eng. 2024, 183, 108622. [CrossRef]
- Shi, C.; Xu, M.; Zhu, Z.; Zhang, W.; Zhang, M.; Tang, J. GraphAF: a Flow-based Autoregressive Model for Molecular Graph Generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2020.
- Luo, Y.; Wan, Z.; Chen, Y.; Mai, G.; Chung, F.; Larson, K. TransFlower: An Explainable Transformer-Based Model with Flow-to-Flow Attention for Commuting Flow Prediction. CoRR arXiv:2402.15398 2024, [2402.15398]. [CrossRef]
- Feng, W.; Wang, L.; Lin, Z.; Zhu, Y.; Wang, H.; Dong, J.; Bai, R.; Wang, H.; Zhou, J.; Peng, W.; et al. Generation of 3D molecules in pockets via a language model. Nature Machine Intelligence 2024, 6, 62–73.
- Lu, S.; Lin, H.; Yao, L.; Gao, Z.; Ji, X.; E, W.; Zhang, L.; Ke, G. Uni-3DAR: Unified 3D Generation and Understanding via Autoregression on Compressed Spatial Tokens. CoRR arXiv:2503.16278 2025, [2503.16278]. [CrossRef]
- Zholus, A.; Kuznetsov, M.; Schutski, R.; Shayakhmetov, R.; Polykovskiy, D.; Chandar, S.; Zhavoronkov, A. BindGPT: A Scalable Framework for 3D Molecular Design via Language Modeling and Reinforcement Learning. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2025, pp. 26083–26091. [CrossRef]
- Luo, Y.; Ji, S. An Autoregressive Flow Model for 3D Molecular Geometry Generation from Scratch. In Proceedings of the The Tenth Int. Conf. Learn. Represent., ICLR, 2022.
- Zhang, W.; Zhang, L.; Pfoser, D.; Zhao, L. Disentangled Dynamic Graph Deep Generation. In Proceedings of the SIAM Int. Conf. Data Mining, SDM, 2021, pp. 738–746. [CrossRef]
- Gebauer, N.W.A.; Gastegger, M.; Schütt, K. Symmetry-adapted generation of 3d point sets for the targeted discovery of molecules. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2019, pp. 7564–7576.
- Holden, D.; Saito, J.; Komura, T. A deep learning framework for character motion synthesis and editing. ACM Trans. Graphics 2016, 35, 138:1–138:11. [CrossRef]
- Kipf, T.N.; Welling, M. Variational Graph Auto-Encoders. CoRR arXiv:1611.07308 2016, [1611.07308].
- Hu, Z.; Dong, Y.; Wang, K.; Chang, K.; Sun, Y. GPT-GNN: Generative Pre-Training of Graph Neural Networks. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, 2020, pp. 1857–1867. [CrossRef]
- Jin, W.; Barzilay, R.; Jaakkola, T.S. Junction Tree Variational Autoencoder for Molecular Graph Generation. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2018, Vol. 80, pp. 2328–2337.
- Liu, Q.; Allamanis, M.; Brockschmidt, M.; Gaunt, A.L. Constrained Graph Variational Autoencoders for Molecule Design. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2018, pp. 7806–7815.
- Madhawa, K.; Ishiguro, K.; Nakago, K.; Abe, M. GraphNVP: An Invertible Flow Model for Generating Molecular Graphs, 2019, [arXiv:stat.ML/1905.11600].
- Jin, W.; Yang, K.; Barzilay, R.; Jaakkola, T. Learning Multimodal Graph-to-Graph Translation for Molecular Optimization, 2019, [arXiv:cs.LG/1812.01070].
- Hou, Z.; Liu, X.; Cen, Y.; Dong, Y.; Yang, H.; Wang, C.; Tang, J. GraphMAE: Self-Supervised Masked Graph Autoencoders. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, 2022, pp. 594–604. [CrossRef]
- Lin, Z.; Zhang, Y.; Duan, L.; Ou-Yang, L.; Zhao, P. MoVAE: A Variational AutoEncoder for Molecular Graph Generation. In Proceedings of the SIAM Int. Conf. Data Mining, SDM, 2023, pp. 514–522. [CrossRef]
- Mitton, J.; Senn, H.M.; Wynne, K.; Murray-Smith, R. A Graph VAE and Graph Transformer Approach to Generating Molecular Graphs. CoRR arXiv:2104.04345 2021, [2104.04345].
- Kusner, M.J.; Paige, B.; Hernández-Lobato, J.M. Grammar Variational Autoencoder. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2017, Vol. 70, pp. 1945–1954.
- Wei, G.; Huang, Y.; Duan, C.; Song, Y.; Du, Y. Navigating Chemical Space with Latent Flows. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2024.
- Satorras, V.G.; Hoogeboom, E.; Fuchs, F.; Posner, I.; Welling, M. E(n) Equivariant Normalizing Flows. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2021, pp. 4181–4192.
- Song, Y.; Gong, J.; Xu, M.; Cao, Z.; Lan, Y.; Ermon, S.; Zhou, H.; Ma, W. Equivariant Flow Matching with Hybrid Probability Transport for 3D Molecule Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2023.
- Lippe, P.; Gavves, E. Categorical Normalizing Flows via Continuous Transformations. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2021.
- Song, Y.; Gong, J.; Qu, Y.; Zhou, H.; Zheng, M.; Liu, J.; Ma, W. Unified Generative Modeling of 3D Molecules via Bayesian Flow Networks. CoRR arXiv:2403.15441 2024, [2403.15441]. [CrossRef]
- Irwin, R.; Tibo, A.; Janet, J.P.; Olsson, S. Semlaflow-efficient 3d molecular generation with latent attention and equivariant flow matching. In Proceedings of the Int. Conf. Artif. Intell. Stat., AISTATS, 2025.
- Xu, M.; Luo, S.; Bengio, Y.; Peng, J.; Tang, J. Learning Neural Generative Dynamics for Molecular Conformation Generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2021.
- Dunn, I.; Koes, D.R. Mixed Continuous and Categorical Flow Matching for 3D De Novo Molecule Generation. CoRR arXiv:2404.19739 2024, [2404.19739]. [CrossRef]
- Hong, H.; Lin, W.; Tan, K. Accelerating 3D Molecule Generation via Jointly Geometric Optimal Transport. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2025.
- Ma, C.; Yang, Q.; Gao, X.; Zhang, X. DEMO: Disentangled Molecular Graph Generation via an Invertible Flow Model. In Proceedings of the Proc. Int. Conf. Inf. Knowledge Manage., CIKM, 2022, pp. 1420–1429. [CrossRef]
- Pandey, M.; Subbaraj, G.; Cherkasov, A.; Ester, M.; Bengio, E. Pretraining Generative Flow Networks with Inexpensive Rewards for Molecular Graph Generation. CoRR arXiv:2503.06337 2025, [2503.06337]. [CrossRef]
- Eijkelboom, F.; Bartosh, G.; Naesseth, C.A.; Welling, M.; van de Meent, J. Variational Flow Matching for Graph Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2024.
- Hassan, M.; Shenoy, N.; Lee, J.; Stärk, H.; Thaler, S.; Beaini, D. ET-Flow: Equivariant Flow-Matching for Molecular Conformer Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2024.
- Henter, G.E.; Alexanderson, S.; Beskow, J. MoGlow: probabilistic and controllable motion synthesis using normalising flows. ACM Trans. Graphics 2020, 39, 236:1–236:14. [CrossRef]
- Kuznetsov, M.; Polykovskiy, D. MolGrow: A Graph Normalizing Flow for Hierarchical Molecular Generation. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2021, pp. 8226–8234. [CrossRef]
- Cao, N.D.; Kipf, T. MolGAN: An implicit generative model for small molecular graphs. CoRR arXiv:1805.11973 2018, [1805.11973].
- Yamada, M.; Sugiyama, M. Molecular Graph Generation by Decomposition and Reassembling. CoRR arXiv:2302.00587 2023, [2302.00587]. [CrossRef]
- Park, J.; Ahn, J.; Choi, J.; Kim, J. Mol-AIR: Molecular Reinforcement Learning with Adaptive Intrinsic Rewards for Goal-Directed Molecular Generation. J. Chem. Inf. Model. 2025, 65, 2283–2296. [CrossRef]
- Zhang, L.; Zhao, L.; Qin, S.; Pfoser, D. TG-GAN: Deep Generative Models for Continuously-time Temporal Graph Generation. CoRR arXiv:2005.08323 2020, [2005.08323].
- Guimaraes, G.L.; Sánchez-Lengeling, B.; Farias, P.L.C.; Aspuru-Guzik, A. Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models. CoRR arXiv:1705.10843 2017, [1705.10843].
- Zhang, O.; Huang, Y.; Cheng, S.; Yu, M.; Zhang, X.; Lin, H.; Zeng, Y.; Wang, M.; Wu, Z.; Zhao, H.; et al. FragGen: towards 3D geometry reliable fragment-based molecular generation. Chem. Sci. 2024, 15, 19452–19465.
- Pölsterl, S.; Wachinger, C. Adversarial Learned Molecular Graph Inference and Generation. In Proceedings of the ECML-PKDD, 2020, Vol. 12458, pp. 173–189. [CrossRef]
- You, J.; Liu, B.; Ying, Z.; Pande, V.S.; Leskovec, J. Graph Convolutional Policy Network for Goal-Directed Molecular Graph Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2018, pp. 6412–6422.
- Zhou, D.; Zheng, L.; Han, J.; He, J. A Data-Driven Graph Generative Model for Temporal Interaction Networks. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, New York, NY, USA, 2020; pp. 401–411. [CrossRef]
- Ling, C.; Cao, H.; Zhao, L. STGEN: Deep Continuous-Time Spatiotemporal Graph Generation. In Proceedings of the ECML-PKDD, 2022, Vol. 13715, pp. 340–356. [CrossRef]
- He, X.; Fu, D.; Tong, H.; Maciejewski, R.; He, J. Temporal Heterogeneous Graph Generation with Privacy, Utility, and Efficiency. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2025.
- Zhang, M.; Cai, Z.; Pan, L.; Hong, F.; Guo, X.; Yang, L.; Liu, Z. MotionDiffuse: Text-Driven Human Motion Generation With Diffusion Model. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 4115–4128. [CrossRef]
- Xu, M.; Powers, A.S.; Dror, R.O.; Ermon, S.; Leskovec, J. Geometric latent diffusion models for 3d molecule generation. In Proceedings of the Proc. Int. Conf. Machin. Learn., ICML. PMLR, 2023, pp. 38592–38610.
- Kose, O.D.; Shen, Y. FairWire: Fair Graph Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2024.
- Li, M.; Kreacic, E.; Potluru, V.K.; Li, P. GraphMaker: Can Diffusion Models Generate Large Attributed Graphs? Trans. Mach. Learn. Res. 2024, 2024.
- Hoogeboom, E.; Satorras, V.G.; Vignac, C.; Welling, M. Equivariant Diffusion for Molecule Generation in 3D. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2022, Vol. 162, pp. 8867–8887.
- Liu, M.; Yan, K.; Oztekin, B.; Ji, S. GraphEBM: Molecular Graph Generation with Energy-Based Models. CoRR arXiv:2102.00546 2021, [2102.00546].
- Huang, H.; Sun, L.; Du, B.; Lv, W. Conditional Diffusion Based on Discrete Graph Structures for Molecular Graph Generation. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2023, pp. 4302–4311. [CrossRef]
- Chen, Z.; Peng, B.; Ning, X.; et al. Shape-conditioned 3D Molecule Generation via Equivariant Diffusion Models. In Proceedings of the NeurIPS, GenBio Workshop, 2023.
- Niu, C.; Song, Y.; Song, J.; Zhao, S.; Grover, A.; Ermon, S. Permutation Invariant Graph Generation via Score-Based Generative Modeling. In Proceedings of the Int. Conf. Artif. Intell. Stat., AISTATS, 2020, Vol. 108, pp. 4474–4484.
- Bao, F.; Zhao, M.; Hao, Z.; Li, P.; Li, C.; Zhu, J. Equivariant Energy-Guided SDE for Inverse Molecular Design. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2023.
- Jung, H.; Park, Y.; Schmid, L.; Jo, J.; Lee, D.; Kim, B.; Yun, S.Y.; Shin, J. Conditional synthesis of 3d molecules with time correction sampler. Adv. neural inf. proces. syst., NeurIPS 2024, 37, 75914–75941.
- Huang, L.; Zhang, H.; Xu, T.; Wong, K.C. Mdm: Molecular diffusion model for 3d molecule generation. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2023, Vol. 37, pp. 5105–5112.
- Schneuing, A.; Harris, C.; Du, Y.; Didi, K.; Jamasb, A.; Igashov, I.; Du, W.; Gomes, C.; Blundell, T.L.; Lio, P.; et al. Structure-based drug design with equivariant diffusion models. Nature Computational Science 2024, 4, 899–909.
- Han, X.; Shan, C.; Shen, Y.; Xu, C.; Yang, H.; Li, X.; Li, D. Training-free multi-objective diffusion model for 3d molecule generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2023.
- Morehead, A.; Cheng, J. Geometry-complete diffusion for 3D molecule generation and optimization. Communications Chemistry 2024, 7, 150.
- Huang, L.; Xu, T.; Yu, Y.; Zhao, P.; Chen, X.; Han, J.; Xie, Z.; Li, H.; Zhong, W.; Wong, K.C.; et al. A dual diffusion model enables 3D molecule generation and lead optimization based on target pockets. Nature Communications 2024, 15, 2657.
- Wang, Y.; Rossi, R.A.; Park, N.; Chen, H.; Ahmed, N.K.; Trivedi, P.; Dernoncourt, F.; Koutra, D.; Derr, T. A Large-scale Training Paradigm for Graph Generative Models. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2025.
- Rong, C.; Ding, J.; Liu, Z.; Li, Y. Complexity-aware Large Scale Origin-Destination Network Generation via Diffusion Model. CoRR arXiv:2306.04873 2023, [2306.04873]. [CrossRef]
- Liu, Z.; Luo, Y.; Huang, H.; Zhang, E.; Li, S.; Fang, J.; Shi, Y.; Wang, X.; Kawaguchi, K.; Chua, T. NExT-Mol: 3D Diffusion Meets 1D Language Modeling for 3D Molecule Generation. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2025.
- Ninniri, M.; Podda, M.; Bacciu, D. Classifier-Free Graph Diffusion for Molecular Property Targeting. In Proceedings of the ECML-PKDD, 2024, Vol. 14944, pp. 318–335. [CrossRef]
- Xu, C.; Wang, H.; Wang, W.; Zheng, P.; Chen, H. Geometric-facilitated denoising diffusion model for 3D molecule generation. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2024, Vol. 38, pp. 338–346.
- Leskovec, J.; Chakrabarti, D.; Kleinberg, J.M.; Faloutsos, C.; Ghahramani, Z. Kronecker Graphs: An Approach to Modeling Networks. J. Mach. Learn. Res. 2010, 11, 985–1042. [CrossRef]
- Perra, N.; Gonçalves, B.; Pastor-Satorras, R.; Vespignani, A. Activity driven modeling of time varying networks. Sci. Rep. 2012, 2, 469.
- Hadian, A.; Nobari, S.; Minaei-Bidgoli, B.; Qu, Q. ROLL: Fast In-Memory Generation of Gigantic Scale-free Networks. In Proceedings of the Proc. ACM SIGMOD Int. Conf. Manage. Data, SIGMOD, 2016, pp. 1829–1842. [CrossRef]
- Wang, C.; Wang, B.; Huang, B.; Song, S.; Li, Z. FastSGG: Efficient Social Graph Generation Using a Degree Distribution Generation Model. In Proceedings of the Proc. Int. Conf. Data. Eng., ICDE, 2021, pp. 564–575. [CrossRef]
- Park, H.; Kim, M. TrillionG: A Trillion-scale Synthetic Graph Generator using a Recursive Vector Model. In Proceedings of the Proc. ACM SIGMOD Int. Conf. Manage. Data, SIGMOD, 2017, pp. 913–928. [CrossRef]
- Maekawa, S.; Sasaki, Y.; Fletcher, G.; Onizuka, M. GenCAT: Generating attributed graphs with controlled relationships between classes, attributes, and topology. Inf. Syst. 2023, 115, 102195. [CrossRef]
- Wang, L.; Zhang, J.; Yang, H.; Chen, Z.; Tang, J.; Zhang, Z.; Chen, X.; Lin, Y.; Sun, H.; Song, R.; et al. User Behavior Simulation with Large Language Model-based Agents. ACM Trans. Inf. Syst. 2025, 43, 55:1–55:37. [CrossRef]
- Zhang, J.; Hou, Y.; Xie, R.; Sun, W.; McAuley, J.J.; Zhao, W.X.; Lin, L.; Wen, J. AgentCF: Collaborative Learning with Autonomous Language Agents for Recommender Systems. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW, 2024, pp. 3679–3689. [CrossRef]
- Ji, J.; Li, Y.; Liu, H.; Du, Z.; Wei, Z.; Qi, Q.; Shen, W.; Lin, Y. SRAP-Agent: Simulating and Optimizing Scarce Resource Allocation Policy with LLM-based Agent. In Proceedings of the Conf. Empir. Methods Nat. Lang. Process., Find. EMNLP, 2024, pp. 267–293. [CrossRef]
- Liu, Y.; Song, Z.; Zhang, J.; Zhang, X.; Chen, X.; Yan, R. The Stepwise Deception: Simulating the Evolution from True News to Fake News with LLM Agents. CoRR arXiv:2410.19064 2024.
- Ren, R.; Qiu, P.; Qu, Y.; Liu, J.; Zhao, X.; Wu, H.; Wen, J.; Wang, H. BASES: Large-scale Web Search User Simulation with Large Language Model based Agents. In Proceedings of the Conf. Empir. Methods Nat. Lang. Process., Find. EMNLP, 2024, pp. 902–917. [CrossRef]
- Gao, C.; Lan, X.; Lu, Z.; Mao, J.; Piao, J.; Wang, H.; Jin, D.; Li, Y. S3: Social-network Simulation System with Large Language Model-Empowered Agents. CoRR arXiv:2307.14984 2023, [2307.14984]. [CrossRef]
- Yang, Z.; Zhang, Z.; Zheng, Z.; Jiang, Y.; Gan, Z.; Wang, Z.; Ling, Z.; Chen, J.; Ma, M.; Dong, B.; et al. OASIS: Open Agent Social Interaction Simulations with One Million Agents, 2024, [arXiv:cs.CL/2411.11581].
- Park, J.S.; Popowski, L.; Cai, C.J.; Morris, M.R.; Liang, P.; Bernstein, M.S. Social Simulacra: Creating Populated Prototypes for Social Computing Systems. In Proceedings of the Proc. Annu. ACM Symp. User Interface Softw. Technol., UIST, 2022, pp. 74:1–74:18. [CrossRef]
- Rossetti, G.; Stella, M.; Cazabet, R.; Abramski, K.; Cau, E.; Citraro, S.; Failla, A.; Improta, R.; Morini, V.; Pansanella, V. Y Social: an LLM-powered Social Media Digital Twin. CoRR arXiv:2408.00818 2024, [2408.00818]. [CrossRef]
- Kingma, D.P.; Dhariwal, P. Glow: Generative Flow with Invertible 1x1 Convolutions. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2018, pp. 10236–10245.
- Shehzad, A.; Xia, F.; Abid, S.; Peng, C.; Yu, S.; Zhang, D.; Verspoor, K. Graph Transformers: A Survey. CoRR arXiv:2407.09777 2024.
- Zhang, Z.; Liu, S.; Liu, Z.; Zhong, R.; Cai, Q.; Zhao, X.; Zhang, C.; Liu, Q.; Jiang, P. Llm-powered user simulator for recommender system. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2025, Vol. 39, pp. 13339–13347.
- Dinh, L.; Sohl-Dickstein, J.; Bengio, S. Density estimation using Real NVP. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2017.
- Jin, W.; Barzilay, R.; Jaakkola, T.S. Hierarchical Generation of Molecular Graphs using Structural Motifs. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2020, Vol. 119, pp. 4839–4848.
- Gamage, A.; Chien, E.; Peng, J.; Milenkovic, O. Multi-MotifGAN (MMGAN): Motif-Targeted Graph Generation And Prediction. In Proceedings of the Proc. Int. Conf. Acoust. Speech. Signal. Process., ICASSP, 2020, pp. 4182–4186. [CrossRef]
- Wang, C.; Xue, N.; Pradhan, S. A Transition-based Algorithm for AMR Parsing. In Proceedings of the Conf. North Am. Chapter Assoc. Comput. Linguist.: Hum. Lang. Technol., NAACL-HLT, 2015, pp. 366–375. [CrossRef]
- Chen, B.; Sun, L.; Han, X. Sequence-to-Action: End-to-End Semantic Graph Generation for Semantic Parsing. In Proceedings of the Proc. Annu. Meet. Assoc. Comput Linguist., ACL, 2018, pp. 766–777. [CrossRef]
- Edunov, S.; Logothetis, D.; Wang, C.; Ching, A.; Kabiljo, M. Generating Synthetic Social Graphs with Darwini. In Proceedings of the Proc. Int. Conf. Distrib. Comput. Syst., ICDCS, 2018, pp. 567–577. [CrossRef]
- Tenorio, V.M.; Zilberstein, N.; Segarra, S.; Marques, A.G. Graph Guided Diffusion: Unified Guidance for Conditional Graph Generation. CoRR arXiv:2505.19685 2025, [2505.19685]. [CrossRef]
- Zhang, D.C.; Yang, M.; Ying, R.; Lauw, H.W. Text-Attributed Graph Representation Learning: Methods, Applications, and Challenges. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW, 2024, pp. 1298–1301. [CrossRef]
- Ramakrishnan, R.; Dral, P.O.; Rupp, M.; von Lilienfeld, O.A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 2014, 1.
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768.
- Polykovskiy, D.; Zhebrak, A.; Sanchez-Lengeling, B.; Golovanov, S.; Tatanov, O.; Belyaev, S.; Kurbanov, R.; Artamonov, A.; Aladinskiy, V.; Veselov, M.; et al. Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models. Front. Pharmacol. 2020.
- Zdrazil, B.; Felix, E.; Hunter, F.; Manners, E.J.; Blackshaw, J.; Corbett, S.; de Veij, M.; Ioannidis, H.; Lopez, D.M.; Mosquera, J.; et al. The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res. 2023, 52, D1180–D1192, [https://academic.oup.com/nar/article-pdf/52/D1/D1180/55040046/gkad1004.pdf]. [CrossRef]
- Hachmann, J.; Olivares-Amaya, R.; Atahan-Evrenk, S.; Amador-Bedolla, C.; Sánchez-Carrera, R.S.; Gold-Parker, A.; Vogt, L.; Brockway, A.M.; Aspuru-Guzik, A. The Harvard Clean Energy Project: Large-Scale Computational Screening and Design of Organic Photovoltaics on the World Community Grid. The Journal of Physical Chemistry Letters 2011, 2, 2241–2251, [. [CrossRef]
- Tran-Nguyen, V.K.; Jacquemard, C.; Rognan, D. LIT-PCBA: An Unbiased Data Set for Machine Learning and Virtual Screening. Journal of Chemical Information and Modeling 2020, 60, 4263–4273. PMID: 32282202, . [CrossRef]
- Borgwardt, K.M.; Ong, C.S.; Schönauer, S.; Vishwanathan, S.; Smola, A.J.; Kriegel, H.P. Protein function prediction via graph kernels. Bioinformatics 2005, 21, i47–i56.
- Schomburg, I.; Chang, A.; Ebeling, C.; Gremse, M.; Heldt, C.; Huhn, G.; Schomburg, D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004, 32, D431–D433.
- Golomb, S.W. Polyominoes: puzzles, patterns, problems, and packings; Vol. 16, Princeton University Press, 1996.
- Dobson, P.D.; Doig, A.J. Distinguishing enzyme structures from non-enzymes without alignments. J. Mol. Biol. 2003, 330, 771–783.
- Bureau., U.C. Longitudinal Employer-Household Dynamics.
- Li, Y.; Yu, R.; Shahabi, C.; Liu, Y. Diffusion Convolutional Recurrent Neural Network: Data-Driven Traffic Forecasting. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2018.
- Savva, M.; Chang, A.X.; Hanrahan, P.; Fisher, M.; Nießner, M. PiGraphs: learning interaction snapshots from observations. ACM Trans. Graph. 2016, 35, 139:1–139:12. [CrossRef]
- Ionescu, C.; Papava, D.; Olaru, V.; Sminchisescu, C. Human3.6M: Large Scale Datasets and Predictive Methods for 3D Human Sensing in Natural Environments. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 36, 1325–1339. [CrossRef]
- Gao, Y.; Zhao, L. Incomplete Label Multi-Task Ordinal Regression for Spatial Event Scale Forecasting. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2018, pp. 2999–3006. [CrossRef]
- Harper, F.M.; Konstan, J.A. The MovieLens Datasets: History and Context. ACM Trans. Interact. Intell. Syst. 2016, 5, 19:1–19:19. [CrossRef]
- Thompson, R.; Knyazev, B.; Ghalebi, E.; Kim, J.; Taylor, G.W. On Evaluation Metrics for Graph Generative Models. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2022.
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524.
- Liu, G.; Xu, J.; Luo, T.; Jiang, M. Graph Diffusion Transformers for Multi-Conditional Molecular Generation. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2024.
- Jiang, B.; Zhang, Z.; Lin, D.; Tang, J.; Luo, B. Semi-Supervised Learning With Graph Learning-Convolutional Networks. In Proceedings of the Proc. IEEE Comput. Soc. Conf. Comput. Vision Pattern Recognit., CVPR, 2019, pp. 11313–11320. [CrossRef]
- Morris, C.; Ritzert, M.; Fey, M.; Hamilton, W.L.; Lenssen, J.E.; Rattan, G.; Grohe, M. Weisfeiler and Leman Go Neural: Higher-Order Graph Neural Networks. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2019, pp. 4602–4609. [CrossRef]
- Wang, C.; Kantarcioglu, M. Graph Generative Models Evaluation with Masked Autoencoder. In Proceedings of the Trends and Applications in Knowledge Discovery and Data Mining, PAKDD, 2025, Vol. 15835, pp. 137–148. [CrossRef]
- Hosseini, R.; Simini, F.; Vishwanath, V.; Willett, R.; Hoffmann, H. Quality Measures for Dynamic Graph Generative Models. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2025.
- Peng, J.; Ji, J.; Lei, R.; Wei, Z.; Liu, Y.; Hong, C. GDGB: A Benchmark for Generative Dynamic Text-Attributed Graph Learning, 2025, [arXiv:cs.AI/2507.03267].
- Wang, L.; Wang, Y.; Ni, B.; Zhao, Y.; Wang, H.; Ma, Y.; Derr, T. SaVe-TAG: Semantic-aware Vicinal Risk Minimization for Long-Tailed Text-Attributed Graphs. CoRR arXiv:2410.16882 2024.
- Liu, B.; Qi, G. LLM-CG: Large language model-enhanced constraint graph for distantly supervised relation extraction. Neurocomputing 2025, 655, 131426. [CrossRef]
- Yirik, M.A.; Steinbeck, C. Chemical graph generators. PLoS Comput. Biol. 2021, 17, e1008504.
- Zhang, Z.; Xu, M.; Jamasb, A.; Chenthamarakshan, V.; Lozano, A.; Das, P.; Tang, J. Protein representation learning by geometric structure pretraining. CoRR arXiv:2203.06125 2022.
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500.
- Kong, X.; Zhang, Z.; Zhang, Z.; Jiao, R.; Ma, J.; Huang, W.; Liu, K.; Liu, Y. UniMoMo: Unified Generative Modeling of 3D Molecules for De Novo Binder Design. CoRR arXiv:2503.19300 2025.
- Kong, X.; Jia, Y.; Huang, W.; Liu, Y. Full-atom peptide design with geometric latent diffusion. Adv. neural inf. proces. syst., NeurIPS 2024, 37, 74808–74839.
- Guo, X.; Du, Y.; Tadepalli, S.; Zhao, L.; Shehu, A. Generating tertiary protein structures via interpretable graph variational autoencoders. Bioinformatics Advances 2021, 1, vbab036.
- Rahman, T.; Du, Y.; Zhao, L.; Shehu, A. Generative adversarial learning of protein tertiary structures. Molecules 2021, 26, 1209.
- Hu, Y.; Tan, Y.; Han, A.; Zheng, L.; Hong, L.; Zhou, B. Secondary structure-guided novel protein sequence generation with latent graph diffusion. In Proceedings of the IEEE Int. Conf. on Bioinformatics and Biomedicine, BIBM. IEEE, 2024, pp. 31–41.
- Ingraham, J.; Garg, V.; Barzilay, R.; Jaakkola, T. Generative models for graph-based protein design. Advances in neural information processing systems 2019, 32.
- Ahn, S.; Chen, B.; Wang, T.; Song, L. Spanning Tree-based Graph Generation for Molecules. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2022.
- Zhou, D.; Zheng, L.; Xu, J.; He, J. Misc-GAN: A Multi-scale Generative Model for Graphs. Frontiers Big Data 2019, 2, 3. [CrossRef]
- Ninniri, M.; Podda, M.; Bacciu, D. Graph Diffusion that can Insert and Delete. arXiv preprint arXiv:2506.15725 2025.
- Lim, J.; Ryu, S.; Kim, J.W.; Kim, W.Y. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminf. 2018, 10, 31.
- Assouel, R.; Ahmed, M.; Segler, M.H.S.; Saffari, A.; Bengio, Y. DEFactor: Differentiable Edge Factorization-based Probabilistic Graph Generation. CoRR arXiv:1811.09766 2018, [1811.09766].
- Diamant, N.L.; Tseng, A.M.; Chuang, K.V.; Biancalani, T.; Scalia, G. Improving Graph Generation by Restricting Graph Bandwidth. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2023, Vol. 202, pp. 7939–7959.
- Fu, X.; Gao, Y.; Wei, Y.; Sun, Q.; Peng, H.; Li, J.; Li, X. Hyperbolic Geometric Latent Diffusion Model for Graph Generation. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2024.
- US Census Bureau. Longitudinal Employer-Household Dynamics (LEHD). U.S. Census Bureau, 2019. Version: 2019.
- Martinkus, K.; Loukas, A.; Perraudin, N.; Wattenhofer, R. SPECTRE: Spectral Conditioning Helps to Overcome the Expressivity Limits of One-shot Graph Generators. In Proceedings of the Int. Conf. Machin. Learn., ICML, 17-23 Jul 2022, Vol. 162, pp. 15159–15179.
- Bradonjić, M.; Hagberg, A.; Percus, A.G. Giant component and connectivity in geographical threshold graphs. In Proceedings of the International Workshop on Algorithms and Models for the Web-Graph. Springer, 2007, pp. 209–216.
- Du, Y.; Guo, X.; Cao, H.; Ye, Y.; Zhao, L. Disentangled Spatiotemporal Graph Generative Models. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2022, pp. 6541–6549. [CrossRef]
- Bacciu, D.; Micheli, A.; Podda, M. Edge-based sequential graph generation with recurrent neural networks. Neurocomputing 2020, 416, 177–189. [CrossRef]
- Kawai, W.; Mukuta, Y.; Harada, T. GRAM: Scalable Generative Models for Graphs with Graph Attention Mechanism. CoRR arxiv:1906.01861 2019, [1906.01861].
- Fan, S.; Huang, B. Attention-Based Graph Evolution. In Proceedings of the Advances in Knowledge Discovery and Data Mining - 24th Pacific-Asia Conference, PAKDD; Lauw, H.W.; Wong, R.C.; Ntoulas, A.; Lim, E.; Ng, S.; Pan, S.J., Eds. Springer, 2020, Vol. 12084, Lecture Notes in Computer Science, pp. 436–447. [CrossRef]
- Liu, J.; Kumar, A.; Ba, J.; Kiros, J.; Swersky, K. Graph Normalizing Flows. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2019, pp. 13556–13566.
- Adamic, L.A.; Glance, N. The political blogosphere and the 2004 US election: divided they blog. In Proceedings of the Proc. of the 3rd international workshop on Link discovery, 2005, pp. 36–43.
- Wu, M.; Chen, X.; Liu, L. EDGE++: Improved Training and Sampling of EDGE. CoRR arXiv:2310.14441 2023, [2310.14441]. [CrossRef]
- Tang, J.; Zhang, J.; Yao, L.; Li, J.; Zhang, L.; Su, Z. Arnetminer: extraction and mining of academic social networks. In Proceedings of the Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., KDD, 2008, pp. 990–998.
- Xirogiannopoulos, K.; Khurana, U.; Deshpande, A. GraphGen: Exploring Interesting Graphs in Relational Data. Proc. VLDB Endow. 2015, 8, 2032–2035. [CrossRef]
- Jure, L. Snap datasets: Stanford large network dataset collection. Retrieved December 2021 from http://snap. stanford. edu/data 2014.
- Leskovec, J.; Mcauley, J. Learning to discover social circles in ego networks. Adv. neural inf. proces. syst., NeurIPS 2012, 25.
- McCallum, A.K.; Nigam, K.; Rennie, J.; Seymore, K. Automating the construction of internet portals with machine learning. Information Retrieval 2000, 3, 127–163.
- Sen, P.; Namata, G.; Bilgic, M.; Getoor, L.; Galligher, B.; Eliassi-Rad, T. Collective classification in network data. AI magazine 2008, 29, 93–93.
- Trivedi, R.; Yang, J.; Zha, H. GraphOpt: Learning Optimization Models of Graph Formation. In Proceedings of the Int. Conf. Machin. Learn., ICML, 2020, Vol. 119, pp. 9603–9613.
- Klimt, B.; Yang, Y. The enron corpus: A new dataset for email classification research. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW. Springer, 2004, pp. 217–226.
- Kunegis, J. Konect: the koblenz network collection. In Proceedings of the Proc. Int. Conf. World Wide Web, WWW, 2013, pp. 1343–1350.
- Chandrashekar, M.; Cottam, J.A. Graph Generation with a Focusing Lexicon. In Proceedings of the Proc. Int. Conf. Big Data, Big Data, 2019, pp. 4928–4931. [CrossRef]
- Babalola, K.O.; Jennings, O.B.; Urdiales, E.; DeBardelaben, J.A. Statistical Methods for Generating Synthetic Email Data Sets. In Proceedings of the Proc. Int. Conf. Big Data, Big Data, 2018, pp. 3986–3990. [CrossRef]
- Zhang, X.; Liu, Y.; Wang, J.; Hu, Z.; Chen, X.; Yan, R. SAGraph: A Large-Scale Social Graph Dataset with Comprehensive Context for Influencer Selection in Marketing. CoRR arXiv:2403.15105 2024.
- Maiorana, Z.; Morales Henry, P.; Weintraub, J. metoo Digital Media Collection - Twitter Dataset, 2020. [CrossRef]
- Mou, X.; Wei, Z.; Huang, X. Unveiling the Truth and Facilitating Change: Towards Agent-based Large-scale Social Movement Simulation. In Proceedings of the Proc. Annu. Meet. Assoc. Comput Linguist., Find.ACL, 2024, pp. 4789–4809. [CrossRef]
- Mou, X.; Qian, C.; Liu, W.; Huang, X.; Wei, Z. EcoLANG: Efficient and Effective Agent Communication Language Induction for Social Simulation. CoRR arXiv:2505.06904 2025, [2505.06904]. [CrossRef]
- Chang, R.C.; Rao, A.; Zhong, Q.; Wojcieszak, M.; Lerman, K. # RoeOverturned: Twitter dataset on the abortion rights controversy. In Proceedings of the Proc. Int. AAAI Conf. on Web and Social Media, ICWSM, 2023, Vol. 17, pp. 997–1005.
- Zhang, X.; Lin, J.; Sun, L.; Qi, W.; Yang, Y.; Chen, Y.; Lyu, H.; Mou, X.; Chen, S.; Luo, J.; et al. ElectionSim: Massive Population Election Simulation Powered by Large Language Model Driven Agents. CoRR arXiv:2410.20746 2024, [2410.20746]. [CrossRef]
- Zhang, A.; Chen, Y.; Sheng, L.; Wang, X.; Chua, T. On Generative Agents in Recommendation. In Proceedings of the Proc. Int. ACM SIGIR Conf. Res. Dev. Inf. Retr., SIGIR, 2024, pp. 1807–1817. [CrossRef]
- Hou, Y.; Li, J.; He, Z.; Yan, A.; Chen, X.; McAuley, J. Bridging Language and Items for Retrieval and Recommendation. CoRR arXiv:2403.03952 2024.
- Liu, Y.; Liu, W.; Gu, X.; Rui, Y.; He, X.; Zhang, Y. LMAgent: A Large-scale Multimodal Agents Society for Multi-user Simulation. CoRR arXiv:2412.09237 2024, [2412.09237]. [CrossRef]
- Kang, W.C.; McAuley, J. Self-attentive sequential recommendation. In Proceedings of the Proc. IEEE Int. Conf. Data Min. ICDM. IEEE, 2018, pp. 197–206.
- Huang, X.; Lian, J.; Lei, Y.; Yao, J.; Lian, D.; Xie, X. Recommender AI Agent: Integrating Large Language Models for Interactive Recommendations. CoRR arXiv:2308.16505 2023, [2308.16505]. [CrossRef]
- Cantador, I.; Brusilovsky, P.; Kuflik, T. Second workshop on information heterogeneity and fusion in recommender systems (HetRec2011). In Proceedings of the Proc. ACM Conf. Recomm. Syst., ACM RecSys, 2011, pp. 387–388.
- Ozsoy, M.G. Multilingual Prompts in LLM-Based Recommenders: Performance Across Languages. CoRR arXiv:2409.07604 2024, [2409.07604]. [CrossRef]
- Liao, J.; Li, S.; Yang, Z.; Wu, J.; Yuan, Y.; Wang, X.; He, X. LLaRA: Large Language-Recommendation Assistant. In Proceedings of the Proc. Int. ACM SIGIR Conf. Res. Dev. Inf. Retr., SIGIR, 2024, pp. 1785–1795. [CrossRef]
- Zheng, K.; Sun, Q.; Xu, C.; Yu, P.; Guo, Q. Towards a Unified Paradigm: Integrating Recommendation Systems as a New Language in Large Models. CoRR arXiv:2412.16933 2024, [2412.16933]. [CrossRef]
- Yang, Y.; Zhang, S.; Zhang, C.; Yu, J.J.Q. Origin-Destination Matrix Prediction via Hexagon-based Generated Graph. In Proceedings of the IEEE Conf. Intell. Transport Syst. Proc., ITSC, 2021, pp. 1399–1404. [CrossRef]
- Rong, C.; Ding, J.; Liu, Y.; Li, Y. A Large-scale Benchmark Dataset for Commuting Origin-destination Matrix Generation. CoRR arXiv:2407.15823 2024, [2407.15823]. [CrossRef]
- Li, M.; Chen, S.; Zhao, Y.; Zhang, Y.; Wang, Y.; Tian, Q. Dynamic Multiscale Graph Neural Networks for 3D Skeleton Based Human Motion Prediction. In Proceedings of the Proc. IEEE Comput. Soc. Conf. Comput. Vision Pattern Recognit., CVPR, 2020, pp. 211–220. [CrossRef]
- Yan, S.; Li, Z.; Xiong, Y.; Yan, H.; Lin, D. Convolutional Sequence Generation for Skeleton-Based Action Synthesis. In Proceedings of the Proc. IEEE Int. Conf. Comput. Vis., ICCV, 2019, pp. 4393–4401. [CrossRef]
- Yin, W.; Yin, H.; Kragic, D.; Björkman, M. Graph-based Normalizing Flow for Human Motion Generation and Reconstruction. In Proceedings of the IEEE Int. Conf. Robot Hum. Interact. Commun., RO-MAN, 2021, pp. 641–648. [CrossRef]
- Pareja, A.; Domeniconi, G.; Chen, J.; Ma, T.; Suzumura, T.; Kanezashi, H.; Kaler, T.; Schardl, T.B.; Leiserson, C.E. EvolveGCN: Evolving Graph Convolutional Networks for Dynamic Graphs. In Proceedings of the Proc. AAAI Conf. Artif. Intell., AAAI, 2020, pp. 5363–5370. [CrossRef]
- Yu, L.; Sun, L.; Du, B.; Lv, W. Towards better dynamic graph learning: New architecture and unified library. In Proceedings of the Proc. Adv. neural inf. proces. syst., NeurIPS, 2023, Vol. 36, pp. 67686–67700.
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; Anadkat, S.; et al. Gpt-4 technical report. CoRR arXiv:2303.08774 2023.
- Chen, X.; Wang, Y.; He, J.; Du, Y.; Hassoun, S.; Xu, X.; Liu, L. Graph Generative Pre-trained Transformer. In Proceedings of the Proc. Int. Conf. Machin. Learn., ICML, 2025.
- Shakarian, P.; Bhatnagar, A.; Aleali, A.; Shaabani, E.; Guo, R. The independent cascade and linear threshold models. In Diffusion in social networks; 2015; pp. 35–48.
- Gao, C.; Zheng, Y.; Li, N.; Li, Y.; Qin, Y.; Piao, J.; Quan, Y.; Chang, J.; Jin, D.; He, X.; et al. A survey of graph neural networks for recommender systems: Challenges, methods, and directions. ACM Trans. on Recommender Systems 2023, 1, 1–51.
- Ebrat, D.; Paradalis, E.; Rueda, L. Lusifer: LLM-based user simulated feedback environment for online recommender systems. CoRR arXiv:2405.13362 2024.
- Corecco, N.; Piatti, G.; Lanzendörfer, L.A.; Fan, F.X.; Wattenhofer, R. SUBER: An RL Environment with Simulated Human Behavior for Recommender Systems. ECAI 2024 2024, 392, 2210–2217.
- Chen, L.; Dai, Q.; Zhang, Z.; Feng, X.; Zhang, M.; Tang, P.; Chen, X.; Zhu, Y.; Dong, Z. RecUserSim: A Realistic and Diverse User Simulator for Evaluating Conversational Recommender Systems. In Proceedings of the Companion Proc. ACM Web Conf., 2025, pp. 133–142.
- Chen, Y.; Yao, Q.; Zhang, J.; Cheng, J.; Bian, Y. Hierarchical Graph Tokenization for Molecule-Language Alignment. In Proceedings of the Proc. Int. Conf. Machin. Learn., ICML, 2025.
- Zhang, Y.; Ye, G.; Yuan, C.; Han, B.; Huang, L.K.; Yao, J.; Liu, W.; Rong, Y. Atomas: Hierarchical alignment on molecule-text for unified molecule understanding and generation. CoRR arXiv:2404.16880 2024.
- Wang, Z.; Chen, Y.; Ma, P.; Yu, Z.; Wang, J.; Liu, Y.; Ye, X.; Sakurai, T.; Zeng, X. Image-based generation for molecule design with SketchMol. Nature Machine Intelligence 2025, 7, 244–255.
- Zhou, G.; Gao, Z.; Ding, Q.; Zheng, H.; Xu, H.; Wei, Z.; Zhang, L.; Ke, G. Uni-Mol: A Universal 3D Molecular Representation Learning Framework. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2023.
- Liu, S.; Nie, W.; Wang, C.; Lu, J.; Qiao, Z.; Liu, L.; Tang, J.; Xiao, C.; Anandkumar, A. Multi-modal molecule structure-text model for text-based retrieval and editing. Nature Machine Intelligence 2023, 5, 1447–1457.
- O’Bray, L.; Horn, M.; Rieck, B.; Borgwardt, K.M. Evaluation Metrics for Graph Generative Models: Problems, Pitfalls, and Practical Solutions. In Proceedings of the Int. Conf. Learn. Represent., ICLR, 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).