1. Introduction
The biological mechanisms underlying cellular aging are a central focus of gerontological research. Human mesenchymal stromal/stem cells (MSCs), with their capacity for self-renewal and multilineage differentiation, exhibit profound functional alterations with advancing age. These age-related changes, including diminished proliferative rates, impaired differentiation potential, and the acquisition of a senescence-associated secretory phenotype (SASP), render MSCs a powerful paradigm for dissecting the molecular hallmarks of cellular senescence.
A critical, yet often underexplored, aspect of cellular aging is the remodeling of the nuclear envelope. The LINC (Linker of Nucleoskeleton and Cytoskeleton) complex, a conserved molecular bridge spanning the nuclear envelope, is fundamental to nuclear structural integrity, chromatin organization, and mechanosensitive signaling. Within this complex, the Sad1 and UNC84 domain-containing protein 2 (SUN2), a transmembrane protein of the inner nuclear membrane encoded by SUN2, plays a pivotal role in anchoring the nuclear lamina to the cytoskeleton. While previous studies have established a global landscape of age-related transcriptional changes, the specific role of LINC complex components, such as SUN2, as biomarkers of senescence remains to be systematically validated.
Therefore, the present study was designed to investigate the relationship between SUN2 expression and donor age in human MSCs. By analyzing a comprehensive MSC transcriptomic dataset, we aim to quantify the correlation between SUN2 mRNA levels and chronological age. This analysis will critically assess the potential of SUN2 to serve as a reliable and quantifiable biomarker for the aging state of MSCs, thereby contributing to the identification of novel targets for interventions aimed at mitigating age-associated stem cell dysfunction.
2. Materials and Methods
2.1. Data Source
Gene expression data were obtained from the Stemformatics database (Dataset: Alves et al., 2012). The dataset comprises microarray expression profiles of MSCs isolated from donors of varying ages. The study by Alves et al. characterized donor age-related gene signatures in MSCs derived from bone marrow aspirates.
2.2. Gene Identification: Ensembl Gene Nomenclature
The Ensembl database provides stable gene identifiers (ENSG IDs) that are independent of gene symbol changes and facilitate cross-species genomic comparisons. Each ENSG ID uniquely identifies a specific genomic locus and its associated transcripts. ENSG00000100242 corresponds to the human SUN2 gene located on chromosome 22q13.33. Use of Ensembl IDs ensures consistent gene identification across diverse datasets and bioinformatics platforms.
2.3. Statistical Analysis
A rigorous quantitative analysis was undertaken to investigate the potential of SUN2 (ENSG00000 100242) as an age-associated biomarker. Initially, the raw mRNA expression values for the SUN2 transcript were isolated from the compiled MSC dataset. A preliminary descriptive statistical analysis was executed to determine the central tendency (mean), dispersion (standard deviation), and range (minimum, maximum) of the expression data across all samples. To formally test the hypothesis of a linear relationship with age, a parametric assessment was performed using Pearson’s correlation coefficient (r), providing a measure of both the strength and direction of the association. To further elucidate this relationship and construct a predictive model, a simple linear regression was fitted, with SUN2 expression as the dependent variable and donor age as the independent variable. This regression model facilitated the visualization of the expression trajectory and the estimation of the rate of expression change per unit of age. Statistical significance for all analyses was established a priori at a 95% confidence level ().
3. Results
3.1. Expression Statistics
Analysis of SUN2 expression in MSCs yielded a mean expression level of
(
), with values ranging from
to
(
Table 1). These values represent normalized log-transformed expression data.
3.2. Correlation with Age
Pearson correlation analysis revealed a significant negative correlation between SUN2 expression and donor age (
,
). This finding indicates that SUN2 expression consistently declines with advancing chronological age in MSCs (
Figure 1).
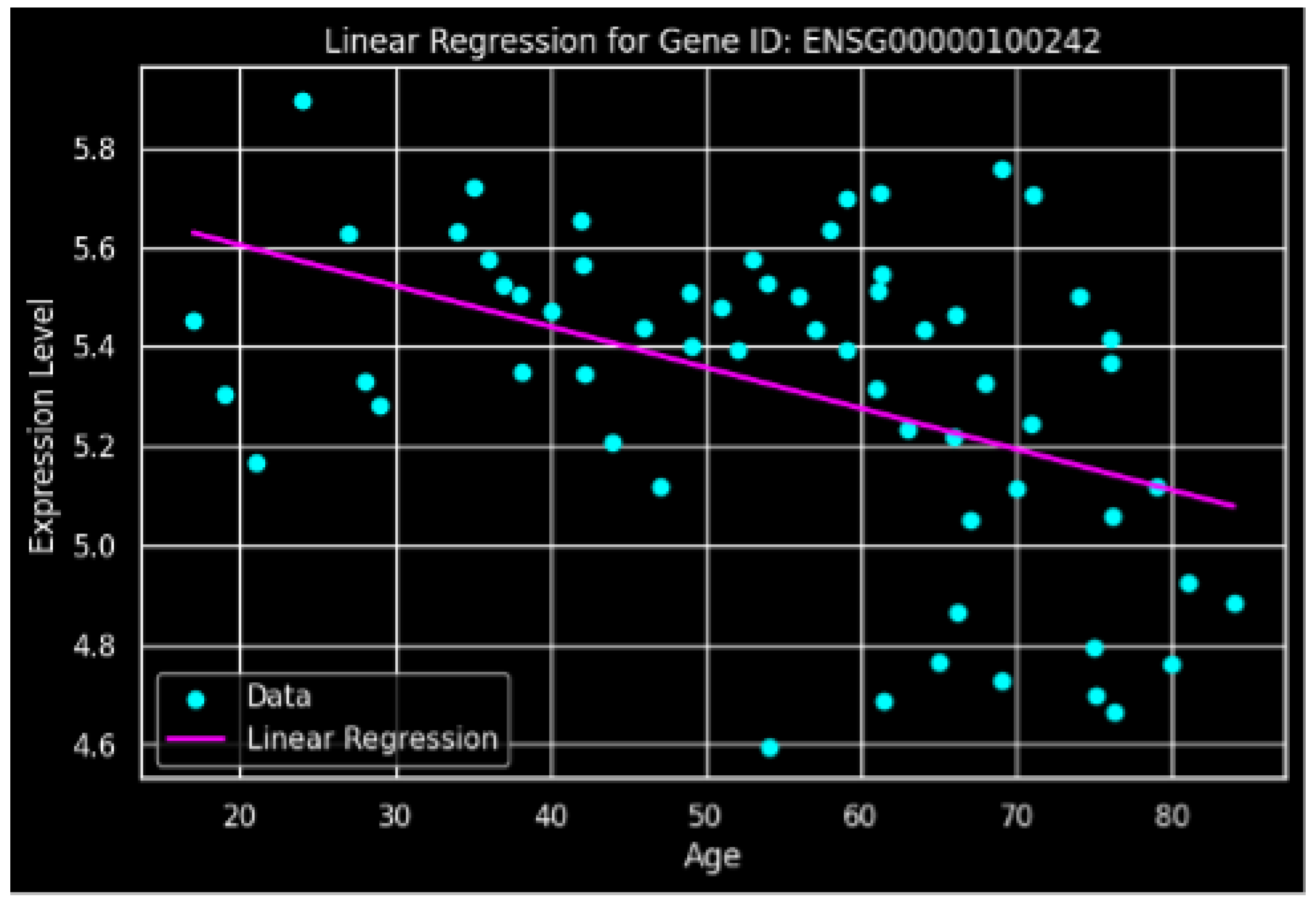
3.3. Linear Regression Analysis
Linear regression modeling of the age-expression relationship yielded an intercept of
and a slope coefficient of
(
Figure 2). The negative slope confirms the inverse relationship, indicating an approximate decline of
expression units per year of chronological age. The regression model supports the statistical significance of the age-dependent decline.
Table 2.
Linear Regression Parameters.
Table 2.
Linear Regression Parameters.
| Gene ID |
Intercept |
Slope |
Correlation (r) |
p-value |
| ENSG00000100242 |
5.769 |
-0.0082 |
-0.451 |
0.00027 |
4. Discussion
This study demonstrates a significant age-related decline in SUN2 expression in human MSCs. The negative correlation (, ) indicates that approximately 20% of the variance in SUN2 expression can be attributed to chronological age. The linear regression slope of suggests a gradual but consistent downregulation over the lifespan.
SUN2 is an essential component of the LINC complex, mediating connections between the nucleoskeleton and cytoskeleton. This structural framework is critical for nuclear positioning, mechanical signal transduction, and chromatin organization. Studies by Meinke et al. (2014) demonstrated that SUN2 variants associated with muscular dystrophy disrupt nuclear-cytoskeletal connections and impair myonuclear organization, highlighting the functional importance of SUN2 in maintaining cellular architecture.
The observed age-related decline in SUN2 expression may contribute to multiple aspects of cellular aging. Reduced SUN2 levels could compromise nuclear integrity, impair mechanotransduction pathways, and alter chromatin accessibility, collectively affecting gene expression regulation and cellular function. Age-related gene expression changes in peripheral blood monocytes have been linked to osteoporosis (Li et al., 2019), and dysregulated gene expression profiles are implicated in Alzheimer’s disease (İzgi, 2017) and Parkinson’s disease pathogenesis (Smajic, 2022). The progressive downregulation of SUN2 in MSCs may reflect broader mechanisms of cellular senescence, including epigenetic modifications, altered transcriptional regulation, or compensatory responses to accumulated cellular damage. Furthermore, age-related changes in articular cartilage mechanopathophysiology involve complex gene expression alterations (Houtman et al., 2021), suggesting that SUN2 downregulation may contribute to musculoskeletal aging phenotypes.
The consistent negative correlation and statistical significance position SUN2 as a candidate biomarker for biological aging in MSCs. Validation of this finding in independent cohorts and functional studies elucidating the mechanistic consequences of SUN2 downregulation are warranted. Future investigations should examine whether interventions that maintain or restore SUN2 expression can ameliorate age-related cellular dysfunction.
Limitations
This study utilized cross-sectional data from a single MSC dataset. Longitudinal studies tracking SUN2 expression changes within individual donors over time would strengthen causal inferences. Additionally, protein-level validation of SUN2 expression and functional assays examining LINC complex integrity in aged MSCs are needed to confirm the biological significance of the observed transcriptional changes.
5. Conclusion
SUN2 expression demonstrates a statistically significant, age-associated decrease in human MSCs. The observed negative correlation with chronological age, coupled with SUN2’s established function in maintaining nuclear-cytoskeletal integrity and modulating gene expression, posits it as a viable molecular indicator of cellular senescence. These results advance the mechanistic understanding of MSC aging at the molecular level and may guide the development of targeted interventions aimed at mitigating age-related decline in regenerative potential.
References
- Alves H, van Ginkel J, Groen N, Hulsman M, Mentink A, Reinders M, van Blitterswijk C, de Boer J. A mesenchymal stromal cell gene signature for donor age. PLoS One, 2012; 7, e42908. [CrossRef] [PubMed]
- İzgi, H. (2017). Meta analysis of alzheimer’s disease at the gene expression level [M.S. - Master of Science]. Middle East Technical University.
- Li L, Wang XQ, Liu XT, Guo R, Zhang RD. Integrative analysis reveals key mRNAs and lncRNAs in monocytes of osteoporotic patients. Math Biosci Eng 2019, 16, 5947–5971. [CrossRef] [PubMed]
- Houtman, E. , Tuerlings, M., Riechelman, J., Suchiman, E. (H. E. D.), van der Wal, R., Nelissen, R. G. H. H., Mei, H., Ramos, Y. F. M., Coutinho de Almeida, R., & Meulenbelt, I. (2021). Elucidating mechano-pathology of osteoarthritis: transcriptome-wide differences in mechanically stressed aged human cartilage explants. Arthritis Research & Therapy, 23, 215. [CrossRef]
- Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, Shackleton S. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet 2014, 10, e1004605. [CrossRef] [PubMed]
- Smajic, S. (2022). The role of neuromelanin in dopaminergic neuron demise and inflammation in idiopathic Parkinson’s disease [Doctoral dissertation, University of Luxembourg]. ORBilu. https://orbilu.uni.lu/handle/10993/52611.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).