Submitted:
06 November 2025
Posted:
07 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- We propose PsoSubstanceRank, a novel computational framework that deeply integrates the generative power of Large Language Models with comprehensive multi-source biomedical Knowledge Graphs for precise natural substance recommendation in psoriasis.
- We develop a sophisticated mechanistic path reasoning and scoring module within the knowledge graph, enabling multi-dimensional validation, detailed mechanistic explanation, and robust prioritization of natural substance-gene interactions.
- We significantly enhance the interpretability and reliability of natural substance recommendations for psoriasis, providing high-confidence hypotheses with clear mechanistic pathways, thereby reducing the subsequent experimental validation workload.
2. Related Work
2.1. Large Language Models for Biomedical Hypothesis Generation and Drug Discovery
2.2. Knowledge Graphs and LLM-KG Fusion for Mechanistic Biomedical Reasoning
3. Method
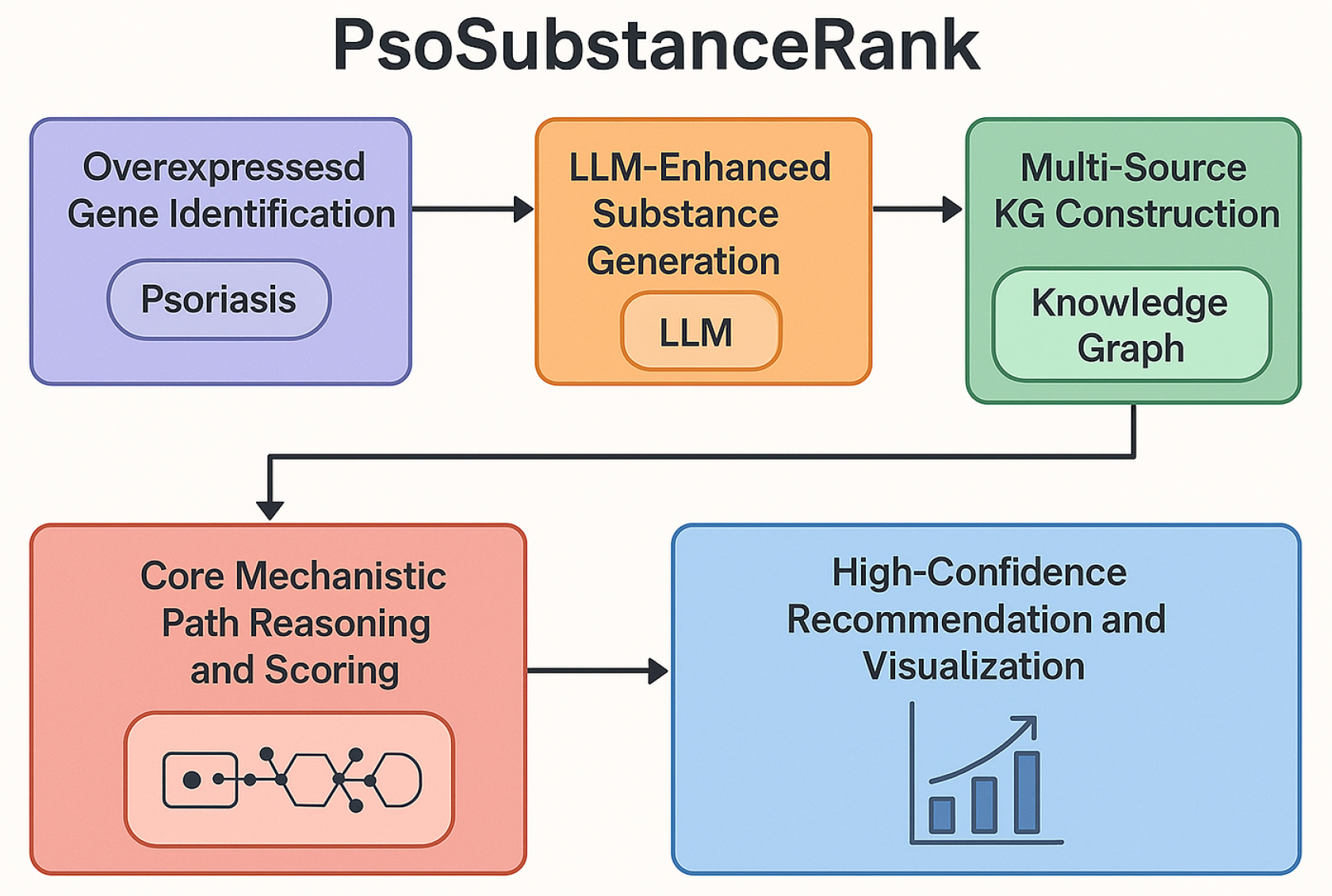
3.1. Psoriasis Overexpressed Gene Identification Module
- Data Acquisition: We utilize the publicly available gene expression microarray dataset GSE30999 from the Gene Expression Omnibus (GEO) database. This dataset comprises 170 skin biopsy samples, including both lesional and non-lesional skin, collected from 85 individuals with moderate-to-severe psoriasis. The paired nature of lesional and non-lesional samples from the same individuals allows for robust intra-subject comparisons, minimizing inter-individual variability.
- Data Preprocessing: Raw microarray data undergoes a rigorous sequence of quality control (QC) procedures. This includes initial assessment of raw intensity distributions, detection and removal of outlier samples based on array hybridization quality metrics, and evaluation of data integrity using standard bioinformatics tools. Subsequently, data normalization is performed using established methods, such as the Robust Multi-array Average (RMA) algorithm for Affymetrix arrays. This process involves background correction, probe-level summarization, and quantile normalization to ensure comparability of gene expression levels across all samples and to mitigate technical variations.
- Differential Expression Analysis: A comprehensive differential expression analysis is conducted to compare gene expression profiles between psoriatic lesional skin and non-lesional control skin. Statistical methods, specifically the empirical Bayes method implemented in the limma R package, are employed. This approach is particularly effective for microarray data with relatively small sample sizes, borrowing information across genes to improve the precision of variance estimates. To identify genes with significantly altered expression, we apply stringent statistical criteria. We define overexpressed genes as those exhibiting a statistically significant upregulation with an adjusted p-value < 0.05 (after Benjamini-Hochberg correction for multiple testing) and a fold change > 1.5, indicating a substantial increase in expression in lesional skin compared to non-lesional skin.
- Output: The module yields a refined list of core overexpressed genes in psoriasis. This list is typically prioritized based on statistical significance (smallest adjusted p-value) and magnitude of fold change, identifying the most prominent therapeutic targets for subsequent exploration.
3.2. LLM-Enhanced Natural Substance Generation Module
- Input: The primary input to this module is the refined list of psoriasis overexpressed genes derived from the Psoriasis Overexpressed Gene Identification Module. Each gene identifier (e.g., official gene symbol) is processed individually.
- Prompt Engineering: To guide the LLMs toward generating biologically plausible and mechanistically relevant hypotheses, we design sophisticated prompt strategies. These prompts are meticulously engineered to incorporate explicit constraints regarding desired mechanistic actions and contextual information.
- LLM Platforms: We utilize state-of-the-art Large Language Models to ensure broad knowledge coverage, diverse perspectives, and robust generative capabilities. Specifically, we employ OpenAI ChatGPT (GPT-4 model) and Google Gemini Advanced. By using multiple LLM platforms, we aim to cross-validate suggestions and harness the unique strengths and knowledge bases of each model.
- Output: For each overexpressed gene, the LLMs generate a set of candidate natural substances. Each candidate is accompanied by a preliminary description of its potential mechanism of action relevant to the target gene or the broader disease context of psoriasis. This output forms the initial pool of substance-gene hypotheses to be rigorously validated.
3.3. Multi-Source Biomedical Knowledge Graph Construction Module
-
Data Sources: The KG integrates information from a wide array of established biomedical databases and literature repositories, chosen for their complementary coverage of biological entities and relationships:
- Gene-Disease/Pathway Data: Resources like KEGG, Reactome, and Gene Ontology (GO) provide curated information on metabolic and signaling pathways, disease associations, and gene functions, crucial for understanding biological context.
- Substance-Gene/Protein Interactions: Databases such as ChEMBL, DrugBank, STITCH, and BIND offer extensive data on small molecule-protein interactions, drug targets, and chemical-protein associations, forming the basis for direct mechanistic links.
- Substance-Disease Associations: The Comparative Toxicogenomics Database (CTD) is integrated to provide insights into chemical-disease relationships, including therapeutic, diagnostic, and toxic effects.
- Natural Substance Information: PubChem serves as a primary source for chemical structures and properties of natural substances, while the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) offers ethnomedicinal context and pharmacokinetic data for traditional natural products.
- Literature Text Mining: Semantic relations between genes, substances, pathways, and diseases are extracted from a vast corpus of PubMed abstracts and full-text articles using advanced natural language processing (NLP) techniques. This enriches the KG with novel and emerging associations not yet curated in structured databases.
- KG Structure and Integration: Heterogeneous data from these diverse sources are integrated into a unified graph structure. Nodes in the KG represent various biomedical entities, including genes, proteins, natural substances, diseases (e.g., Psoriasis), biological pathways, and molecular functions. Edges represent the diverse relationships between these entities (e.g., "regulates," "interacts with," "participates in," "associated with," "inhibits," "activates"). Automated methods, such as named entity recognition (NER) and relation extraction (RE) algorithms, are employed for data parsing, entity linking, and relation identification from text, complemented by semi-automated curation to ensure high data quality and consistency across sources.
- Storage and Querying: The constructed KG is stored in a dedicated graph database system (e.g., Neo4j). This choice facilitates efficient storage, retrieval, and complex graph traversal queries, which are essential for mechanistic path reasoning and evidence aggregation. Graph databases inherently support the representation of complex, interconnected data and offer optimized query performance for relationship-centric data models.
- Output: A rich and comprehensive knowledge graph encompassing a vast network of biomedical entities and their relationships, serving as the factual backbone for PsoSubstanceRank.
3.4. Mechanistic Path Reasoning and Scoring Module (PsoSubstanceRank Core)
- Direct Interaction Path Validation: We search for direct known interactions between substance S and gene G in the KG. For an overexpressed gene, we specifically prioritize inhibitory or down-regulatory interactions, as these are desired for therapeutic modulation. The direct interaction score, , quantifies the strength and nature of these immediate connections:where is the set of direct edges (interactions) between substance S and gene G in the KG. The term reflects the confidence, experimental evidence level, and the specific nature of the interaction edge e (e.g., a higher weight for a experimentally validated inhibitory interaction compared to a computationally predicted activating interaction).
- Indirect Pathway Modulation Validation: This component assesses whether substance S can indirectly influence the expression or activity of target gene G through known biological pathways (e.g., inflammatory or immune pathways relevant to psoriasis) or if S is associated with pathways in which G participates. The indirect pathway score, , is computed by analyzing paths of length greater than one connecting S and G via intermediate entities such as proteins, other genes, or biological pathways. We consider the shortest path lengths and the biological relevance of intermediate nodes:where is the set of relevant indirect paths between S and G. is the number of edges in path p, penalizing longer, less direct paths. quantifies the biological significance of the path’s intermediate nodes and edges, assigning higher values to paths that traverse entities highly implicated in psoriasis pathogenesis or known to be modulated by S.
- Literature Evidence Quantification: We quantify the textual evidence supporting the association between S and G (or their related entities) by leveraging co-occurrence counts within the literature corpus integrated into the KG. This score provides empirical support from published research. The literature evidence score, , is calculated as:where represents the frequency of S and G (or their synonyms) co-appearing within a defined textual window (e.g., sentences or abstracts) within the mined literature corpus. The logarithmic transformation smooths the score and mitigates the impact of extremely high co-occurrence counts.
- Negative Evidence and Toxicity Screening: We incorporate a critical penalty for substances known to have significant toxicity, adverse side effects, or negative associations with the target gene or the disease itself. This safety score, , is derived from structured databases like CTD and through comprehensive literature mining for negative reports:where is a quantified measure of substance S’s inherent toxicity (e.g., based on dosage, reported side effects, or regulatory warnings), and represents the strength or frequency of adverse connections between S and psoriasis. and are weighting coefficients reflecting the relative importance of general toxicity versus disease-specific negative associations.
- Comprehensive Priority Scoring: The final priority score for each "natural substance-overexpressed gene" pair is computed as a weighted sum of these individual scores. This aggregate score provides a holistic assessment, balancing therapeutic potential with safety considerations:where are empirically determined weighting coefficients. These coefficients are optimized to reflect the relative importance of direct interactions, indirect pathways, literature support, and safety considerations, respectively, and can be tuned using validation data or expert domain knowledge.
3.5. High-Confidence Natural Substance Recommendation and Visualization Module
- Recommendation Ranking: All "natural substance-overexpressed gene" pairs, along with their calculated comprehensive priority scores , are sorted in descending order. This provides an ordered list from the most promising to the least promising candidates.
- High-Confidence Filtering: A dynamic or fixed threshold is applied to the ranked list to filter for only the top-tier, high-confidence recommendations. This threshold can be determined empirically based on the distribution of scores, by selecting the top X% of recommendations, or by setting a minimum score cutoff. This step ensures that only the most promising and robust candidates are presented for further consideration, minimizing noise.
- Mechanistic Explanation and Visualization: For each selected high-confidence recommendation, detailed mechanistic explanation paths are extracted from the knowledge graph. These paths illustrate the precise molecular interactions, intermediate entities, and biological pathways through which the natural substance is hypothesized to modulate the target overexpressed gene or the broader disease phenotype. These extracted paths can be visualized as subgraphs, enhancing the interpretability and verifiability of the recommendations by providing a clear rationale for each suggestion.
- Output: The module delivers a prioritized list of high-confidence natural substance recommendations specifically tailored for psoriasis overexpressed genes. Each recommendation is accompanied by a comprehensive, interpretable, and verifiable mechanistic explanation derived from the integrated knowledge graph, providing actionable insights for researchers and clinicians.
4. Experiments
4.1. Experimental Setup
4.2. Baseline Methods
- LLM Direct Recommendation (LLM-Direct): This baseline mirrors the approach of existing preliminary studies, such as that by Saed Sayad et al. [2]. It involves directly querying a Large Language Model (e.g., GPT-4) to recommend natural substances for identified overexpressed genes without any subsequent structured validation or sophisticated filtering. Recommendations are primarily based on the linguistic patterns and general knowledge aggregated during the LLM’s training.
- LLM Recommendation with Simple Literature Keyword Filtering (LLM-Filter): This improved baseline builds upon LLM-Direct by adding a rudimentary post-processing step. After receiving initial recommendations from the LLM, a simple keyword-based literature search is performed (e.g., on PubMed) to filter out recommendations that lack any direct co-occurrence of the natural substance and the target gene (or related terms) in published literature. This method provides a basic level of evidence validation but lacks deep mechanistic reasoning or multi-dimensional scoring.
4.3. Evaluation Metrics
- High-Confidence Recommendation Ratio (%): This metric quantifies the percentage of recommended natural substance-gene pairs that are validated to be reliable. Reliability is determined through rigorous multi-source knowledge graph validation, strong mechanistic support, robust literature evidence, and high expert consensus. A higher ratio indicates a greater proportion of trustworthy hypotheses for subsequent experimental work.
- Average Mechanistic Interpretability (1-5 Score): This score, assigned by domain experts (e.g., dermatologists, pharmacologists), evaluates the clarity, specificity, and biological plausibility of the proposed mechanism of action for each recommended substance-gene interaction. A score of 5 represents a highly clear, detailed, and biologically reasonable mechanistic pathway, while 1 indicates a vague or unsubstantiated mechanism.
- Novelty Score (1-5 Score): This metric assesses the extent to which the recommended substances represent novel candidates that are less commonly discussed in existing literature, yet possess strong mechanistic support from our knowledge graph. A score of 5 indicates a highly novel and promising candidate, while 1 suggests a well-known or trivial recommendation. This metric helps identify new therapeutic avenues.
- Validation Workload (Relative Value): This qualitative metric estimates the relative effort and resources required for subsequent experimental validation (e.g., in vitro or in vivo studies) of the recommended substances. Recommendations with higher confidence, clearer mechanisms, and better safety profiles are expected to lead to a lower experimental failure rate, thus reducing the overall validation workload.
4.4. Results and Discussion
4.4.1. Effectiveness of PsoSubstanceRank
4.4.2. Human Evaluation Results
4.5. Analysis of LLM-KG Synergy
4.6. Impact of Mechanistic Path Reasoning
4.7. Case Study: Top Recommended Substances for a Key Psoriasis Gene
4.8. Robustness and Parameter Sensitivity
5. Conclusions
References
- Xu, J.; Ju, D.; Li, M.; Boureau, Y.L.; Weston, J.; Dinan, E. Bot-Adversarial Dialogue for Safe Conversational Agents. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 2950–2968. [CrossRef]
- Labrak, Y.; Bazoge, A.; Morin, E.; Gourraud, P.A.; Rouvier, M.; Dufour, R. BioMistral: A Collection of Open-Source Pretrained Large Language Models for Medical Domains. In Proceedings of the Findings of the Association for Computational Linguistics: ACL 2024. Association for Computational Linguistics, 2024, pp. 5848–5864. [CrossRef]
- Zhou, Y.; Li, X.; Wang, Q.; Shen, J. Visual In-Context Learning for Large Vision-Language Models. In Proceedings of the Findings of the Association for Computational Linguistics, ACL 2024, Bangkok, Thailand and virtual meeting, August 11-16, 2024. Association for Computational Linguistics, 2024, pp. 15890–15902.
- Zhou, Y.; Shen, J.; Cheng, Y. Weak to strong generalization for large language models with multi-capabilities. In Proceedings of the The Thirteenth International Conference on Learning Representations, 2025.
- Zhou, Y.; Song, L.; Shen, J. Improving Medical Large Vision-Language Models with Abnormal-Aware Feedback. arXiv preprint arXiv:2501.01377, arXiv:2501.01377 2025.
- Long, Q.; Wu, Y.; Wang, W.; Pan, S.J. Does in-context learning really learn? rethinking how large language models respond and solve tasks via in-context learning. arXiv preprint arXiv:2404.07546, arXiv:2404.07546 2024.
- Huang, X.; Wang, Z.; Liu, X.; Tian, Y.; Leng, Q. Towards Interpretable and Consistent Multi-Step Mathematical Reasoning in Large Language Models. Available at SSRN 5680042 2025. [Google Scholar]
- Long, Q.; Chen, J.; Liu, Z.; Chen, N.F.; Wang, W.; Pan, S.J. Reinforcing Compositional Retrieval: Retrieving Step-by-Step for Composing Informative Contexts. arXiv preprint arXiv:2504.11420, arXiv:2504.11420 2025.
- Huang, J.; Chang, K.C.C. Towards Reasoning in Large Language Models: A Survey. In Proceedings of the Findings of the Association for Computational Linguistics: ACL 2023. Association for Computational Linguistics; 2023; pp. 1049–1065. [Google Scholar] [CrossRef]
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Safety-Critical Multi-Agent MCTS for Mixed Traffic Coordination at Unsignalized Intersections. IEEE Transactions on Intelligent Transportation Systems, 2025; 1–15. [Google Scholar] [CrossRef]
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Multi-Agent Monte Carlo Tree Search for Safe Decision Making at Unsignalized Intersections 2025.
- Tian, Z.; Lin, Z.; Zhao, D.; Zhao, W.; Flynn, D.; Ansari, S.; Wei, C. Evaluating scenario-based decision-making for interactive autonomous driving using rational criteria: A survey. arXiv preprint arXiv:2501.01886, arXiv:2501.01886 2025.
- Long, Q.; Wang, M.; Li, L. Generative Imagination Elevates Machine Translation. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2021, pp. 5738–5748.
- Wang, Q.; Li, M.; Wang, X.; Parulian, N.; Han, G.; Ma, J.; Tu, J.; Lin, Y.; Zhang, R.H.; Liu, W.; et al. COVID-19 Literature Knowledge Graph Construction and Drug Repurposing Report Generation. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies: Demonstrations. Association for Computational Linguistics, 2021, pp. 66–77. [CrossRef]
- Zhang, N.; Chen, M.; Bi, Z.; Liang, X.; Li, L.; Shang, X.; Yin, K.; Tan, C.; Xu, J.; Huang, F.; et al. CBLUE: A Chinese Biomedical Language Understanding Evaluation Benchmark. In Proceedings of the Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2022, pp. 7888–7915. [CrossRef]
- Liu, F.; Shareghi, E.; Meng, Z.; Basaldella, M.; Collier, N. Self-Alignment Pretraining for Biomedical Entity Representations. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 4228–4238. [CrossRef]
- Qi, T.; Wu, F.; Wu, C.; Yang, P.; Yu, Y.; Xie, X.; Huang, Y. HieRec: Hierarchical User Interest Modeling for Personalized News Recommendation. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 5446–5456. [CrossRef]
- Huang, X.; Zhao, C.; Li, X.; Feng, C.; Zhang, W. GAM-CoT Transformer: Hierarchical Attention Networks for Anomaly Detection in Blockchain Transactions. INNO-PRESS: Journal of Emerging Applied AI 2025, 1. [Google Scholar] [CrossRef]
- Barbaresi.; Adrien. Trafilatura: A Web Scraping Library and Command-Line Tool for Text Discovery and Extraction. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing: System Demonstrations. Association for Computational Linguistics, 2021, pp. 122–131. [CrossRef]
- Tian, Y.; Xu, S.; Cao, Y.; Wang, Z.; Wei, Z. An Empirical Comparison of Machine Learning and Deep Learning Models for Automated Fake News Detection. Mathematics 2025, 13. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, Y.; Song, Y.; Wan, X. Cross-modal Memory Networks for Radiology Report Generation. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 5904–5914. [CrossRef]
- Sun, Y.; Shi, Q.; Qi, L.; Zhang, Y. JointLK: Joint Reasoning with Language Models and Knowledge Graphs for Commonsense Question Answering. In Proceedings of the Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2022, pp. 5049–5060. [CrossRef]
- Lin, B.Y.; Sun, H.; Dhingra, B.; Zaheer, M.; Ren, X.; Cohen, W. Differentiable Open-Ended Commonsense Reasoning. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 4611–4625. [CrossRef]
- Wu, Y.; Zhan, P.; Zhang, Y.; Wang, L.; Xu, Z. Multimodal Fusion with Co-Attention Networks for Fake News Detection. In Proceedings of the Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021. Association for Computational Linguistics; 2021; pp. 2560–2569. [Google Scholar] [CrossRef]
- Tran Phu, M.; Nguyen, T.H. Graph Convolutional Networks for Event Causality Identification with Rich Document-level Structures. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 3480–3490. [CrossRef]
- Tian, Y.; Chen, G.; Song, Y. Aspect-based Sentiment Analysis with Type-aware Graph Convolutional Networks and Layer Ensemble. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 2910–2922. [CrossRef]
- Trivedi, H.; Balasubramanian, N.; Khot, T.; Sabharwal, A. Interleaving Retrieval with Chain-of-Thought Reasoning for Knowledge-Intensive Multi-Step Questions. In Proceedings of the Proceedings of the 61st Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2023, pp. 10014–10037. [CrossRef]
- Wang, P.; Zhu, Z.; Liang, D. Virtual Back-EMF Injection Based Online Parameter Identification of Surface-Mounted PMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2024. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Z.; Feng, Z. Virtual Back-EMF Injection-based Online Full-Parameter Estimation of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Transportation Electrification 2025. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Z.; Liang, D. A Novel Virtual Flux Linkage Injection Method for Online Monitoring PM Flux Linkage and Temperature of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2025. [Google Scholar] [CrossRef]
- Liu, J.; Liu, A.; Lu, X.; Welleck, S.; West, P.; Le Bras, R.; Choi, Y.; Hajishirzi, H. Generated Knowledge Prompting for Commonsense Reasoning. In Proceedings of the Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2022, pp. 3154–3169. [CrossRef]
- Li, Z.; Jin, X.; Guan, S.; Li, W.; Guo, J.; Wang, Y.; Cheng, X. Search from History and Reason for Future: Two-stage Reasoning on Temporal Knowledge Graphs. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 4732–4743. [CrossRef]
| Evaluation Metric | LLM-Direct | LLM-Filter | PsoSubstanceRank (Ours) |
|---|---|---|---|
| High-Confidence Recommendation Ratio (%) | 25% | 40% | 78% |
| (Validated by KG and expert assessment) | |||
| Average Mechanistic Interpretability (1-5) | 2.8 | 3.2 | 4.5 |
| (Expert-assessed clarity and biological rationale) | |||
| Novelty Score (1-5) | 3.5 | 2.5 | 4.0 |
| (Identification of less-explored, mechanistically sound candidates) | |||
| Validation Workload (Relative Value) | High | Medium-High | Low |
| (Estimated effort for subsequent experimental validation) |
| Recommendation (Substance-Gene Pair) | Final Score | ||||
|---|---|---|---|---|---|
| Substance A - Gene X | 0.8 | 0.6 | 0.7 | 0.1 | 2.0 |
| (Strong direct inhibition, moderate indirect pathway modulation) | |||||
| Substance B - Gene Y | 0.2 | 0.9 | 0.5 | 0.05 | 1.55 |
| (Primary indirect modulation via anti-inflammatory pathway) | |||||
| Substance C - Gene Z | 0.5 | 0.3 | 0.9 | 0.2 | 1.5 |
| (Moderate direct interaction, strong literature co-occurrence) |
| Recommended Substance | Final Score | Key Mechanistic Path Identified by KG | Novelty Assessment |
|---|---|---|---|
| Curcumin | 2.15 | Direct inhibition of NF-B activation, which is upstream of IL-17A transcription. | Well-studied |
| (From Curcuma longa) | Indirect modulation of Th17 cell differentiation. | ||
| Resveratrol | 1.98 | Activates Sirtuin 1 (SIRT1), downregulating inflammatory pathways including IL-17A. | Moderate |
| (From Vitis vinifera) | Modulates immune cell function affecting IL-17A production. | ||
| Berberine | 1.82 | Inhibits STAT3 phosphorylation, a key transcription factor for IL-17A expression. | Emerging |
| (From Berberis aristata) | Modulates gut microbiome influencing systemic inflammation. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




