Submitted:
05 November 2025
Posted:
06 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Morphological Response of Stem Cells on Glycated Collagen
2.2. RAGE Expression in ADMSCs During Collagen Adhesion
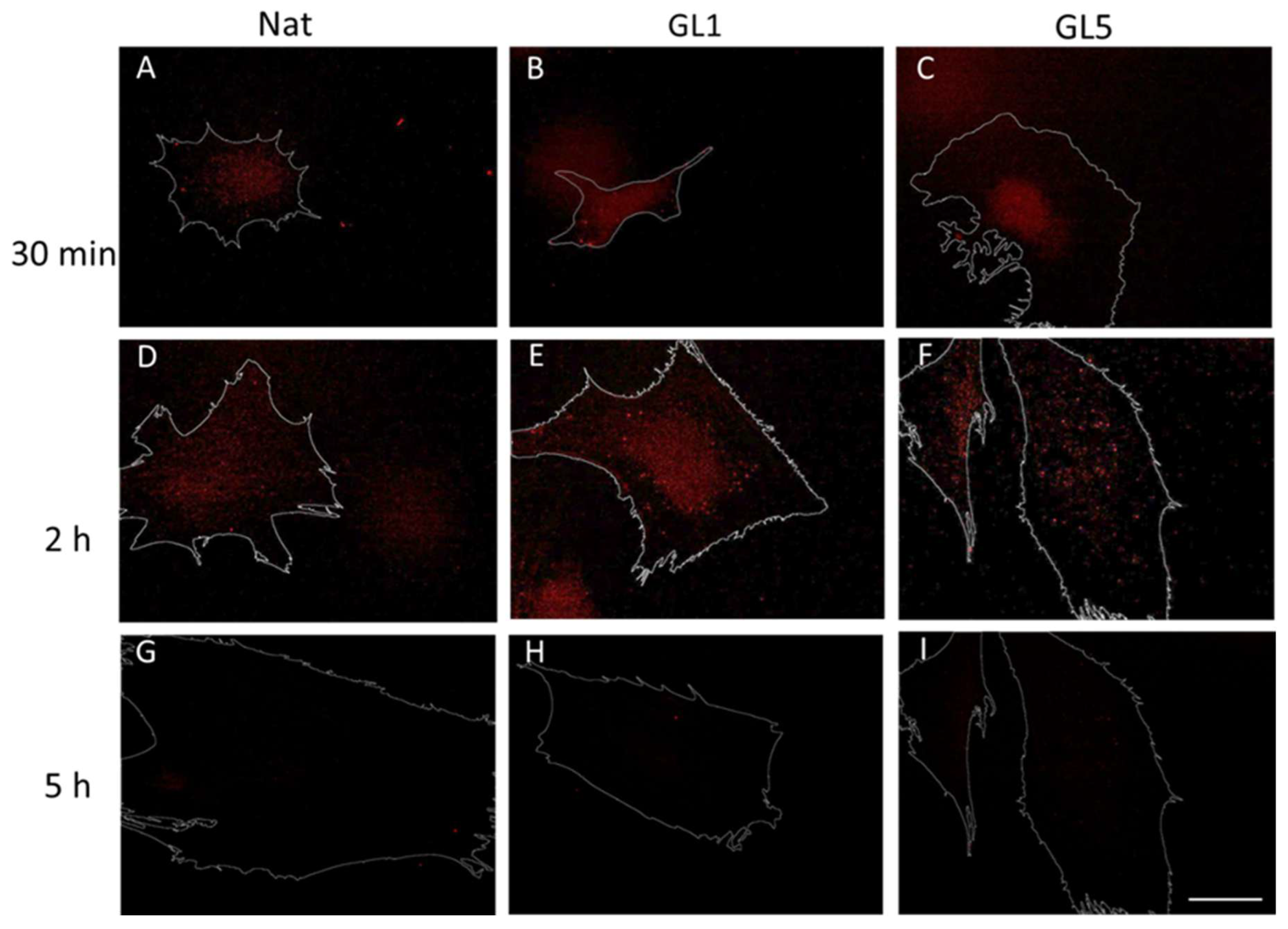
2.3. ADMSC Mechanotransduction from Glycated Collagen
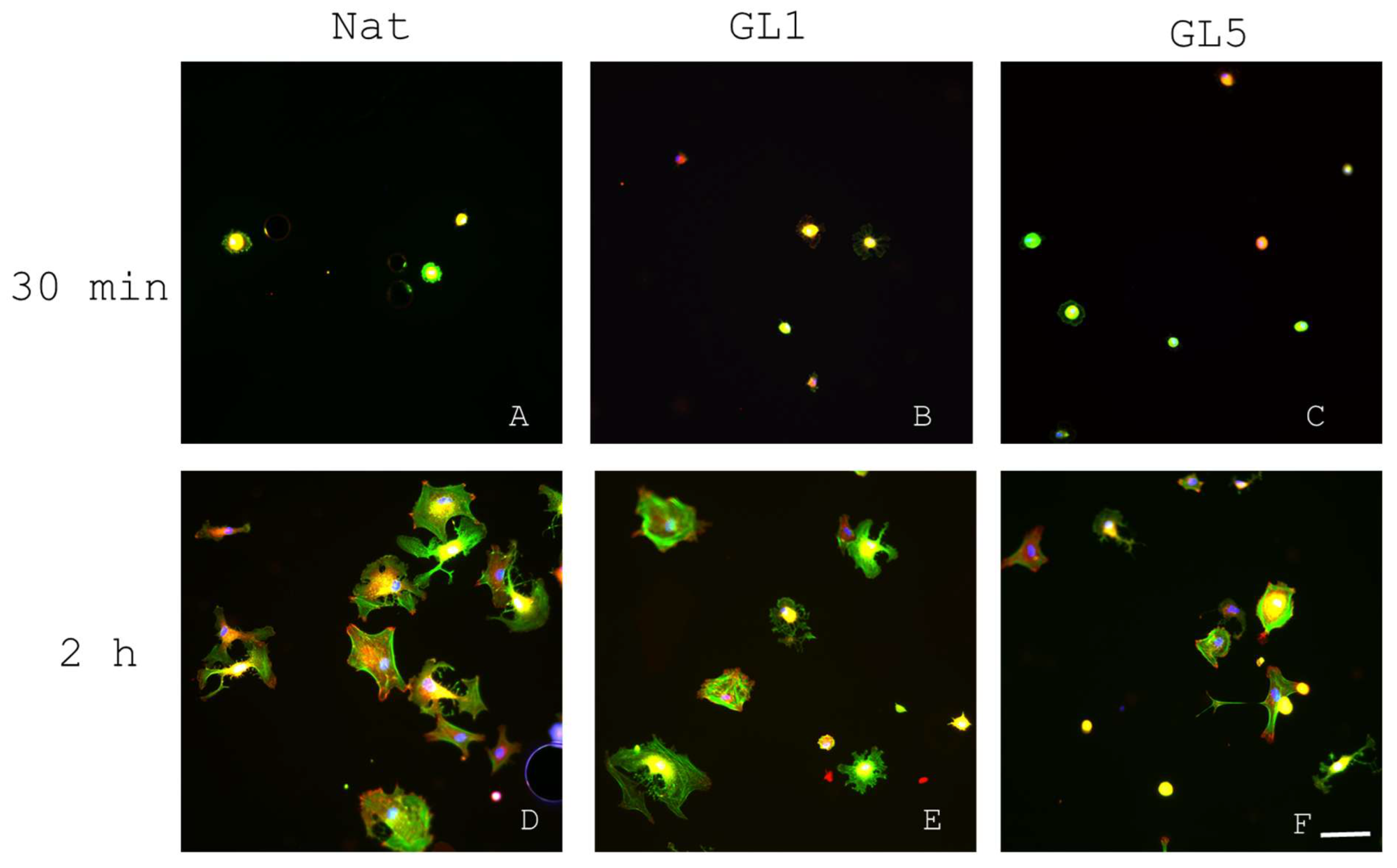
2.3.1. Quantification of Focal Adhesion (FA) Dynamics
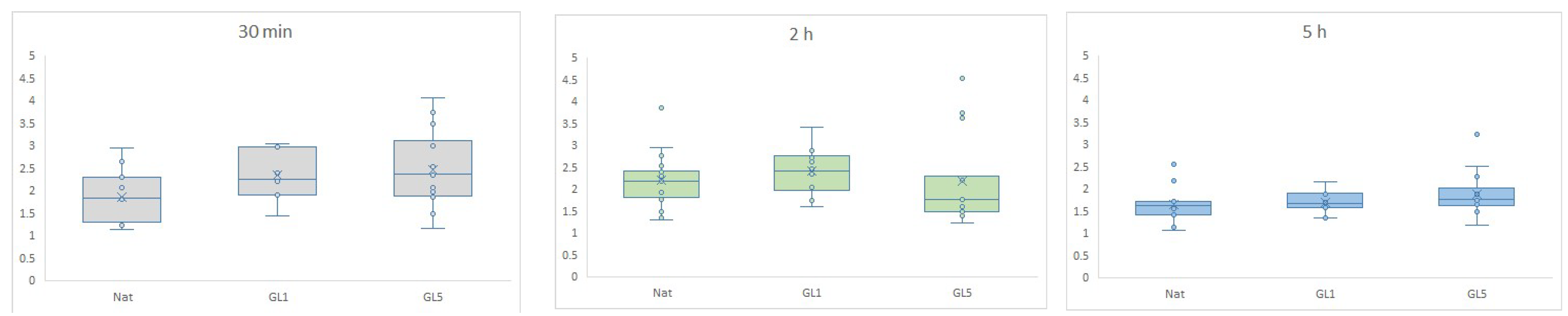
2.3.2. Quantification of YAP/TAZ Activity
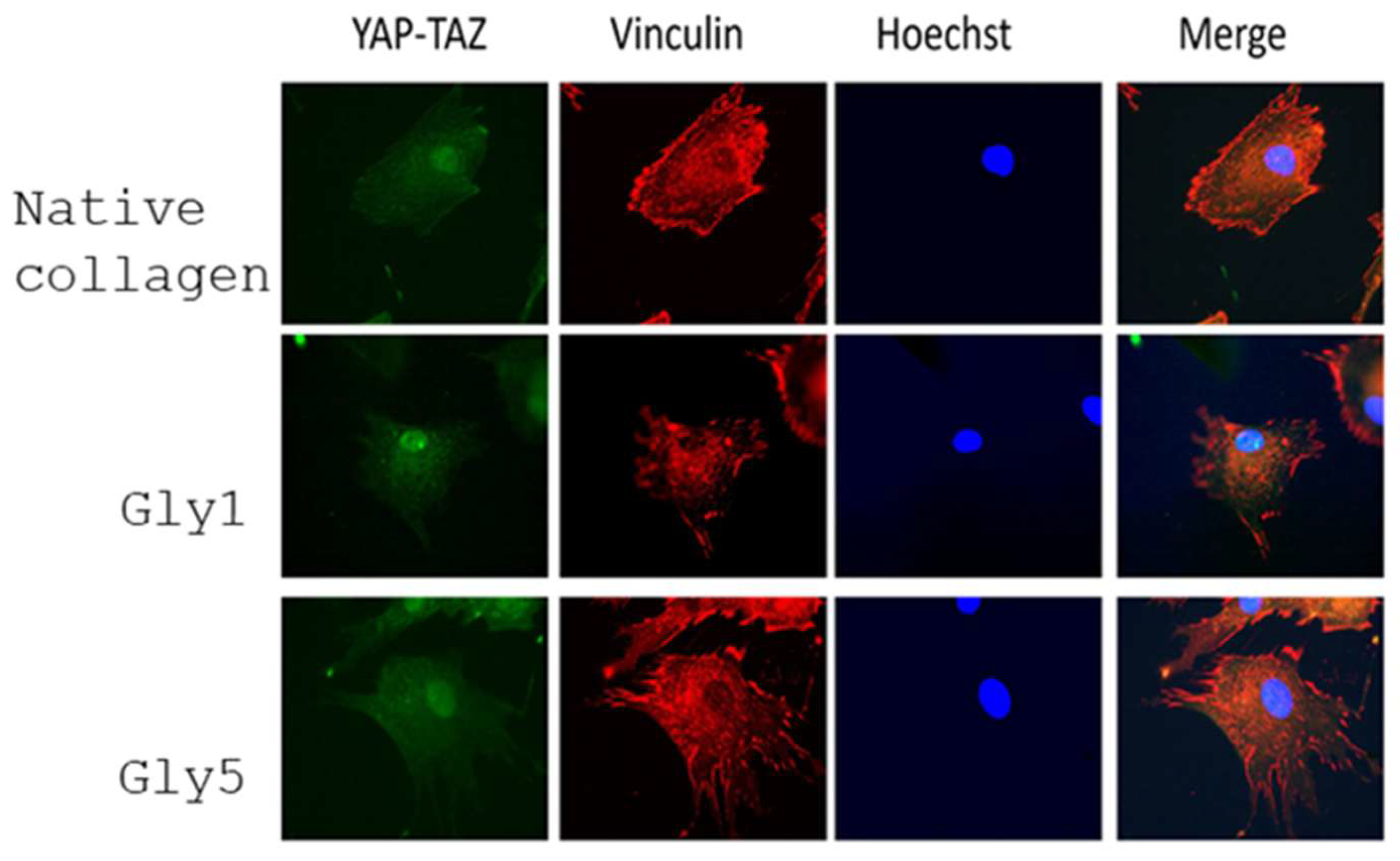
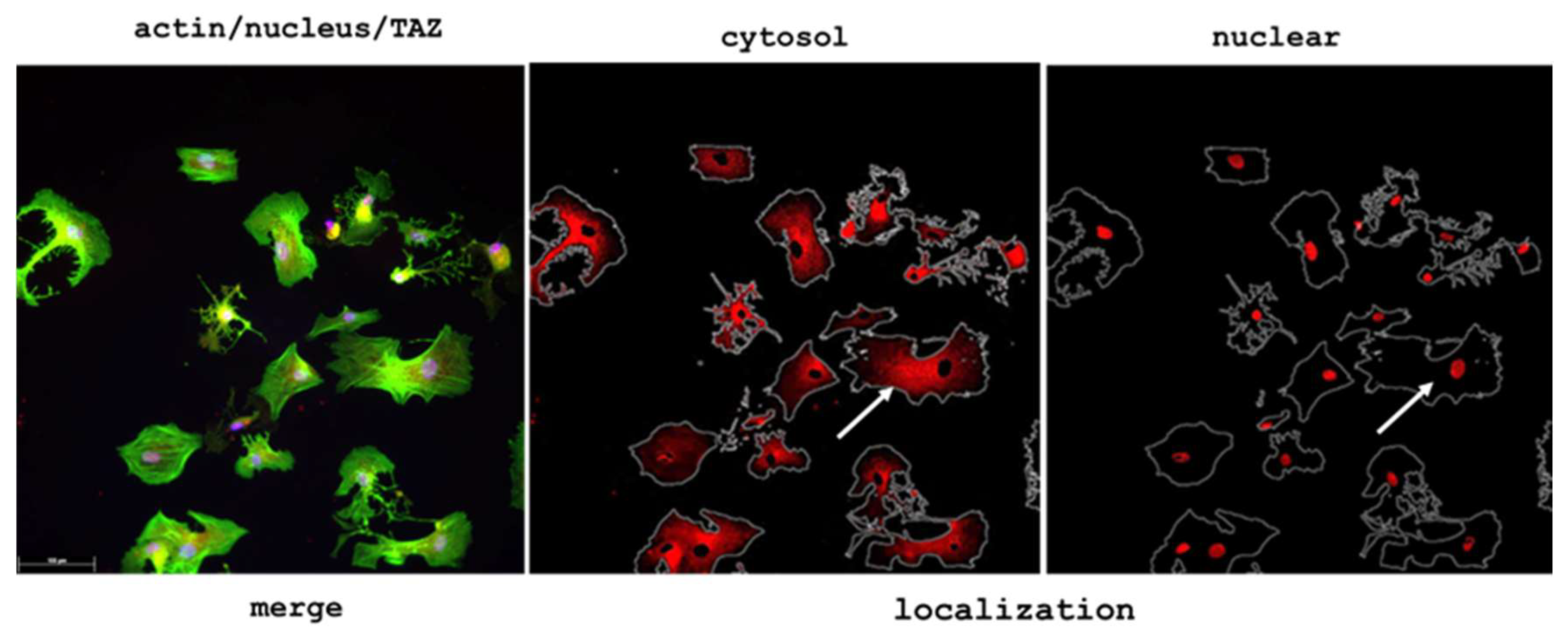
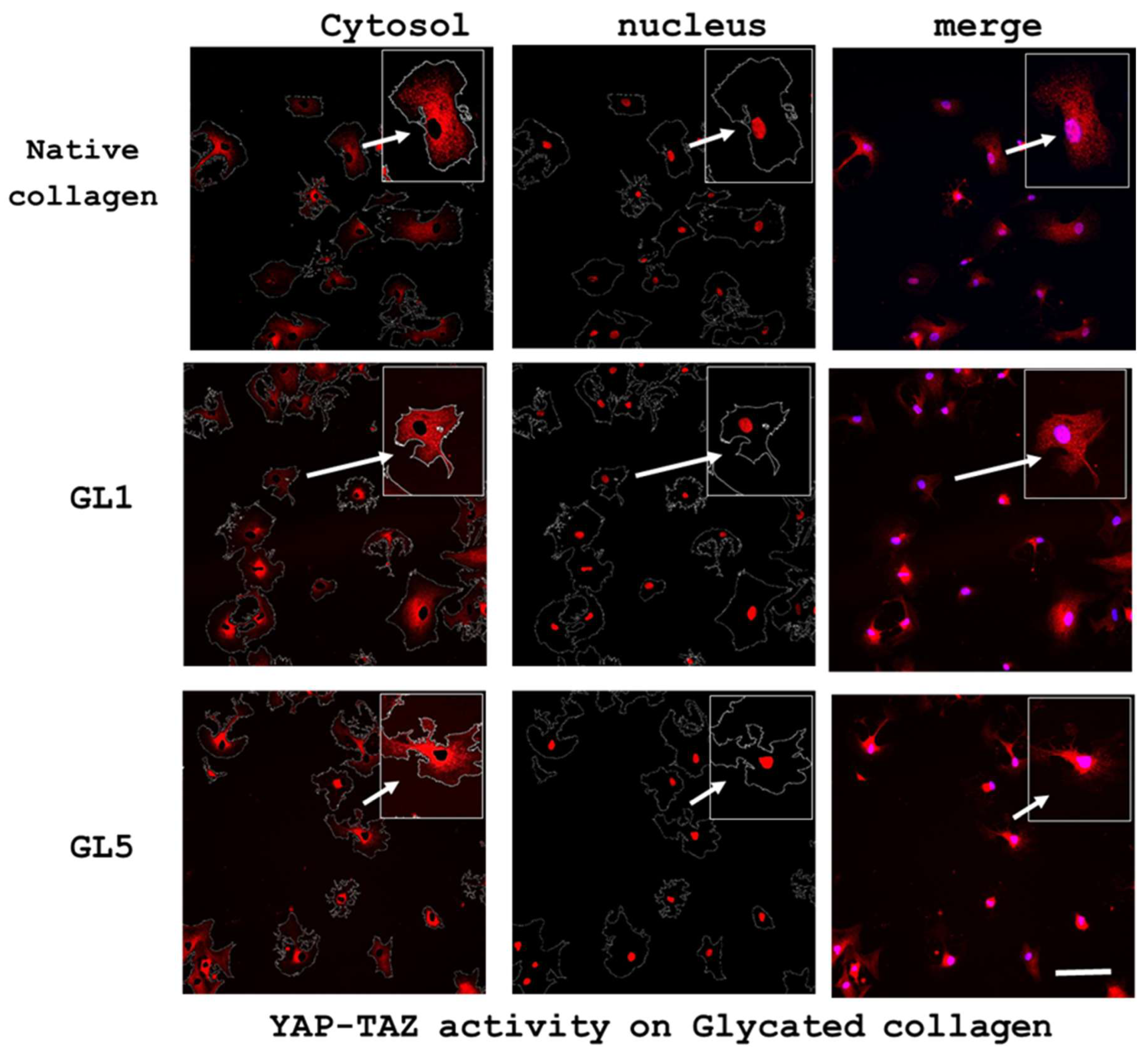
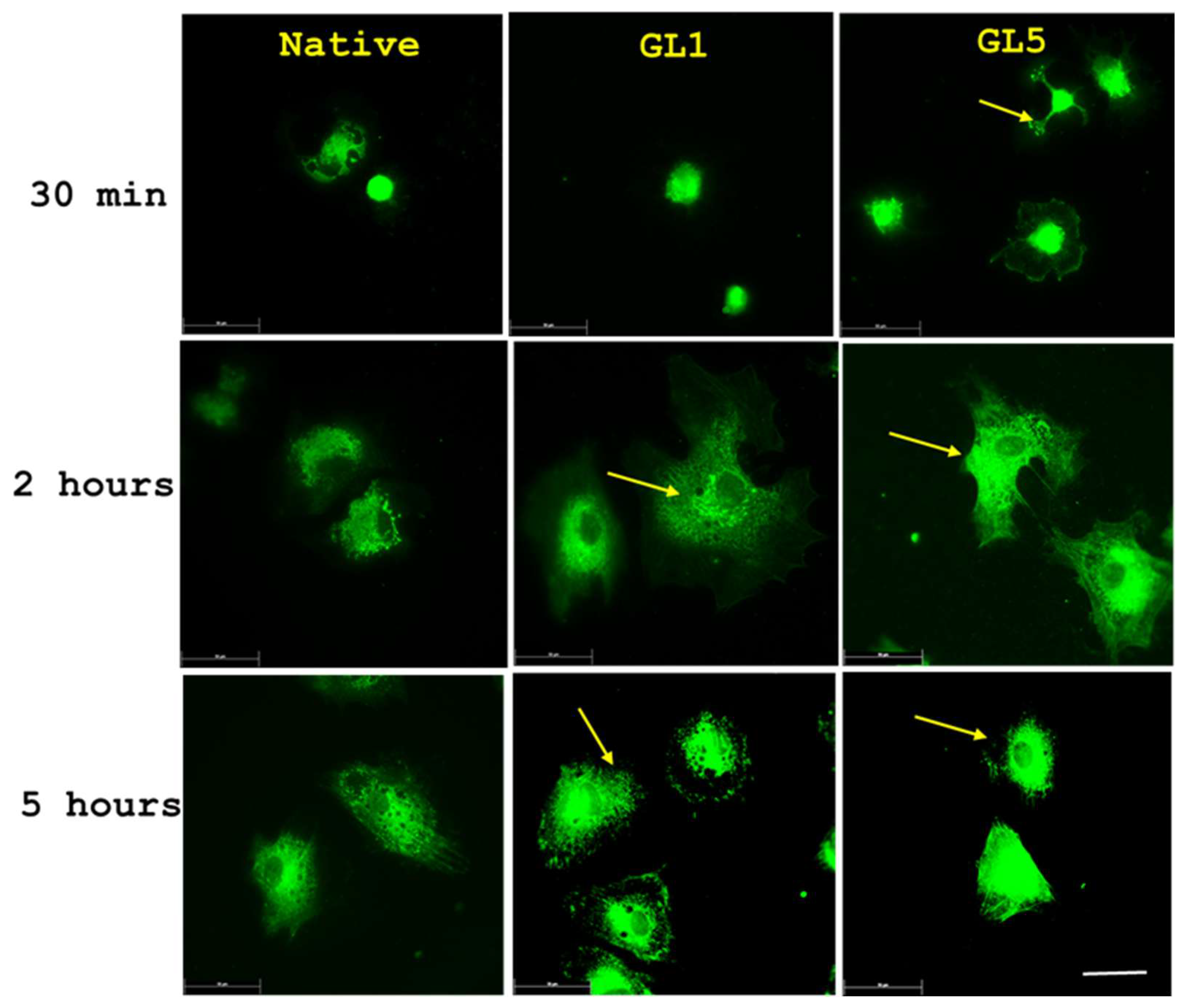
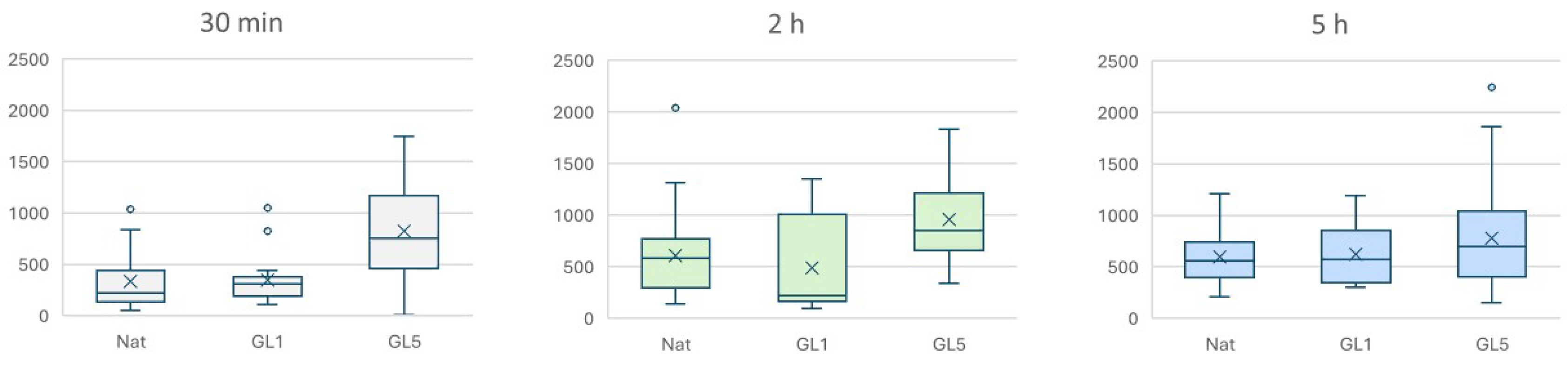
2.4. Morphological Evidence for an Increased Collagen Synthesis of ADMSC on Glycated Collagen
2.5. AFM Studies of Glycated Collagen
3. Discussion
4. Materials and Methods
4.1. Collagen Procedures
4.1.1. Collagen Preparation
4.1.2. Preparation of Glycated Collagen
4.2. Cells
4.2.1. Morphological Studies
4.2.1.1. First Protocol (cell Spreading and FA Formation)
4.2.1.2. Second Protocol (YAP/TAZ Signaling Events)
4.2.1.3. Overall Cell Morphology and Focal Adhesion Formation
4.2.1.4. Quantitative Morphometry Analysis of Raw Format Images by ImageJ
4.2.1.5. Quantification of Intracellular Collagen Content
4.3. Atomic Force Microscopy of Glycated Collagen
4.3.1. Sample Preparation
4.3.2. AFM Imaging
4.3.3. Morphological Characterisation
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD-MSCs | Adipose-Derived Mesenchymal Stem Cells |
| RTC | Rat Tail Collagen |
| FBS | Fetal Bovine Serum |
| MSCs | Mesenchymal Stem Cells |
| ECM | Extracellular Matrix |
| PBS | Phosphate Buffered Saline |
| CSA | Cell Spreading Area |
| CAR | Cell Aspect Ratio |
| CSI | Cell Shape Index |
| RAGE | Receptor for Advanced Glycation End-products |
| YAP | Yes-Associated Protein |
| TAZ | Transcriptional Co-activator with PDZ-binding Motif |
| GL1 | 1 day glycated RTC |
| GL5 | 1 day glycated RTC |
References
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014, 15, 802–812. [CrossRef]
- Mierke, C.T. Extracellular Matrix Cues Regulate Mechanosensing and Mechanotransduction of Cancer Cells. Cells 2024, 13, 96. [CrossRef]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742.
- Klein, E.A.; Yin, L.; Kothapalli, D.; Castagnino, P.; Byfield, F.J.; Xu, T.; Levental, I.; Hawthorne, E.; Janmey, P.A.; Assoian, R.K. Cell-Cycle Control by Physiological Matrix Elasticity and In Vivo Tissue Stiffening. Curr. Biol. 2009, 19, 1511–1518. [CrossRef]
- Wang, L.; You, X.; Zhang, L.; Zhang, C.; Zou, W. Mechanical regulation of bone remodeling. Bone Res. 2022, 10, 1–15. [CrossRef]
- Kozlova, I.; Sytnyk, V. Cell Adhesion Molecules as Modulators of the Epidermal Growth Factor Receptor. Cells 2024, 13, 1919. [CrossRef]
- Li, D.; Zhou, J.; Chowdhury, F.; Cheng, J.; Wang, N.; Wang, F. Role of Mechanical Factors in Fate Decisions of Stem Cells. Regen. Med. 2011, 6, 229–240. [CrossRef]
- Komsa-Penkova, R.; Alexandrova-Watanabe, A.; Todinova, S.; Ivanova, V.; Stoycheva, S.; Temnishki, P.; Dimitrov, B.; Dimitrov, D.; Tonchev, P.; Georgieva, G.; et al. Adhesion of Mesenchymal Stem Cells to Glycated Collagen—Comparative Analysis of Dynamic and Static Conditions. Polymers 2025, 17, 821. [CrossRef]
- Majhy B., Priyadarshinia P., Sen A. K. Effect of surface energy and roughness on cell adhesion and growth – facile surface modification for enhanced cell culture RSC Adv., 2021, 11, 154674.
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [CrossRef]
- Heino, J. Cellular signaling by collagen-binding integrins.Adv Exp Med Biol 2014;819:143-55.
- Zeltz C, Gullberg DJ, The integrin-collagen connection - a glue for tissue repair? Cell Sci. 2016 Mar 15;129(6):1284.
- Komsa-Penkova, R.; Dimitrov, B.; Todinova, S.; Ivanova, V.; Stoycheva, S.; Temnishki, P.; Georgieva, G.; Tonchev, P.; Iliev, M.; Altankov, G. Early Stages of Ex Vivo Collagen Glycation Disrupt the Cellular Interaction and Its Remodeling by Mesenchymal Stem Cells—Morphological and Biochemical Evidence. Int. J. Mol. Sci. 2024, 25, 5795. [CrossRef]
- Myllyharju, J. (2005) Intracellular Post-Translational Modifications of Collagens. In: Brinckmann, J., Notbohm H, Müller, P K (eds) Collagen. Topics in Current Chemistry, vol 247. Springer, Berlin, Heidelberg. [CrossRef]
- Komsa-Penkova, R.; Yordanova, A.; Tonchev, P.; Kyurkchiev, S.; Todinova, S.; Strijkova, V.; Iliev, M.; Dimitrov, B.; Altankov, G. Altered Mesenchymal Stem Cells Mechanotransduction from Oxidized Collagen: Morphological and Biophysical Observations. Int. J. Mol. Sci. 2023, 24, 3635. [CrossRef]
- Mikulíková, K.; Eckhardt, A.; Pataridis, S.; Mikšík, I. Study of posttranslational non-enzymatic modifications of collagen using capillary electrophoresis/mass spectrometry and high performance liquid chromatography/mass spectrometry. J. Chromatogr. A 2007, 1155, 125–133. [CrossRef]
- Li, Z.; Lee, H.; Zhu, C. Molecular mechanisms of mechanotransduction in integrin-mediated cell-matrix adhesion. Exp. Cell Res. 2016, 349, 85–94. [CrossRef]
- Lee, J.-H.; Park, H.-K.; Kim, K.S. Intrinsic and extrinsic mechanical properties related to the differentiation of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2016, 473, 752–757. [CrossRef]
- Sun Z., Guo S. S., Fässler R. Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016).
- Horbett T.A. The role of adsorbed proteins in animal cell adhesion Colloids and Surfaces B: Biointerfaces Vol 2, Issues 1–3, 14 March 1994, Pages 225-240.
- Burridge, K.; Monaghan-Benson, E.; Graham, D.M. Mechanotransduction: from the cell surface to the nucleus via RhoA. Philos. Trans. R. Soc. B: Biol. Sci. 2019, 374, 20180229. [CrossRef]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting integrin pathways: mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 1–42. [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [CrossRef]
- Zinatizadeh, M.R.; Miri, S.R.; Zarandi, P.K.; Chalbatani, G.M.; Rapôso, C.; Mirzaei, H.R.; Akbari, M.E.; Mahmoodzadeh, H. The Hippo Tumor Suppressor Pathway (YAP/TAZ/TEAD/MST/LATS) and EGFR-RAS-RAF-MEK in cancer metastasis. Genes Dis. 2021, 8, 48–60. [CrossRef]
- Halder, G.; Dupont, S.; Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012, 13, 591–600.
- Pobbati, A.V.; Hong, W. A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy. Theranostics 2020, 10, 3622–3635. [CrossRef]
- Sessa, L.; Gatti, E.; Zeni, F.; Antonelli, A.; Catucci, A.; Koch, M.; Pompilio, G.; Fritz, G.; Raucci, A.; E Bianchi, M. The Receptor for Advanced Glycation End-Products (RAGE) Is Only Present in Mammals, and Belongs to a Family of Cell Adhesion Molecules (CAMs). PLOS ONE 2014, 9, e86903. [CrossRef]
- Ramasamy, R.; Yan, S.F.; Schmidt, A.M. Receptor for AGE (RAGE): signaling mechanisms in the pathogenesis of diabetes and its complications. Ann. New York Acad. Sci. 2011, 1243, 88–102. [CrossRef]
- Murillo, J.; Wang, Y.; Xu, X.; Klebe, R.J.; Chen, Z.; Zardeneta, G.; Pal, S.; Mikhailova, M.; Steffensen, B. Advanced Glycation of Type I Collagen and Fibronectin Modifies Periodontal Cell Behavior. J. Periodontol. 2008, 79, 2190–2199. [CrossRef]
- Dandia, H.; Makkad, K.; Tayalia, P. Glycated collagen – a 3D matrix system to study pathological cell behavior. Biomater. Sci. 2019, 7, 3480–3488. [CrossRef]
- Crop, M.J.; Baan, C.C.; Korevaar, S.S.; Ijzermans, J.N.M.; Pescatori, M.; Stubbs, A.P.; Van Ijcken, W.F.J.; Dahlke, M.H.; Eggenhofer, E.; Weimar, W.; et al. Inflammatory conditions affect gene expression and function of human adipose tissue-derived mesenchymal stem cells. Clin. Exp. Immunol. 2010, 162, 474–486. [CrossRef]
- Franz, C.M.; Muller, D.J. (2011). Studying Collagen Self-Assembly by Time-Lapse High-Resolution Atomic Force Microscopy. In: Braga, P., Ricci, D. (eds) Atomic Force Microscopy in Biomedical Research. Methods in Molecular Biology, vol 736. Humana Press. [CrossRef]
- Sloseris, D.; Forde, N.R. AGEing of collagen: The effects of glycation on collagen’s stability, mechanics and assembly. Matrix Biol. 2024, 135, 153–160. [CrossRef]
- Johnson, R.; Halder, G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2013, 13, 63–79. [CrossRef]
- Janmey P. A., Wells R. G., Assoian R. K., McCulloch C. A., From tissue mechanics to transcription factors. Differentiation 86, 112–120 (2013).
- Komatsu et al., 2018. Type I collagen deposition via osteoinduction ameliorates YAP/TAZ activity in MSC/ECM complexes Stem Cell Research & Therapy.
- Olson, L.C.; Nguyen, T.M.; Heise, R.L.; Boyan, B.D.; Schwartz, Z.; McClure, M.J. Advanced Glycation End Products Are Retained in Decellularized Muscle Matrix Derived from Aged Skeletal Muscle. Int. J. Mol. Sci. 2021, 22, 8832. [CrossRef]
- Komsa-Penkova, R.; Stavreva, G.; Belemezova, K.; Kyurkchiev, S.; Todinova, S.; Altankov, G. Mesenchymal Stem-Cell Remodeling of Adsorbed Type-I Collagen—The Effect of Collagen Oxidation. Int. J. Mol. Sci. 2022, 23, 3058. [CrossRef]
- Komsa-Penkova, R.; Spirova, R.; Bechev, B. Modification of Lowrys Method for Collagen Concentration Measurement. J. Biochem. Biophys. Methods 1996, 32, 33–43. [CrossRef]
- Kuznetsova, N.; Leikin, S. Does the Triple Helical Domain of Type I Collagen Encode Molecular Recognition and Fiber Assembly while Telopeptides Serve as Catalytic Domains?. J. Biol. Chem. 1999, 274, 36083–36088. [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; A Cimini, B.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 2018, 16, e2005970. [CrossRef]
- Stirling, D.R.; E Carpenter, A.; A Cimini, B. CellProfiler Analyst 3.0: accessible data exploration and machine learning for image analysis. Bioinformatics 2021, 37, 3992–3994. [CrossRef]
- Boudaoud, A.; Burian, A.; Borowska-Wykręt, D.; Uyttewaal, M.; Wrzalik, R.; Kwiatkowska, D.; Hamant, O. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 2014, 9, 457–463. [CrossRef]
- Antonio, P.D.; Lasalvia, M.; Perna, G.; Capozzi, V. Scale-independent roughness value of cell membranes studied by means of AFM technique. Biochim. et Biophys. Acta (BBA) - Biomembr. 2012, 1818, 3141–3148. [CrossRef]
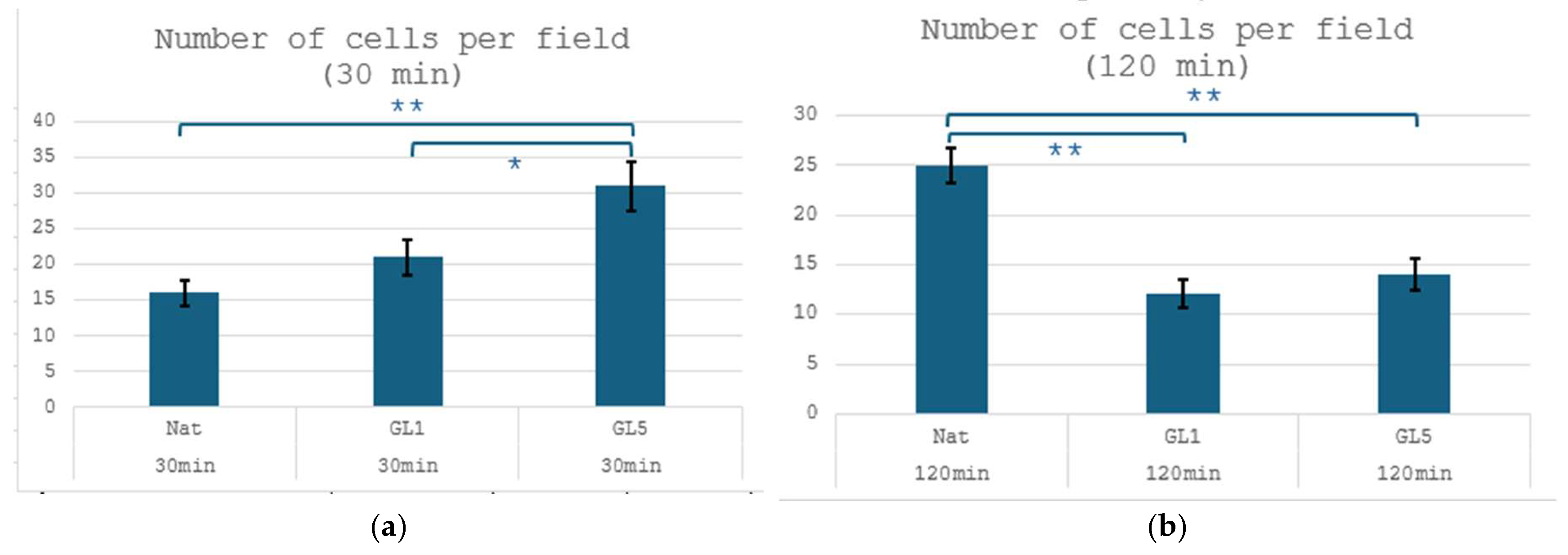
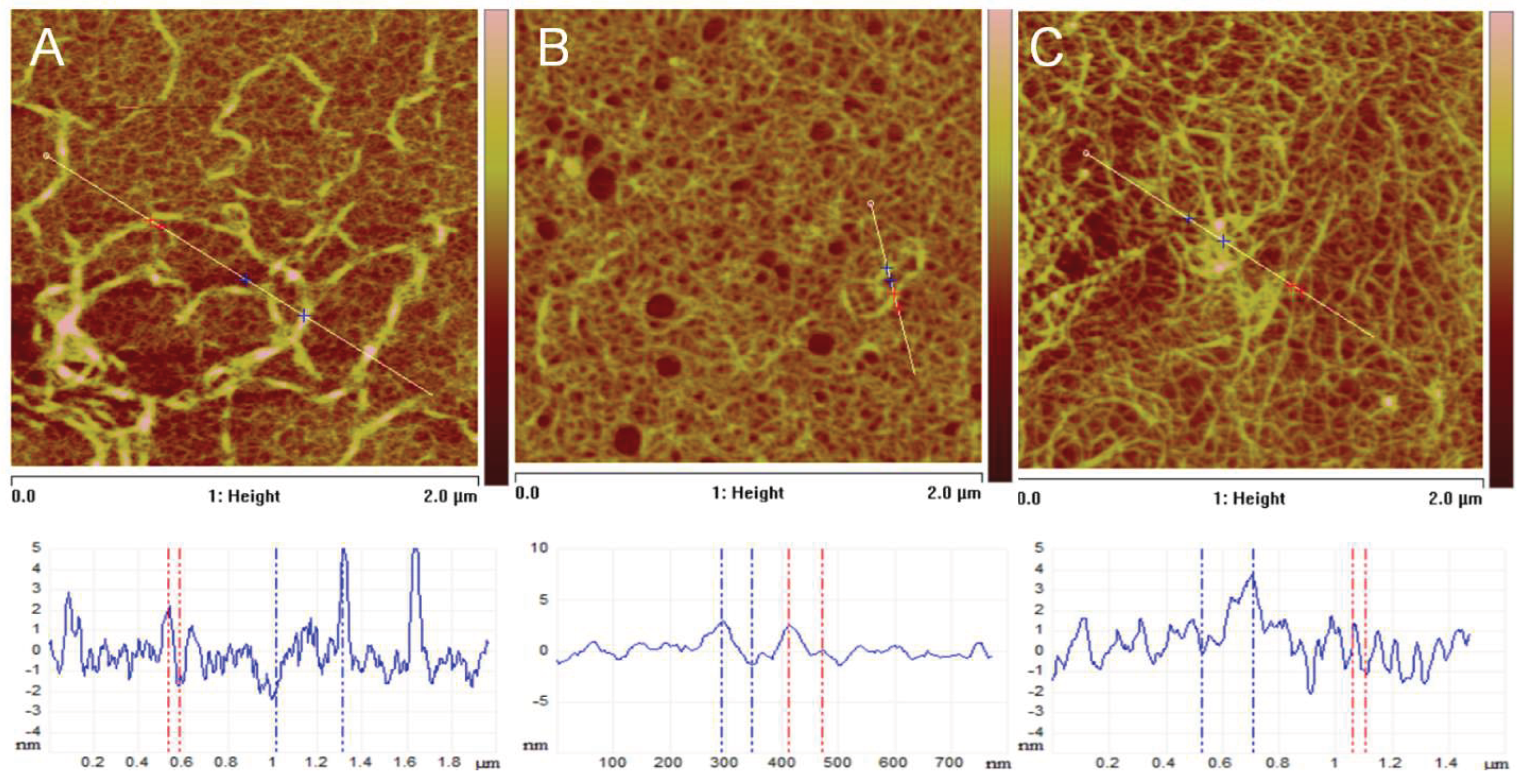
| Parameter | Incubation Time | Nat | GL1 | GL5 |
| CSA (μm2) | 30 min | 3165 ± 1926 (n = 11) | 1337 ± 762 (n = 11) | 1326 ± 907 (n = 14) |
| 2 h | 7522 ± 3767 (n = 20) | 5939 ± 4382 (n = 9) | 5454 ± 2717 (n = 12) | |
| 5 h | 7097 ± 3814 (n = 11) | 6663 ± 2440 (n = 12) | 6508 ± 2037 (n = 14) | |
| Perimeter (μm) | 30 min | 225 ± 67 | 144 ± 48 | 136 ± 55 |
| 2 h | 468 ± 206 | 409 ± 216 | 544 ± 198 | |
| 5 h | 607 ± 220 | 724 ± 184 | 854 ± 343 | |
| CSI | 30 min | 0.73 ± 0.18 | 0.79 ± 0.14 | 0.87 ± 0.13 |
| 2 h | 0.48 ± 0.17 | 0.56 ± 0.18 | 0.38 ± 0.12 | |
| 5 h | 0.28 ± 0.17 | 0.23 ± 0.08 | 0.17 ± 0.07 |
| Incubation Time | Substrate Type | Number of FAs per Cell | Total Area of FАs per Cell |
| 30 min | Nat | 1 ± 1 | 3.57 ± 0.39 µm2 |
| 30 min | GL1 | 2 ± 1 | 15.14 ± 7.65 µm2 |
| 30 min | GL5 | 2 ± 2 | 15.75 ± 5.04 µm2 |
| 2 h | Nat | 98 ± 29 | 1407.46 ± 542.01 µm2 |
| 2 h | GL1 | 60 ± 12 | 782.08 ± 267.41 µm2 |
| 2 h | GL5 | 51 ± 16 | 492.60 ± 102.39 µm2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





