Submitted:
04 November 2025
Posted:
06 November 2025
You are already at the latest version
Abstract
Keywords:
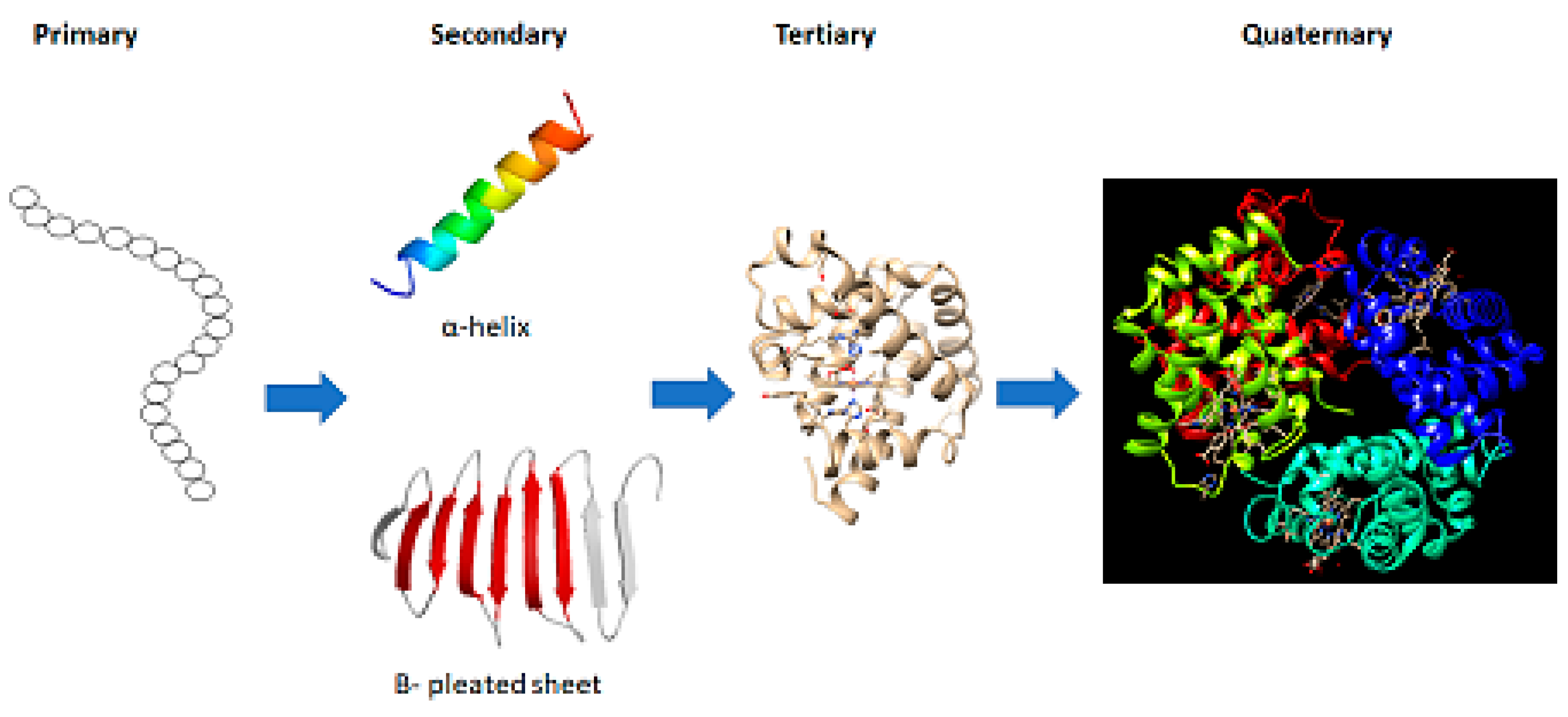
Introduction
Advantages and Future Perspectives
Conclusion
References
- J (1981) The anatomy and taxonomy of protein structure. Adv Protein Chem 34:167.
- Branden C, Tooze J (1991) Introduction to protein structures. Garland Publishing, New York.
- Kolodny R et al (2013) On the universe of protein folds. Annu Rev Biophys 42:559–582.
- Ouzounis CA et al (2003) Classification schemes for protein structure and function. Nat Rev Genet 4(7):508–519.
- 1 Matthieu Dreher, Marc Piuzzi, Ahmed Turki, Matthieu Chavent, Marc Baaden, Nicolas Férey, Sébastien Limet, Bruno Raffin, Sophie Robert, Interactive Molecular Dynamics: Scaling up to Large Systems, Procedia Computer Science, Volume 18, 2013, Pages 20-29, ISSN 1877-0509. [CrossRef]
- Ratamero EM, Bellini D, Dowson CG, Romer RA. Touching proteins with virtual bare hands: Visualizing protein-drug complexes and their dynamics in self-made virtual reality using gaming hardware. J Comput Aided Mol Des. 2018; 32(6):703–709. PMID: 29882064. [CrossRef]
- Grebner C, Norrby M, Enstrom J, Nilsson I, Hogner A, Henriksson J, et al. 3D-Lab: a collaborative webbased platform for molecular modeling. Future Med Chem. 2016; 8(14):1739–52. PMID: 27577860. [CrossRef]
- Goddard TD, Brilliant AA, Skillman TL, Vergenz S, Tyrwhitt-Drake J, Meng EC, et al. Molecular Visualization on the Holodeck. J Mol Biol. 2018; 430(21):3982–3996. PMID: 29964044. [CrossRef]
- O’Connor M, Deeks HM, Dawn E, Metatla O, Roudaut A, Sutton M, et al. Sampling molecular conformations and dynamics in a multiuser virtual reality framework. Sci Adv. 2018; 4(6):eaat2731. PMID: 29963636. [CrossRef]
- Al-Balushi SM, Al-Hajri SH. Associating animations with concrete models to enhance students’ comprehension of different visual representations in organic chemistry. Chemistry Education Research and Practice. 2014; 15(1):47–58. [CrossRef]
- Matthieu Dreher, Marc Piuzzi, Ahmed Turki, Matthieu Chavent, Marc Baaden, Nicolas Férey, Sébastien Limet, Bruno Raffin, Sophie Robert, Interactive Molecular Dynamics: Scaling up to Large Systems, Procedia Computer Science, Volume 18, 2013, Pages 20-29, ISSN 1877-0509. [CrossRef]
- Cassidy KC, Šefčík J, Raghav Y, Chang A, Durrant JD (2020) ProteinVR: Web-based molecular visualization in virtual reality. PLOS Computational Biology 16(3): e1007747. [CrossRef]
- Daniel B. Roche, Maria T. Buenavista, Liam J. McGuffin, The FunFOLD2 server for the prediction of protein–ligand interactions, Nucleic Acids Research, Volume 41, Issue W1, 1 July 2013, Pages W303–W307. [CrossRef]
- Lee, M., Jeong, Cs. & Kim, D. Predicting and improving the protein sequence alignment quality by support vector regression. BMC Bioinformatics 8, 471 (2007). [CrossRef]
- Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins. 2011;79 Suppl 10(Suppl 10):161-71. Epub 2011 Oct 11. PMID: 21987485; PMCID: PMC3226909. [CrossRef]
- Lambert C, Leonard N, De Bolle X, Depiereux E.ESyPred3D: Prediction of proteins 3D structures.Bioinformatics. 2002 Sep;18(9):1250-1256.
- B. Webb, A. Sali. Comparative Protein Structure Modeling Using Modeller. Current Protocols in Bioinformatics 54, John Wiley & Sons, Inc., 5.6.1-5.6.37, 2016.
- McGuffin, Liam J; Adiyaman, Recep; Maghrabi, Ali H A; Shuid, Ahmad N; Brackenridge, Danielle A; Nealon, John O; Philomina, Limcy S (2019-05-02). "IntFOLD: an integrated web resource for high performance protein structure and function prediction". Nucleic Acids Research. 47 (W1): W408–W413. . ISSN 0305-1048. PMC 6602432. PMID 31045208. [CrossRef]
- J. Á. Ramírez and A. M. V. Bueno, "Learning organic chemistry with virtual reality," 2020 IEEE International Conference on Engineering Veracruz (ICEV), 2020, pp. 1-4. [CrossRef]
- Doak DG, Denyer GS, Gerrard JA, Mackay JP, Allison JR. Peppy: A virtual reality environment for exploring the principles of polypeptide structure. Protein Sci. 2020;29(1):157-168. [CrossRef]
- Tianshu Xu, Venkata VB Yallapragada, Mark Tangney, and Sabin Tabirca. 2021. ProMVR - Protein Multiplayer Virtual Reality Tool. In Proceedings of the 27th ACM Symposium on Virtual Reality Software and Technology (VRST '21). Association for Computing Machinery, New York, NY, USA, Article 91, 1–3. [CrossRef]
- Todd H, Emsley P. Development and assessment of CootVR, a virtual reality computer program for model building. Acta Crystallogr D Struct Biol. 2021 Jan 1;77(Pt 1):19-27. doi: 10.1107/S2059798320013625. Epub 2021 Jan 1. PMID: 33404522; PMCID: PMC7787110.
- UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. Protein Sci. 2021 Jan;30(1):70.
- UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. Protein Sci. 2018 Jan;27(1):14-25.
- ‘Caroline Stefani, Adam Lacy-Hulbert, Thomas Skillman,ConfocalVR: Immersive Visualization for Confocal Microscopy, Journal of Molecular Biology, Volume 430, Issue 21, 2018, Pages 4028-4035,ISSN 0022-2836. (https://www.sciencedirect.com/science/article/pii/S0022283618306648). [CrossRef]
- O’Connor M, Deeks HM, Dawn E, Metatla O, Roudaut A, Sutton M, et al. Sampling molecular conformations and dynamics in a multiuser virtual reality framework. Sci Adv. 2018; 4(6):eaat2731. PMID: 29963636. [CrossRef]
- Bennie SJ, Ranaghan KE, Deeks H, Goldsmith HE, O’Connor MB, Mulholland AJ, et al. Teaching enzyme catalysis using interactive molecular dynamics in virtual reality. Journal of Chemical Education. 2019; 96(11):2488–2496. [CrossRef]
- Ferrell JB, Campbell JP, McCarthy DR, McKay KT, Hensinger M, Srinivasan R, et al. Chemical Exploration with Virtual Reality in Organic Teaching Laboratories. Journal of Chemical Education. 2019; 96(9):1961–1966. [CrossRef]
- O’Connor MB, Bennie SJ, Deeks HM, Jamieson-Binnie A, Jones AJ, Shannon RJ, et al. Interactive molecular dynamics in virtual reality from quantum chemistry to drug binding: An open-source multi-person framework. The Journal of chemical physics. 2019; 150(22):220901. PMID: 31202243. [CrossRef]
- Rajendiran N, Durrant JD. Pyrite: A blender plugin for visualizing molecular dynamics simulations using industry-standard rendering techniques. Journal of Computational Chemistry. 2018; 39(12):748–755. PMID: 29280166. [CrossRef]
| Sl.No. | Software | Author | Link |
| 1 | Protein VR - Durrant lab (13) | Cassidy et al (2020) | http://durrantlab.com/protein-vr/ |
| 2 | PEP Block builder VR (14) | Yallapragada 2021 | https://github.com/TIanshuXu/Pocket-Peptides-PC |
| 3 | Nanome (15) | Ramrez et al 2020 | Learning organic chemistry with virtual reality | IEEE Conference Publication | IEEE Xplore |
| 4 | Raghav (16) | Raghav et al () | APSSP2: Advanced Protein Secondary Structure Prediction Server (osdd.net) |
| 5 | PROtein VR (17) | Cassidy et al (2020) | ProteinVR – Durrant Lab (pitt.edu) |
| 6 | Peppy VR (18) | Peppy VR - Faculty of Science (sydney.edu.au) | |
| 7 | ProMVR (19) | VR-тренажёры PROMVR – oбучение безoпаснoму пoведению на прoизвoдстве | |
| 8 | CootVR (20) | cootVR (hamishtodd1.github.io) | |
| 9 | Chimera X (21) | Peterson et al 2021 | UCSF ChimeraX Home Page |
| 10 | IntFOLD (22) | Liam et al | https://www.reading.ac.uk/bioinf/IntFOLD/ |
| 11 | RaptorX(23) | Peng et al | http://raptorx.uchicago.edu/ |
| 12 | ESyPred3D (24) | Lambert et al 2002 | ESyPred3D submitting form (unamur.be) |
| 13 | FunFOLD2 (25) | Roche et al 2013 | http://www.reading.ac.uk/bioinf/FunFOLD/FunFOLD_form_2_0.html |
| 14 | FORECAST (26) | http://pbil.kaist.ac.kr/forecast | |
| 15 | Capsid (27) | Peng et al 2011 | Projects – CAPSID (loria.fr) |
| 16 | MODELLER (28) | Webb et al (2016) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





