Submitted:
03 November 2025
Posted:
04 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Differences in Soil Physicochemical Properties Among Chinese Fir Plantations with Different Stand Densities
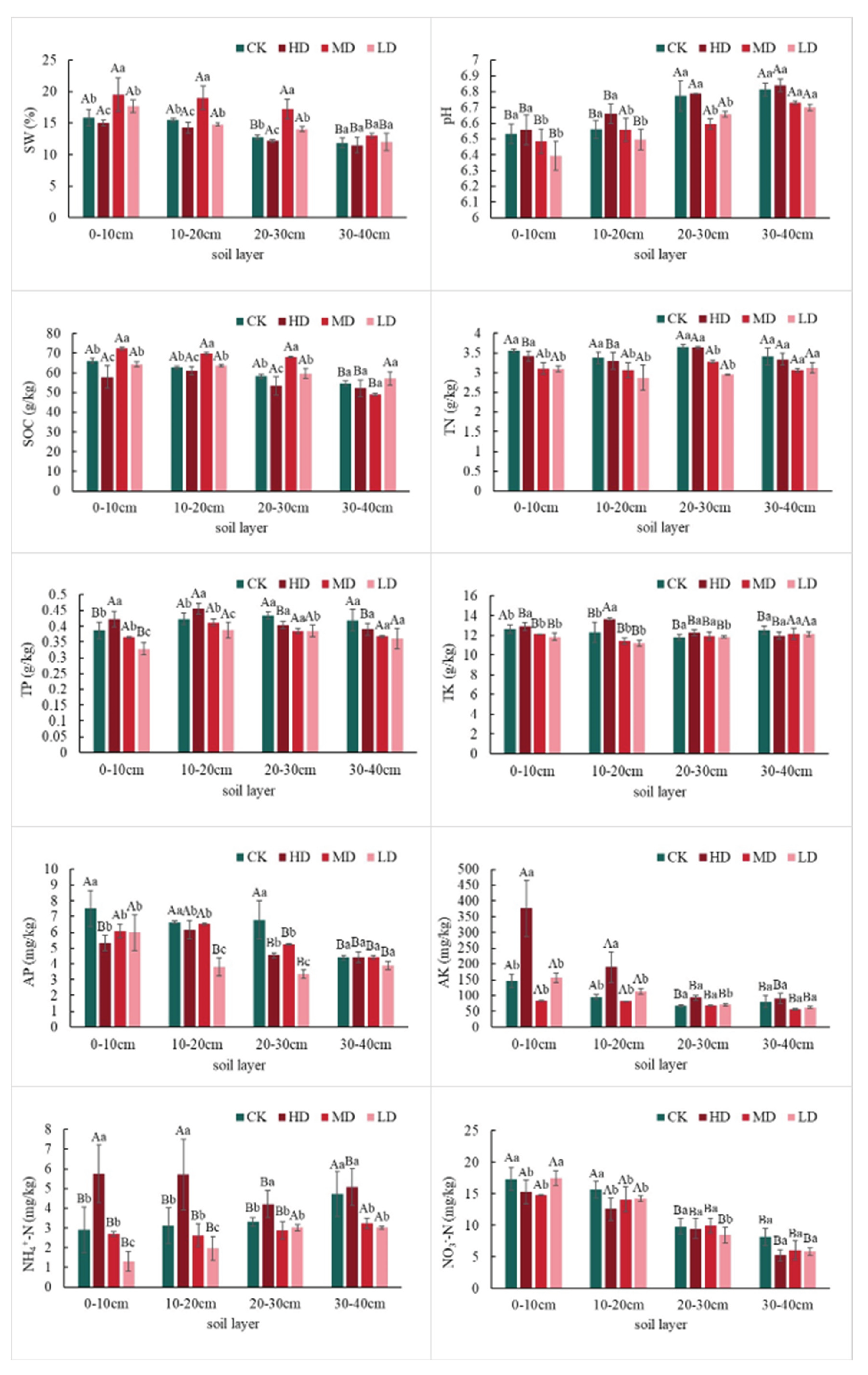
2.1.1. Differences in Soil Nutrient Content
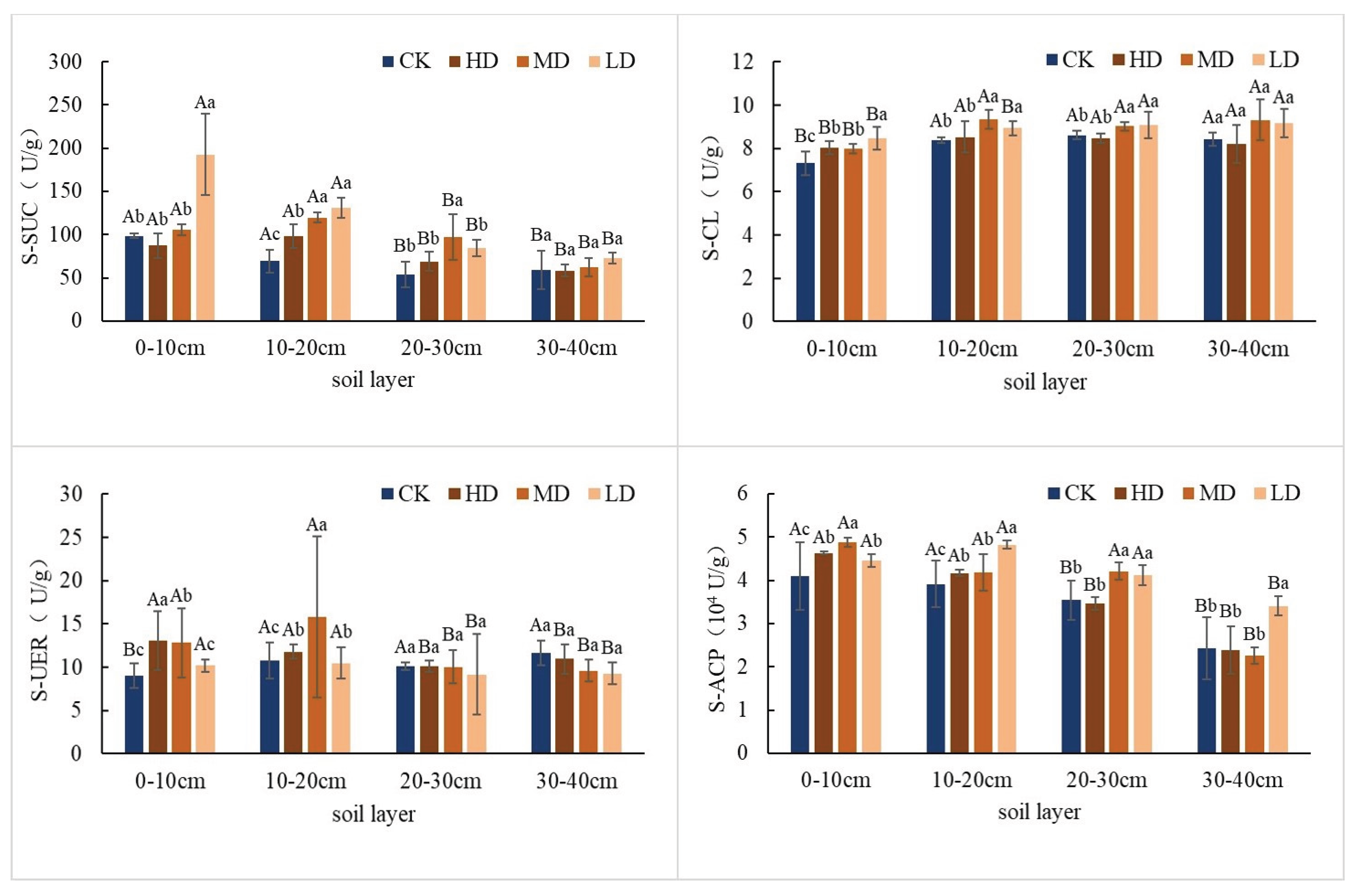
2.1.2. Differences in Soil Enzyme Activity
2.2. Soil Microbial Community Structure in Chinese Fir Plantations Under Different Stand Densities
2.2.1. Differences in Soil Enzyme Activity
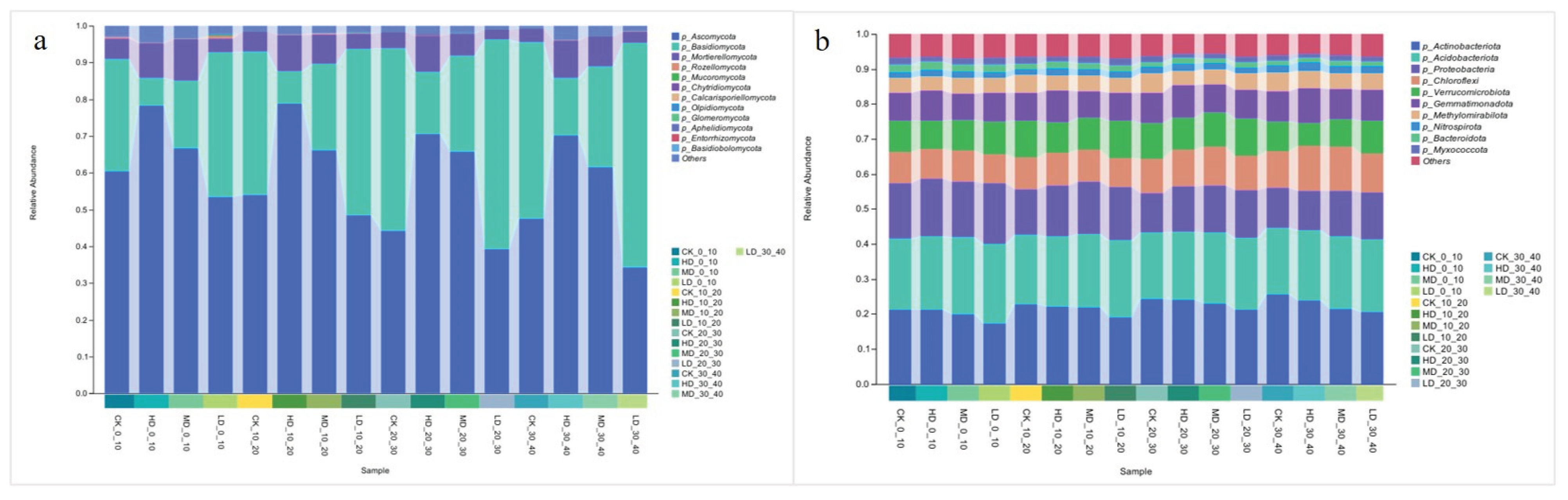
2.2.2. Differences in the Phylum-Level Structure of Soil Microorganisms
2.3. Soil Microbial Diversity Analysis in Larch Plantations Under Different Stand Densities
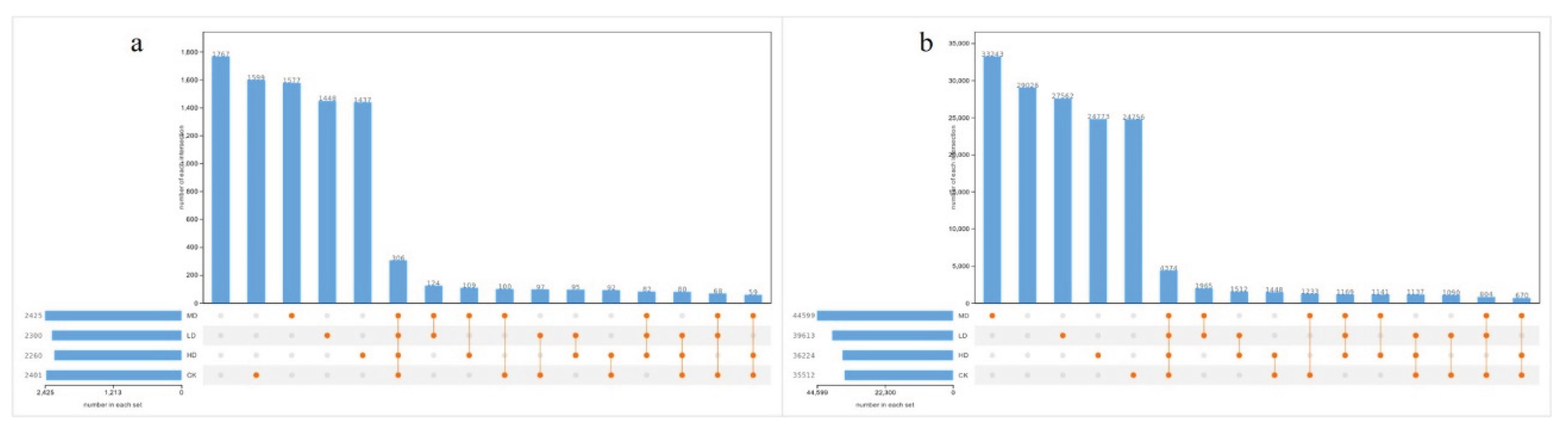
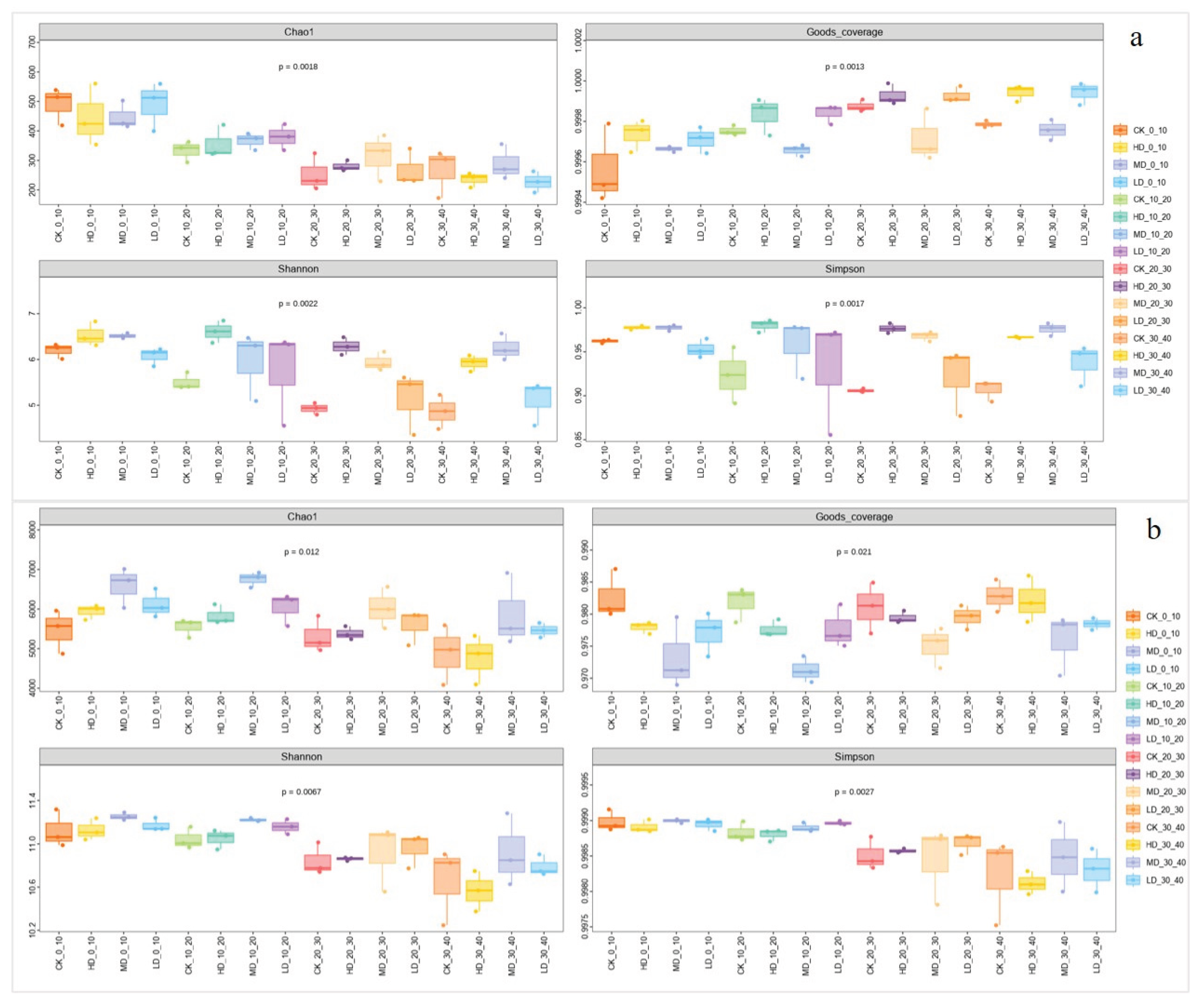
2.3.1. Alpha Diversity Analysis
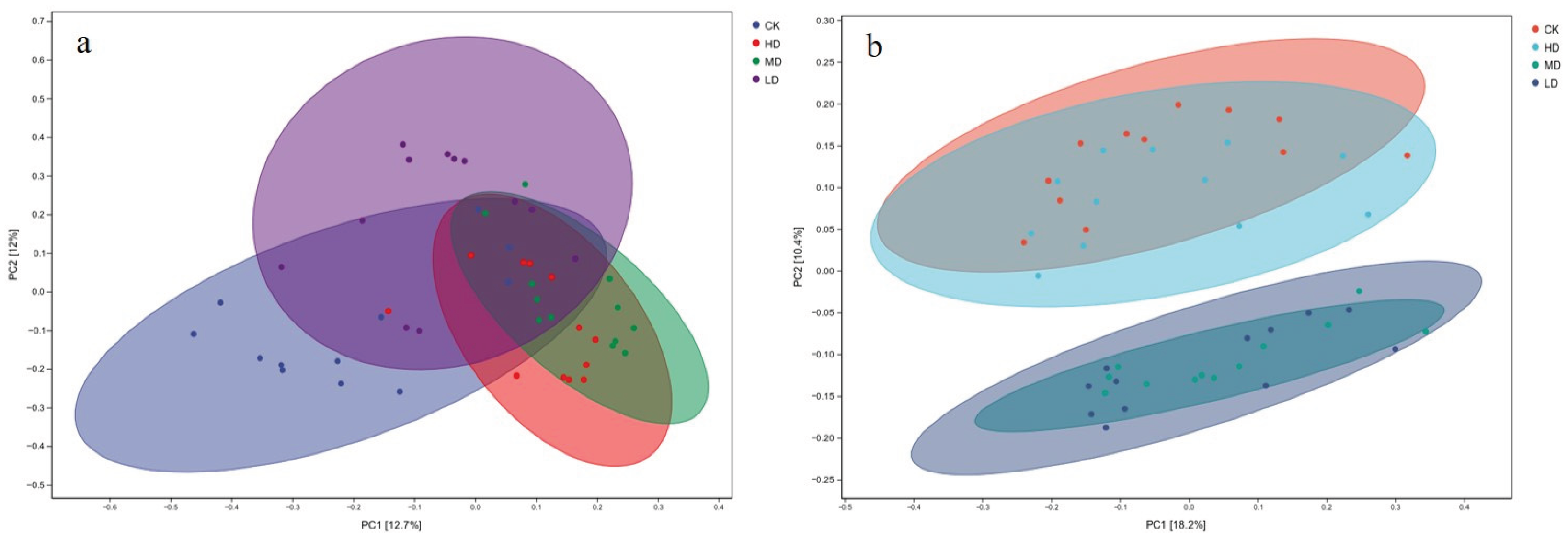
2.3.2. Principal Coordinate Analysis
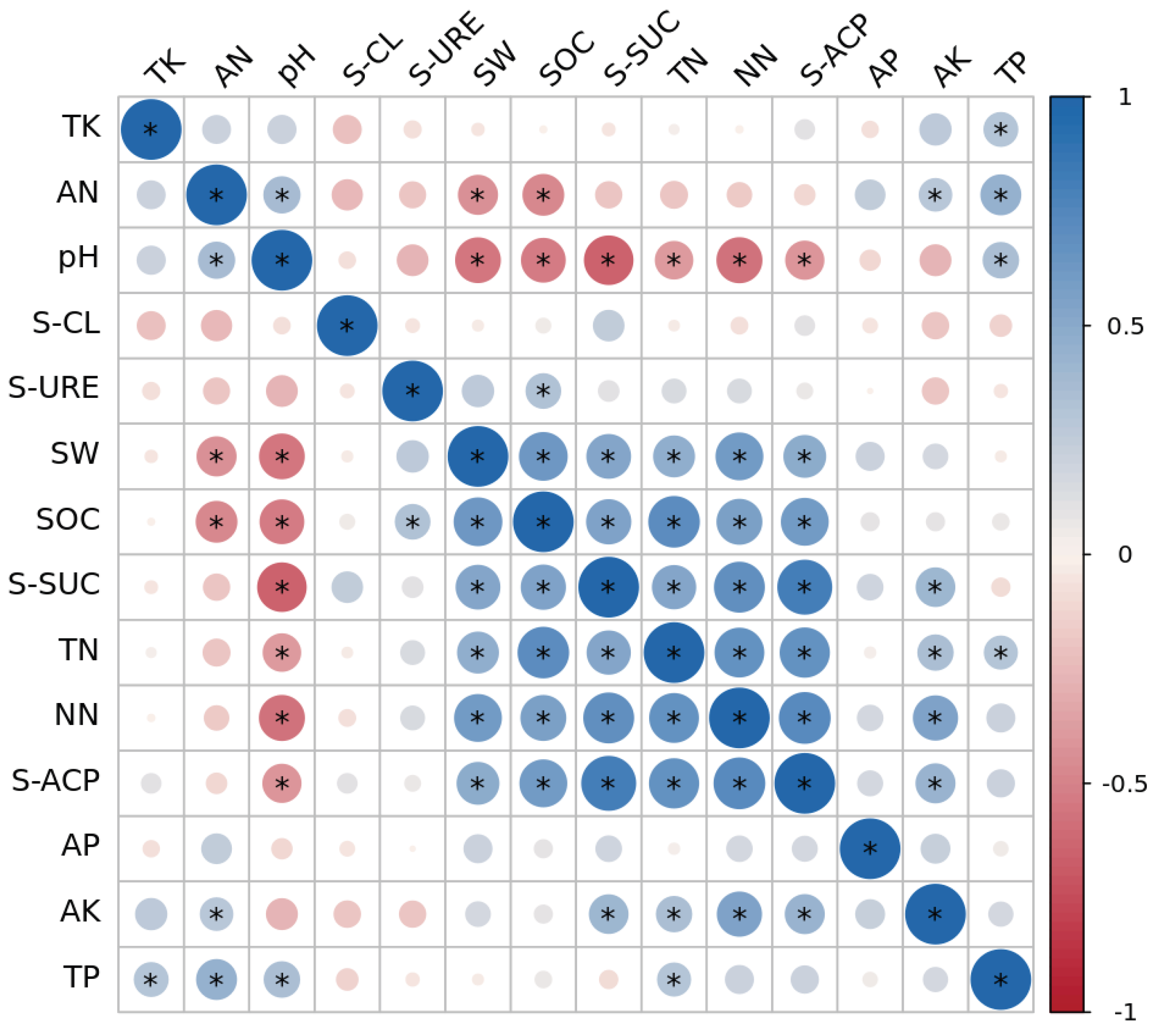
2.4. Correlation Analysis Between Soil Physicochemical Properties and Microbial Communities
3. Discussion
3.1. Effects of Stand Density on Soil Physicochemical Properties
3.2. Effects of Stand Density on Soil Microbial Diversity and Community Composition
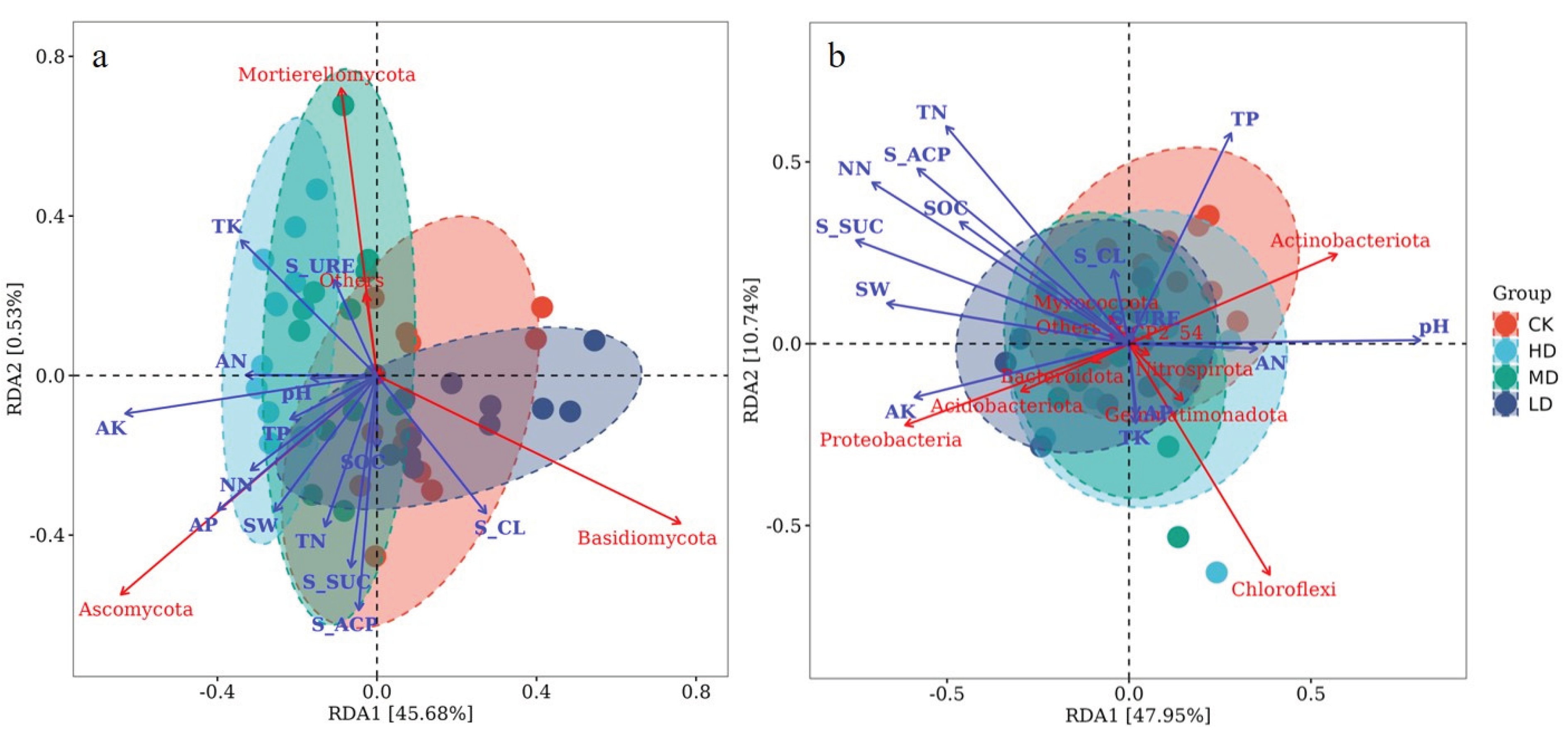
3.3. Relationship Between Soil Microbial Community Structure and Environmental Factors
4. Materials and Methods
4.1. Study Site Description
4.2. Experimental Design and Soil Sampling
| Stand density | Longitude | Latitude | TH (m) | DBH (cm) | UBH (m) | CB (m) |
|---|---|---|---|---|---|---|
| 556 trees ha-1 | 112°04’29’’E | 41°05’11’’N | 15.6±0.35 a | 22.1±0.48 a | 4.2±0.52 b | 4.5±0.31 a |
| 1108 trees ha-1 | 112°03’48’’E | 41°05’36’’N | 15.2±0.88 a | 19.9±0.17 a | 4.4±0.15 b | 4.69±0.81 a |
| 2077 trees ha-1 | 112°04’05’’E | 41°05’23’’N | 14.8±0.15 a | 16.04±0.04 a | 4.9±0.71 b | 3.26±0.37 a |
| 2800 trees ha-1 | 112°03’29’’E | 41°05’33’’N | 14.5±0.12 a | 15.75±0.44 a | 5.3±0.15 a | 2.73±0.64 a |
4.3. Determination of Soil Physicochemical Properties
4.4. DNA Extraction and Illumina Sequencing
4.5. Data Processing g and Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lull, C.; Bautista, I.; Lidón, A.; del Campo, A.D.; González-Sanchis, M.; García-Prats, A. Temporal effects of thinning on soil organic carbon pools, basal respiration and enzyme activities in a Mediterranean Holm oak forest. For. Ecol. Manag. 2020, 464, 118088. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; Huerta-Cepas, J.; Medema, M.H.; Maltz, M.R.; Mundra, S.; Olsson, P.A.; Pent, M.; Põlme, S.; Sunagawa, S.; Ryberg, M. . Tedersoo, L. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Prescott, C.E. Litter decomposition: What controls it and how can we alter it to sequester more carbon in forest soils? Biogeochemistry 2010, 101, 133–149. [Google Scholar] [CrossRef]
- Shen, F.; Jiang, Y.; Yang, Y.; Yang, L. Long-term impacts of stand density on soil fungal and bacterial communities for targeted cultivation of large-diameter Larix olgensis. For. Ecol. Manag. 2025, 591, 122842. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, S.; Li, S.Z.; Deng, Y. Advances in molecular ecology on microbial functional genes of carbon cycle. Microbiol. China 2017, 44, 1676–1689. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, J.H.; Wang, J.Y.; Meng, Z.X.; He, J.; Dong, Z.H.; Liu, K.Q.; Chen, W.Q. Soil microbial richness predicts ecosystem multifunctionality through co-occurrence network complexity in alpine meadow. Acta Ecol. Sin. 2022, 42, 2542–2558. (In Chinese) [Google Scholar] [CrossRef]
- Wang, L.; Deng, D.; Feng, Q.; Xu, Z.; Pan, H.; Li, H. Changes in litter input exert divergent effects on the soil microbial community and function in stands of different densities. Sci. Total Environ. 2022, 845, 157297. [Google Scholar] [CrossRef]
- Xu, Z.; Fan, F.; Lin, Q.; Guo, S.; Li, S.; Zhang, Y.; Feng, Z.; Wang, X.; Rensing, C.; Cao, G.; et al. Effects of Different Stand Densities on the Composition and Diversity of Soil Microbiota in a Cunninghamia lanceolata Plantation. Plants 2025, 14, 98. [Google Scholar] [CrossRef]
- Augusto, L.; Ranger, J.; Binkley, D.; Rothe, A. Impact of several common tree species of European temperate forests on soil fertility. Ann. For. Sci. 2002, 59, 233–253. [Google Scholar] [CrossRef]
- Nave, L.E.; Vance, E.D.; Swanston, C.W.; Curtis, P.S. Harvest impacts on soil carbon storage in temperate forests. For. Ecol. Manag. 2010, 259, 857–866. [Google Scholar] [CrossRef]
- Neilen, A.D.; Chen, C.R.; Parker, B.M.; Faggotter, S.J.; Burford, M.A. Differences in nitrate and phosphorus export between wooded and grassed riparian zones from farmland to receiving waterways under varying rainfall conditions. Sci. Total Environ. 2017, 598, 188–197. [Google Scholar] [CrossRef]
- Li, F.; Wang, Z.; Hou, J.; Li, X.; Wang, D.; Yang, W. The changes in soil organic carbon stock and quality across a subalpine forest successional series. For. Ecosyst. 2024, 11, 100203. [Google Scholar] [CrossRef]
- Fu, M.; Wang, C.; Wang, Y.; Liu, S. Spatiotemporal patterns of soil nitrogen mineralization and nitrification in four temperate forests. Acta Ecol. Sin. 2009, 29, 3747–3758. (In Chinese) [Google Scholar]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Yu, H.; Duan, Y.; Mulder, J.; Dörsch, P.; Zhu, W.; Xu-Ri; Huang, K.; Zheng, Z.; Kang, R.; Wang, C.; Quan, Z.; Zhu, F.; Liu, D.; Peng, S.; Han, S.; Zhang, Y.; Fang, Y. Universal temperature sensitivity of denitrification nitrogen losses in forest soils. Nat. Clim. Chang. 2023, 13, 726–734. [CrossRef]
- Baldrian, P. Distribution of Extracellular Enzymes in Soils: Spatial Heterogeneity and Determining Factors at Various Scales. Soil Sci. Soc. Am. J. 2014, 78, 11–18. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Abarenkov, K. . +48 authors Global diversity and geography of soil fungi. Science 2014, 346. [Google Scholar] [CrossRef]
- Ding, K.; Zhang, Y.T.; Zhang, J.H.; Chai, X.; Zhou, S.S.; Tong, Z.K. Effects of Chinese fir plantations with different densities on understory vegetation and soil microbial community structure. Chin. J. Plant Ecol. 2021, 45, 62–73. [Google Scholar] [CrossRef]
- Yang, A.; Lu, Y.; Zhang, J.; Wu, J.; Xu, J.; Tong, Z. Changes in Soil Nutrients and Acidobacteria Community Structure in Cunninghamia lanceolata Plantations. Sci. Silvae Sin. 2019, 55, 119–127. [Google Scholar] [CrossRef]
- Peng, J.; Liu, H.; Hu, Y.; Sun, Y.; Liu, Q.; Li, J.; Dong, Y. Shift in soil bacterial communities from K- to r-strategists facilitates adaptation to grassland degradation. Land Degrad. Dev. 2022; Article e4304. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in tropical rain forests and coral reefs. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef]
- Tedersoo, L.; Mikryukov, V.; Anslan, S.; Bahram, M.; Khalid, A.N.; Corrales, A.; Agan, A.; Vasco-Palacios, A.M.; Saitta, A.; Antonelli, A.; Rinaldi, A.C.; Verbeken, A.; Sulistyo, B.P.; Tamgnoue, B.; Furneaux, B.; Ritter, C.D.; Nyamukondiwa, C.; Sharp, C.; Marín, C. . Alberdi, A. The Global Forest Fungal Community: Composition, Functions, and Threat Response. Cell 2022, 185, 4250–4267. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I. . Parfrey, L.W. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Erickson, D.W.; Schink, S.J.; Patsalo, V.; Williamson, J.R.; Gerland, U.; Hwa, T. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 2017, 551, 119–123. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Štursová, M.; Žifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Hai, L.; Zhou, M.; Zhao, K.; Hong, G.; Li, Z.; Liu, L.; Gao, X.; Li, Z.; Li, F. Soil Microbial Communities and Their Relationship with Soil Nutrients in Different Density Pinus sylvestris var. mongolica Plantations in the Mu Us Sandy Land. Forests 2025, 16, 547. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J. . Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Camenzind, T.; Hättenschwiler, S.; Treseder, K.K.; Lehmann, A.; Rillig, M.C. Nutrient limitation of soil microbial processes in tropical forests. Ecol. Monogr. 2018, 88, 4–21. [Google Scholar] [CrossRef]
- Zhu, R.; Jin, L.; Sang, Y.; Hu, S.; Wang, B.T.; Jin, F.J. Characterization of potassium-solubilizing fungi, Mortierella spp., isolated from a poplar plantation rhizosphere soil. Arch. Microbiol. 2024, 206, 157. [Google Scholar] [CrossRef]
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, Å.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef]
- Margalef, O.; Sardans, J.; Fernández-Martínez, M.; Molowny-Horas, R.; Janssens, I.A.; Ciais, P. . Peñuelas, J. Global patterns of phosphatase activity in natural soils. Sci. Rep. 2017, 7, 1337. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Oger, P.; Lepleux, C.; Collignon, C.; Frey-Klett, P.; Turpault, M.P. Bacterial weathering and its contribution to nutrient cycling in temperate forest ecosystems. Res. Microbiol. 2011, 162, 820–831. [Google Scholar] [CrossRef]
- Prescott, C.E.; Grayston, S.J. Tree species influence on microbial communities in litter and soil: Current knowledge and research needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Krah, F.-S.; Bässler, C.; Heibl, C.; Soghigian, J.; Schaefer, H.; Hibbett, D.S. Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evol. Biol. 2018, 18, 119. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M. . Tedersoo, L. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Richardson, A.E.; Simpson, R.J. Soil microorganisms mediating phosphorus availability. Plant Physiol. 2011, 156, 989–996. [Google Scholar] [CrossRef]
- Zhou, J.; Guan, D.; Zhou, B.; Zhao, B.; Ma, M.; Qin, J. . Li, J. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 2020, 90, 42–51. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Dai, T.; Zhang, Y.; Tang, Y.; Bai, Y.; Tao, Y.; Huang, B.; Wen, D. Identifying the key taxonomic categories that characterize microbial community diversity using full-scale classification: a case study of microbial communities in the sediments of Hangzhou Bay. FEMS Microbiol. Ecol. 2016, 92, fiw150. [Google Scholar] [CrossRef]
- Naylor, D.; DeGraaf, S.; Purdom, E. . Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J. . Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2012, 7, 2069–2079. [Google Scholar] [CrossRef]
- Arcand, M.M.; Helgason, B.L.; Lemke, R.L. Microbial crop residue decomposition dynamics in organic and conventionally managed soils. Appl. Soil Ecol. 2016, 107, 347–359. [Google Scholar] [CrossRef]
- Liu, C.S.; Zhao, D.F.; Ma, W.J.; Guo, Y.D.; Wang, A.J.; Wang, Q.L.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef]
- Srinivasan, S.; Hoffman, N.G.; Morgan, M.T.; Matsen, F.A.; Fiedler, T.L.; Hall, R.W.; Ross, F.J.; McCoy, C.O.; Bumgarner, R.; Marrazzo, J.M.; Fredricks, D.N. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 2012, 7, e37818. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.H.; Yin, S.X.; Liu, X.; Zhang, W.M.; Gu, T.Y.; Shen, Q.R.; Qiu, H.Z. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; Bai, Y.; Bisanz, J.E.; Bittinger, K.; Brejnrod, A.; Brislawn, C.J.; Brown, C.T.; Callahan, B.J.; Caraballo-Rodríguez, A.M.; Chase, J.; Cope, E.K.; Da Silva, R.; Diener, C.; Dorrestein, P.C.; Douglas, G.M.; Durali, D.M.; Duvallet, C.; Edwardson, C.F.; Ernst, M.; Estaki, M.; Fouquier, J.; Gauglitz, J.M.; Gibbons, S.M.; Gibson, D.L.; Gonzalez, A.; Gorlick, K.; Guo, J.; Hillmann, B.; Holmes, S.; Holste, H.; Huttenhower, C.; Huttley, G.A.; Janssen, S.; Jarmusch, A.K.; Jiang, L.; Kaehler, B.D.; Kang, K.B.; Keefe, C.R.; Keim, P.; Kelley, S.T.; Knights, D.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




