Submitted:
27 October 2025
Posted:
28 October 2025
You are already at the latest version
Abstract
Background and Objectives: Differentiating between subtypes of renal cell carcinoma (RCC) can be challenging due to overlapping immunohistochemical and histomorphological features. Furthermore, despite the evaluation of numerous histopathologic parameters, predicting metastasis and prognosis remains unclear. In this study, we aimed to evaluate the potential of microRNA (miRNA) expression levels in establishing definitive diagnoses, assessing metastatic potential, and determining patient prognosis. Materials and Methods: A total of 35 clear cell RCC (cc-RCC), 11 chromophobe RCC (ch-RCC), 9 papillary RCC (p-RCC) cases, and 9 cc-RCC metastases were retrospectively analyzed. MiRNA-21 and miRNA-210 expression profiles were assessed using non-parametric tests, receiver operating characteristic (ROC) analysis, and Cox regression models. Results: The expression levels of both miRNA-21 and miRNA-210 were significantly elevated in cc-RCC and p-RCC cases. In contrast, miRNA-210 expression levels were significantly reduced in all ch-RCC cases, whereas miRNA-21 expression showed no significant change. Although miRNA-21 expression levels were higher in metastases compared to primary cc-RCC tumors, the difference was not statistically significant (p=0.053). No significant difference was observed in miRNA-210 expression between metastatic and primary cc-RCC tumors (p=0.237). Regardless of histological subtype, an increase in miRNA-210 expression was associated with a significant reduction in overall survival (p=0.03, HR=1.729, 95% CI: 1.043-2.864). Conclusion: The findings suggest that miRNA-21 and miRNA-210 expression levels can help distinguish ch-RCC from cc-RCC and p-RCC. Additionally, miRNA-210, but not miRNA-21, may differentiate cc-RCC from p-RCC. Higher miRNA-210 expression levels were associated with worse overall survival in RCC patients. While miRNA-21 levels were increased in cc-RCC metastases, further studies with larger sample sizes are needed to establish its role in predicting metastasis.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Case Selection and Preparation
2.2. Total RNA Isolation
2.3. Complementary DNA Synthesis from Total RNA
2.4. Evaluation of MiRNA Expression by Quantitative Real-Time PCR
2.5. Normalization of MiRNA Expression and Determination of Expression Profiles
2.6. Statistical Analysis
- The normality of miRNA-21 and miRNA-210 expression levels was assessed using the Kolmogorov-Smirnov test, and homogeneity of variances was evaluated by the Levene test.
- Differences in miRNA expression levels among histological subtypes and between primary tumors and metastases were analyzed using one-way ANOVA, Tukey’s HSD test, and paired t-tests, respectively.
- Correlations in parametric data were assessed with the Pearson correlation test.
- The diagnostic performance of miRNA expression levels for histological subtyping and metastatic potential was evaluated using receiver operating characteristic (ROC) curve analysis.
- The association between overall survival and miRNA expression levels was determined using the Cox regression model.
2.7. Ethical Approval
3. Results
3.1. Clinicopathological Features
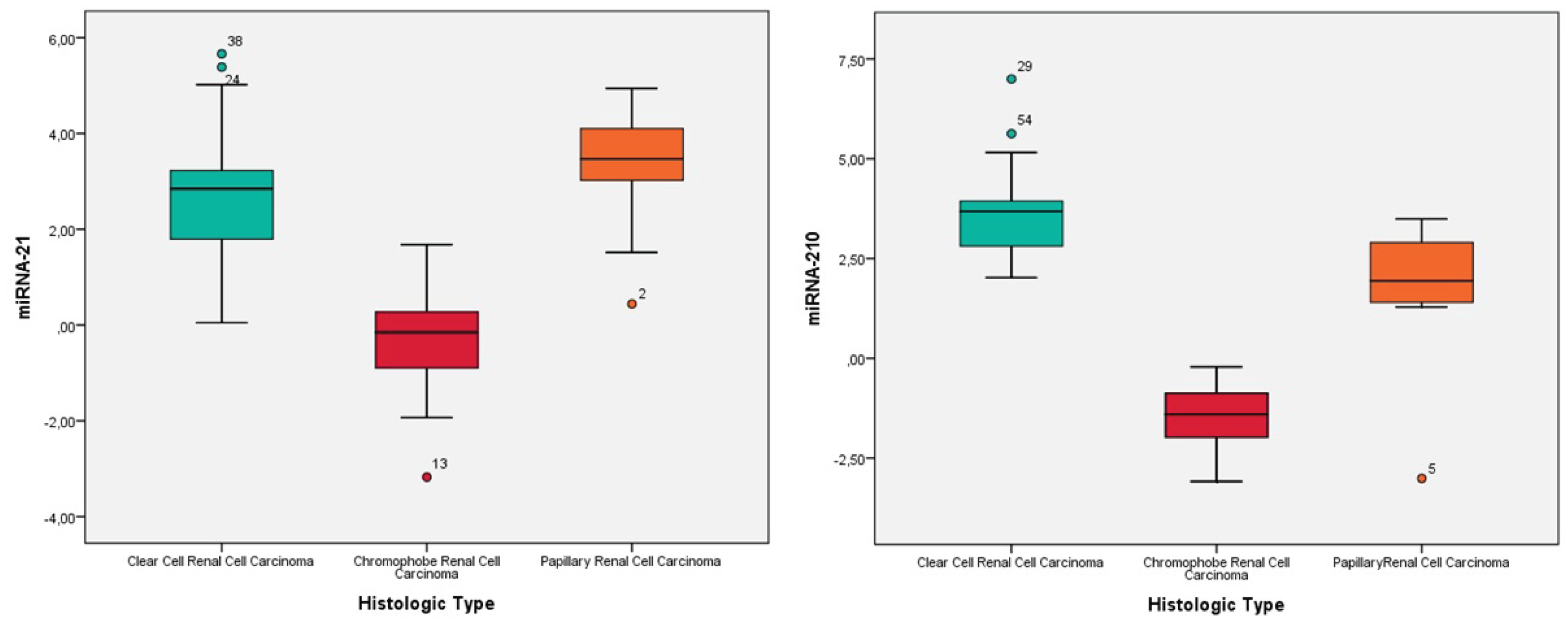
3.2. Expression Levels of MiRNA-21 and MiRNA-210 in Renal Cell Carcinomas
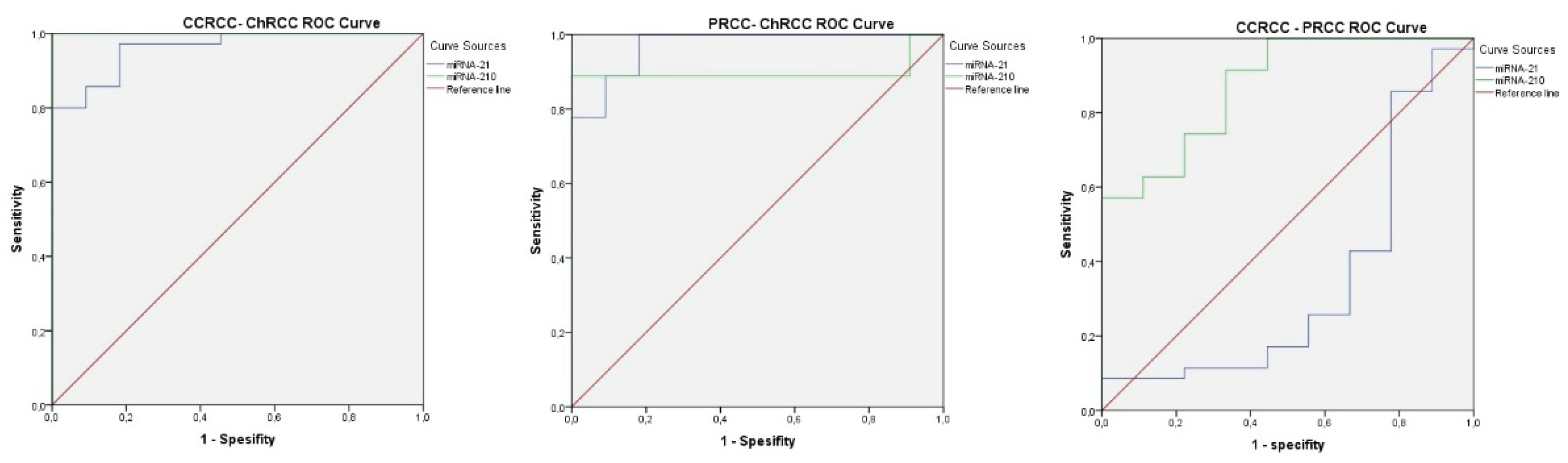
3.3. Histological Subtype Determination Based on MiRNA-21 and MiRNA-210 Expression Levels
- To differentiate cc-RCC from ch-RCC, the cutoff values were 1.72 for miRNA-21 (AUROC = 0.961, 95% CI: 0.909–1.000, p < 0.001) and 0.90 for miRNA-210 (AUROC = 1.000, 95% CI: 1.000–1.000, p < 0.001). Sensitivity and specificity for miRNA-21 and miRNA-210 were 80% and 100%; and 100% and 100%, respectively. (see Figure 2A, Supplementary S4).
- To distinguish p-RCC from ch-RCC, the cutoff values were 0.44 for miRNA-21 (AUROC = 0.970, 95% CI: 0.906–1.000, p < 0.001) and 0.54 for miRNA-210 (AUROC = 0.899, 95% CI: 0.711–1.000, p = 0.003). Sensitivity and specificity for miRNA-21 and miRNA-210 were 100% and 82%, and 88% and 100%, respectively. (see Figure 2B, Supplementary S4).
- To differentiate cc-RCC from p-RCC, the cutoff value for miRNA-210 was 0.54 (AUROC = 0.873, 95% CI: 0.749–0.997, p = 0.001). Sensitivity and specificity were 91% and 67%, respectively. MiRNA-21 did not show a statistically significant distinction (p = 0.150). (see Figure 2C, Supplementary S4).
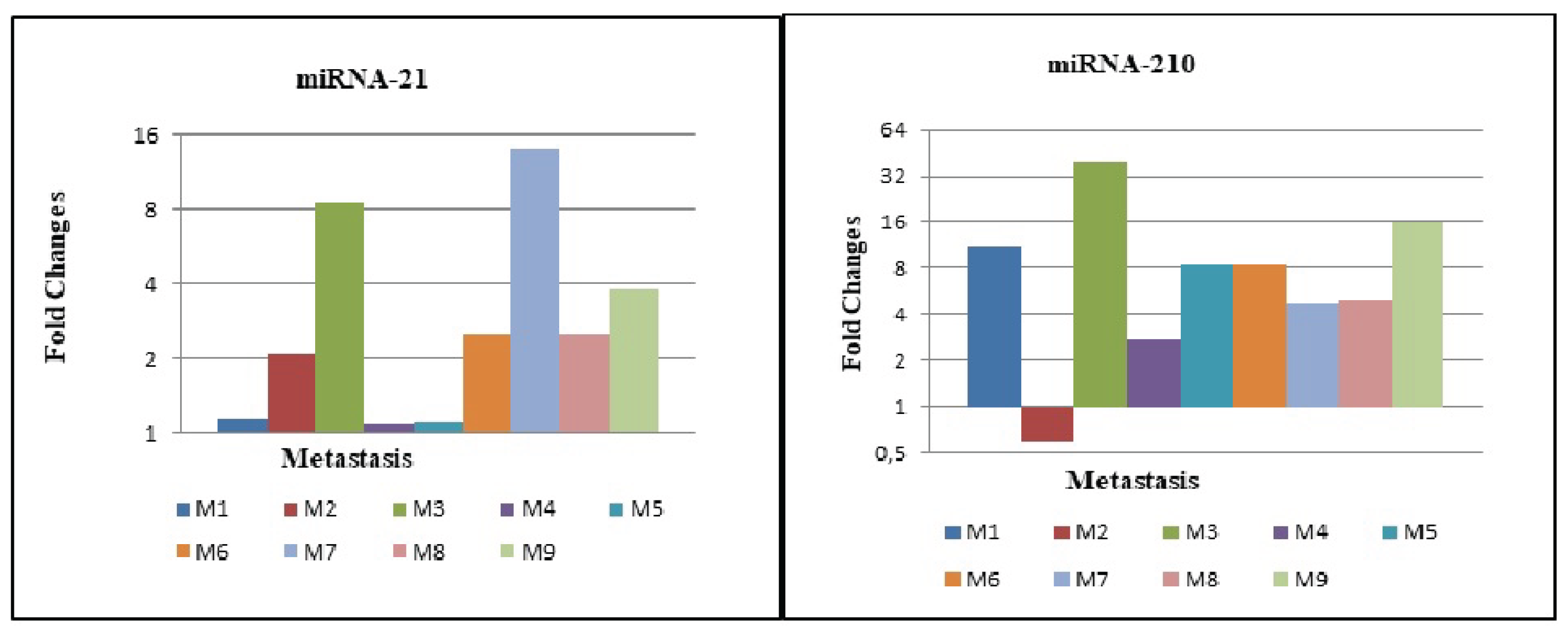
3.4. Expression Levels of MiRNA-21 and MiRNA-210 in Metastatic cc-RCC
3.5. Relationship Between MiRNA-21 and MiRNA-210 Expression Levels and Survival Time
- Clear Cell Renal Cell Carcinoma (ccRCC): 35 patients were included in this group. The mean follow-up time was 87.3 ± 69.7 months, with a mortality rate of 37.1%.
- Chromophobe Renal Cell Carcinoma (chRCC): 11 patients were analyzed in this group. The mean follow-up time was 101.5 ± 30.1 months, with a mortality rate of 9.1%.
- Papillary Renal Cell Carcinoma (pRCC): 9 patients were included in this group. The mean follow-up time was 81.1 ± 51.1 months, with a mortality rate of 33.3%.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moch H, Hoon Tan P, Amin B M, Turajlic S. Renal cell tumours, Introduction. In: Who Classification of Tumors Editorial Board. WHO Classification of Tumours of the Urinary System and Male Genital Organs. 5th Edition ed. Lyon, France: IARC; 2022.
- Zhou M, Hattab MA, Eble JN, Cheng L. Neoplasms of the Kidney. In: Zhou M, Magi-Galluzzi C, editors. Genitourinary Pathology Foundations in Diagnostic Pathology. 2nd Edition ed. Philadephia,PA: Saunders, Elsevier; 2015. p. 306-77.
- Delahunt B, Algaba F, Eble J, Cheville J, Amin MB, Argani P, et al. Papillary renal cell carcinoma. In: Moch H, Humphrey PA, Ulbright TM, Reuter VE, editors. WHO Classification of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC; 2016. p. 23-5.
- Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan F, et al. Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS One. 2014;9(2):e89223. [CrossRef]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999-3004. [CrossRef]
- Heinzelmann J, Unrein A, Wickmann U, Baumgart S, Stapf M, Szendroi A, et al. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: a comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21(3):1046-54. [CrossRef]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-8. [CrossRef]
- McCormick RI, Blick C, Ragoussis J, Schoedel J, Mole DR, Young AC, et al. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer. 2013;108(5):1133-42. [CrossRef]
- Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13(9B):3918-28. [CrossRef]
- Gordanpour A, Nam RK, Sugar L, Seth A. MicroRNAs in prostate cancer: from biomarkers to molecularly-based therapeutics. Prostate Cancer Prostatic Dis. 2012;15(4):314-9. [CrossRef]
- Hong L, Han Y, Zhang Y, Zhang H, Zhao Q, Wu K, et al. MicroRNA-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets. 2013;17(9):1073-80. [CrossRef]
- Meng K, Li Z, Cui X. Three LHPP gene-targeting co-expressed microRNAs (microRNA-765, microRNA-21, and microRNA-144) promote proliferation, epithelial-mesenchymal transition, invasion, and are independent prognostic biomarkers in renal cell carcinomas patients. J Clin Lab Anal. 2021;35:e24077. [CrossRef]
- Fan B, Jin Y, Zhang H, Zhao R, Sun M, Sun M, et al. MicroRNA-21 contributes to renal cell carcinoma cell invasiveness and angiogenesis via the PDCD4/c-Jun (AP-1) signalling pathway. Int J Oncol. 2020;56(1):178-192. [CrossRef]
- Vergho DC, Kneitz S, Kalogirou C, Burger M, Krebs M, Rosenwald A, et al. Impact of miR-21, miR-126 and miR-221 as prognostic factors of clear cell renal cell carcinoma with tumor thrombus of the inferior vena cava. PLoS One. 2014;9(10):e109877. [CrossRef]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647-58. [CrossRef]
- Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, et al. MiRNA-210: A Current Overview. Anticancer Res. 2017;37(12):6511-21. [CrossRef]
- Valera VA, Walter BA, Linehan WM, Merino MJ. Regulatory Effects of microRNA-92 (miR-92) on VHL Gene Expression and the Hypoxic Activation of miR-210 in Clear Cell Renal Cell Carcinoma. J Cancer. 2011;2:515-26. [CrossRef]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101-8.
- Silva-Santos RM, Costa-Pinheiro P, Luis A, Antunes L, Lobo F, Oliveira J, et al. MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer. 2013;109(10):2646-53. [CrossRef]
- Gu L, Li H, Chen L, Ma X, Gao Y, Li X, Zhang Y. MicroRNAs as prognostic molecular signatures in renal cell carcinoma. Oncotarget. 2015;6(6):4536–4549. [CrossRef]
- Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang Y, Shen D. MicroRNA-21 contributes to renal cell carcinoma progression via PTEN/PI3K/AKT pathway modulation. Oncol Rep. 2019;41(1):89–98.
- Petrozza V, et al. Emerging role of secreted miR-210-3p as potential biomarker for clear cell renal cell carcinoma metastasis. Cancer Biomark. 2020;28(4):441–449.
- Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2019;11(16):5730–5739. [CrossRef]
- Wang Z, et al. miRNA-21 promotes renal carcinoma cell invasion by targeting TIMP3 and regulating the MMP2 pathway. RSC Adv. 2017;7(43):27086–27095.
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046-54. [CrossRef]
- Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, Mejia-Guerrero S, et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn. 2012;14(4):385-92. [CrossRef]
- Zaman MS, Shahryari V, Deng G, Thamminana S, Saini S, Majid S, et al. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7(2):e31060. [CrossRef]
- Vergho D, Kneitz S, Rosenwald A, Scherer C, Spahn M, Burger M, et al. Combination of expression levels of miR-21 and miR-126 is associated with cancer-specific survival in clear-cell renal cell carcinoma. BMC Cancer. 2014;14:25. [CrossRef]
- Gunawan RR, Astuti I, Danarto HR. miRNA-21 as High Potential Prostate Cancer Biomarker in Prostate Cancer Patients in Indonesia. Asian Pac J Cancer Prev. 2023;24(3):1095-1099. [CrossRef]
- Speicher MR, Schoell B, du Manoir S, Schrock E, Ried T, Cremer T, et al. Specific loss of chromosomes 1, 2, 6, 10, 13, 17, and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridization. Am J Pathol. 1994;145(2):356-64.
- Gilyazova I, Ivanova E, Izmailov A, Sharifgaliev I, Karunas A, Pudova E, et al. MicroRNA Expression Signatures in Clear Cell Renal Cell Carcinoma: High-Throughput Searching for Key miRNA Markers in Patients from the Volga-Ural Region of Eurasian Continent. Int J Mol Sci. 2023;24(8):6909. [CrossRef]
- Ge YZ, Xin H, Lu TZ, Xu Z, Yu P, Zhao YC, et al. MicroRNA expression profiles predict clinical phenotypes and prognosis in chromophobe renal cell carcinoma. Sci Rep. 2015;5:10328. [CrossRef]
- Vitruk YV, Semko SL, Voylenko OA, Pikul MV, Borikun TV, Zadvornyi TV, et al. Evaluation of response to tyrosine kinase inhibitors in renal cell carcinoma patients based on expression of miR-99b, -144, -210, -222, -302a and -377 in tumor tissue. Exp Oncol. 2021;43(2):98–103. [CrossRef]
- Samaan S, Khella HW, Girgis A, Scorilas A, Lianidou E, Gabril M, et al. miR-210 is a prognostic marker in clear cell renal cell carcinoma. J Mol Diagn. 2015;17(2):136-44. [CrossRef]
- Fridman E, Dotan Z, Barshack I, David MB, Dov A, Tabak S, et al. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn. 2010;12(5):687-96. [CrossRef]
- Lokeshwar SD, Talukder A, Yates TJ, Hennig MJP, Garcia-Roig M, Lahorewala SS, et al. Molecular Characterization of Renal Cell Carcinoma: A Potential Three-MicroRNA Prognostic Signature. Cancer Epidemiol Biomarkers Prev. 2018;27(4):464-72. [CrossRef]
- Gowrishankar B, Ibragimova I, Zhou Y, Slifker MJ, Devarajan K, Al-Saleem T, et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15(3):329–341. [CrossRef]
- Kajdasz A, Majer W, Kluzek K, Sobkowiak J, Milecki T, Derebecka N, et al. Identification of RCC Subtype-Specific microRNAs–Meta-Analysis of High-Throughput RCC Tumor microRNA Expression Data. Cancers (Basel). 2021;13(3):548. [CrossRef]
| HISTOLOGIC TYPE | WHO/ ISUP GRADE | TOTAL | |||||
| 1 | 2 | 3 | 4 | ||||
| Clear Cell RCC* | Pathologic Stage | T1a | 2 | 5 | 6 | 0 | 13 (37,1%) |
| T1b | 0 | 3 | 5 | 0 | 8 (22,9%) | ||
| T2a | 0 | 0 | 1 | 0 | 1 (2,9%) | ||
| T3a | 0 | 1 | 5 | 5 | 11 (31,4%) | ||
| T4 | 0 | 0 | 0 | 2 | 2 (5,7%) | ||
| Total | 2 (5,7%) | 9 (25,7%) | 17 (48,6%) | 7 (20%) | 35 (100%) | ||
| PapillaryRCC* | Pathologic Stage | T1b | 4 | 1 | 0 | 5 (55,6%) | |
| T2a | 0 | 2 | 1 | 3 (33,3%) | |||
| T3a | 1 | 0 | 1 (11,1%) | ||||
| Total | 4 (44,4%) | 4 (44,4%) | 1 (11,2%) | 9 (100%) | |||
| Chromophobe RCC* | Pathologic Stage |
T1a T1b T2a T3a |
3 2 5 1 |
3 (27,3%) | |||
| 2 (18,2%) | |||||||
| 5 (45,4%) | |||||||
| 1 (9,1%) | |||||||
| Total | 11 | 11 (100%) | |||||
| Total | Pathologic Stage | T1a | 2 | 5 | 6 | 0 | 13 (29,5%) |
| T1b | 0 | 7 | 6 | 0 | 13 (29,5%) | ||
| T2a | 0 | 0 | 3 | 1 | 4 (9,1%) | ||
| T3a | 0 | 1 | 6 | 5 | 12 (27,3%) | ||
| T4 | 0 | 0 | 0 | 2 | 2 (4,6%) | ||
| Total | 2 (4,6%) | 13 (29,5%) | 21 (47,7%) | 8 (18,2%) | 44 (100%) | ||
| Histological Type | miRNA-21 expression | N (%) | P value | miRNA-210 expression | N(%) | P value |
|---|---|---|---|---|---|---|
| Clear Cell RCC* | Increased expression | 34(97.1%) | <0,001 | Increased expression | 35(100%) | |
| Unvarying expresison | 1(2.9%) | |||||
| Papillary RCC* | Increased expression | 9 (100%) | Decreased expression | 8(88.9%) | 0.02 | |
| Decresased expression | 1(11.1%) | |||||
| Chromofobe RCC* | Increased expression | 5(45.5%) | 0.763 | Decreased expression | 11 (%100) | |
| Decreased expression | 6(55.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





