Submitted:
21 October 2025
Posted:
22 October 2025
You are already at the latest version
Abstract
Keywords:
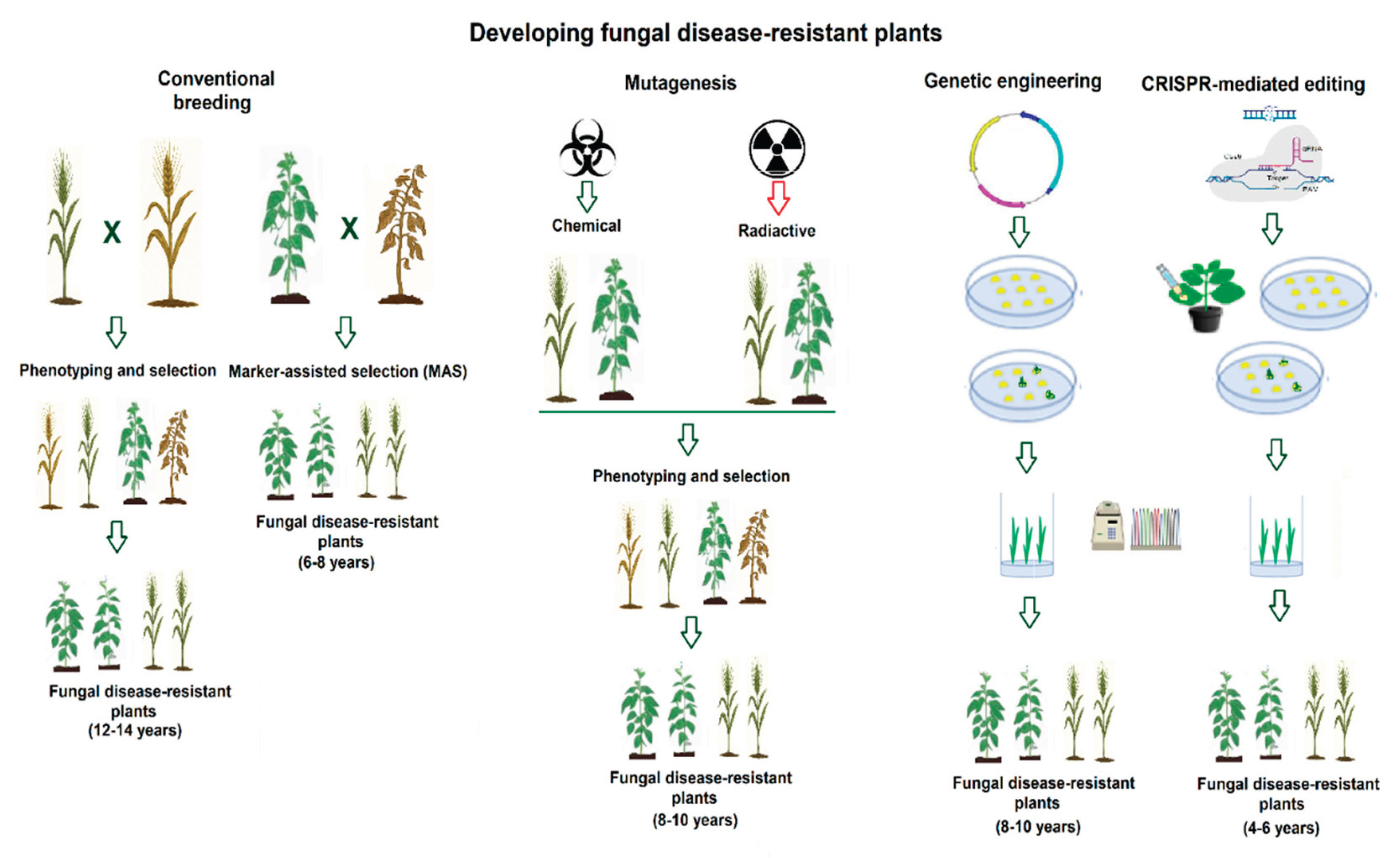
1. Introduction
2. CRISPR–Cas Genome Editing to Improve Fungal-Disease Resistance in Main Cereal Crops
2.1. Wheat
2.2. Rice
| Pathogen | Target Gene | Delivery Method | Type of Editing | References |
| Wheat | ||||
| Fusarium graminearum | Fhb1 | Particle bombardment | deletion mutation | [27] |
| Fusarium graminearum | TaHRC |
Agrobacterium- mediated transformation |
BSMV-mediated gene editing | [28] |
| Fusarium graminearum | TaHRC |
Agrobacterium- mediated transformation |
Knockout | [29] |
| Fusarium graminearum | TaNFXL1 |
Agrobacterium- mediated transformation |
virus-induced gene silencing | [30] |
| Parastagonospora nodorum | Tsn1 and Snn5 | Cas9-RNP mediated editing | Knockout | [32] |
| Parastagonospora nodorum | Tox3 | ribonuclear protein (RNP) complex targeting | Knockout | [33] |
| Blumeria graminis f. sp. tritici | MLO-B1 | Cas9-RNP mediated editing | Deletion | [35] |
| Blumeria graminis f.sp. tritici | TaMLO | Particle bombardment | Knockout | [37] |
| Blumeria graminis f. sp. tritici | TaEDR1 | Particle bombardment | Knockout | [36] |
|
Blumeria graminis f. sp. tritici Puccinia striiformis f. sp. tritici |
TaMKP1 |
Agrobacterium- mediated transformation |
Knockout | [41] |
| Puccinia striiformis f. sp. tritici | TaPsIPK1 | Agrobacterium- | Knockout | [42] |
| Puccinia triticina Erikss | TaGW2 | mediated transformation | Knockout | [43] |
| Rice | ||||
| Magnaporthe oryzae | ALB1 and RSY1 | RNP targeting | RSY1 | [45] |
| Magnaporthe oryzae | OsERF922 | Protoplast transformation | Mutation | [46] |
| Magnaporthe oryzae | OsSEC3A | Protoplast transfection | Mutation | [47] |
| Magnaporthe oryzae | Rei1, Ppg1, Bip1, Bip2, Dbf2 | Protoplast transformation | Mutation | [48] |
| Magnaporthe oryzae | Pi21 |
Agrobacterium- mediated transformation |
Knockout | [49] |
| Magnaporthe oryzae | Pita, Pi21 and ERF922 |
Agrobacterium- mediated transformation |
Knockout | [50] |
| Magnaporthe oryzae | TMS5,Pi21 иXa13 |
Agrobacterium- mediated transformation |
Mutation | [51] |
| Magnaporthe oryzae | Pi21 иOsSULTR3;6 |
Agrobacterium- mediated transformation |
Knockout | [52] |
| Magnaporthe oryzae | Bsr-d1, Pi21иERF922 |
Agrobacterium- mediated transformation |
Knockout | [53] |
| Magnaporthe oryzae | Pi21 |
Agrobacterium- mediated transformation |
Knockout | [54] |
| Magnaporthe oryzae | Pid3 |
Agrobacterium- mediated transformation |
Knockout | [55] |
| Magnaporthe oryzae | Bsr-d1, Perox3 |
Agrobacterium- mediated transformation |
Knockout | [56] |
| Magnaporthe oryzae, Ustilaginoidea virens | MoATG3,MoATG7 иUvPal1 |
Agrobacterium- mediated transformation |
Knockout | [57] |
| Magnaporthe oryzae | rBE5 |
Agrobacterium- mediated transformation |
Base editing | [58] |
2.3. Barley
3. CRISPR-Cas Genome Editing for Improvement Fungal Disease Resistance of Legumes
| Pathogen | Target Gene | Delivery Method | Type of Editing | References |
| Soybean | ||||
| Erysiphe diffusa | GmMLO02, GmMLO19, GmMLO20, GmMLO23 | Agrobacterium-mediated transformation | Knockout | [65] |
| Phytophthora sojae | RXLR Avr4/6 | (PEG)-mediated protoplast transformations | Knockout | [67] |
4. CRISPR-Cas Genome Editing for Improvement Fungal Disease Resistance of Vegetables
4.1. Tomato
4.2. Pepper
4.3. Eggplant
5. Challenges, Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas | CRISPR-associated protein |
| FHB | Fusarium head blight |
| BSMV | Barley mosaic virus |
| VIGS | Virus-induced gene silencing |
| SNB | Septoria nodorum blotch |
| MLO | Powdery mildew locus O |
| CRISPRi | CRISPR interference |
| BYDV | Barley yellow dwarf virus |
| FAO | Food and Agriculture Organization of the United Nations |
| DMR6 | Downy mildew resistance 6 |
| DND1 | Defense no death 1 |
| DCL1 | Dicer-like1 |
| NBS-LRR | Nucleotide-binding site |
| LRR | Leucine-rich repeat |
| XSP10 | Xylem sap protein 10 |
| SlSAMT | Salicylic acid methyltransferase |
| LTP | Lipid transfer protein |
| SA | Salicylic acid |
| MeSA | Methyl salicylate |
| SAM | S-adenosyl-L-methionine |
| SlPG2a | Polygalacturonase |
| SlPL | Pectate lyase |
References
- The United Nations. World population projected to reach 9.8 billion in 2050, and 11.2 billion in 2100. Available online: URL Available online: https://www.un.org/en/desa/world-population-projected-reach-98-billion-2050-and-112-billion-2100. (accessed on 2 October 2025).
- Food and Agriculture Organization. Plant Production and Protection. Available online: URL Available online: https://www.fao.org/plant-production-protection/about/en. (accessed on 2 October 2025).
- Mikaberidze, A.; Gokhale, C.S.; Bargués-Ribera, M.; Verma, P. The Cost of Fungicide Resistance Evolution in Multi-Field Plant Epidemics. PLOS Sustainability and Transformation 2025, 4. [CrossRef]
- Wang, N.; Sundin, G.W.; De La Fuente, L.; Cubero, J.; Tatineni, S.; Brewer, M.T.; Zeng, Q.; Bock, C.H.; Cunniffe, N.J.; Wang, C.; et al. Key Challenges in Plant Pathology in the Next Decade. Phytopathology 2024, 114, 837–842.
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The Most Important Fungal Diseases of Cereals—Problems and Possible Solutions. Agronomy 2021, 11. [CrossRef]
- Sbai, H.; Hajib, A.; Msairi, S.; Amalich, S.; Bouyahya, A.; Lee, L.-H.; Wen Goh, K.; Tabyaoui, M.; Harhar, H. Fungal Infections of Legume Crops: Challenges and Management Approaches. Journal of Agriculture and Food Research 2024, 18, 101447. [CrossRef]
- Tripathi, A.N.; Maurya, S.; Pandey, K.K.; Behera, T.K. Global Scenario of Vegetable Fungal Diseases. Vegetable Science 2024, 51, 54–65. [CrossRef]
- El Afandi, G.; Irfan, M. Pesticides Risk Assessment Review: Status, Modeling Approaches, and Future Perspectives. Agronomy 2024, 14.
- Islam, T.; Danishuddin; Tamanna, N.T.; Matin, M.N.; Barai, H.R.; Haque, M.A. Resistance Mechanisms of Plant Pathogenic Fungi to Fungicide, Environmental Impacts of Fungicides, and Sustainable Solutions. Plants 2024, 13.
- Morgounov, A.; Akin, B.; Demir, L.; Keser, M.; Kokhmetova, A.; Martynov, S.; Orhan, Ş.; Özdemir, F.; Özseven, ̄zzet; Sapakhova, Z.; et al. Yield Gain Due to Fungicide Application in Varieties of Winter Wheat (Triticum Aestivum) Resistant and Susceptible to Leaf Rust. Crop Pasture Sci 2015, 66, 649–659. [CrossRef]
- Madenova, A.; Sapakhova, Z.; Bakirov, S.; Galymbek, K.; Yernazarova, G.; Kokhmetova, A.; Keishilov, Z. Screening of Wheat Genotypes for the Presence of Common Bunt Resistance Genes. Saudi J Biol Sci 2021, 28, 2816–2823. [CrossRef]
- Sun, L.; Lai, M.; Ghouri, F.; Nawaz, M.A.; Ali, F.; Baloch, F.S.; Nadeem, M.A.; Aasim, M.; Shahid, M.Q. Modern Plant Breeding Techniques in Crop Improvement and Genetic Diversity: From Molecular Markers and Gene Editing to Artificial Intelligence—A Critical Review. Plants 2024, 13.
- Gao, M.; Hao, Z.; Ning, Y.; He, Z. Revisiting Growth–Defence Trade-Offs and Breeding Strategies in Crops. Plant Biotechnol J 2024, 22, 1198–1205.
- Ahmad, S.; Wei, X.; Sheng, Z.; Hu, P.; Tang, S. CRISPR/Cas9 for Development of Disease Resistance in Plants: Recent Progress, Limitations and Future Prospects. Brief Funct Genomics 2018, 19, 26–39.
- Ma, L.; Kong, F.; Sun, K.; Wang, T.; Guo, T. From Classical Radiation to Modern Radiation: Past, Present, and Future of Radiation Mutation Breeding. Front Public Health 2021, 9.
- Permyakova, N. V.; Deineko, E. V. Crop Improvement: Comparison of Transgenesis and Gene Editing. Horticulturae 2024, 10.
- Ahmad, S.; Rana, I.A.; Folta, K.M.; Lorenzo, C.D.; Khan, S.H. Editorial: Gene Editing to Achieve Zero Hunger. Front Genome Ed 2025, 7. [CrossRef]
- Waites, J.; Achary, V.M.M.; Syombua, E.D.; Hearne, S.J.; Bandyopadhyay, A. CRISPR-Mediated Genome Editing of Wheat for Enhancing Disease Resistance. Front Genome Ed 2025, 7.
- Lau, C.-H.; Tin, C.; Suh, Y. CRISPR-Based Strategies for Targeted Transgene Knock-in and Gene Correction. Fac Rev 2020, 9. [CrossRef]
- Sapakhova, Z.; Kanat, R.; Choi, K.; Daurov, D.; Daurova, A.; Zhambakin, K.; Shamekova, M. CRISPR-Cas Gene Editing Technology in Potato. Int J Mol Sci 2025, 26. [CrossRef]
- Rynjah, D.; Sandhanam, K.; Bhattacharjee, B.; Deka, B.; Newar, A.; Kalita, T.; Nath, J.; Ahmed, A.B.; Sahu, R.K.; Das, T. CRISPR/Cas9 Gene Editing Systems for Enhancing Secondary Metabolite Biosynthesis via Reproductive Tissue Modification. Discover Plants 2025, 2, 245. [CrossRef]
- Kokhmetova, A.; Kremneva, O.; Volkova, G.; Atishova, M.; Sapakhova, Z. Evaluation of Wheat Cultivars Growing in Kazakhstan and Russia for Resistance to Tan Spot. Journal of Plant Pathology 2017, 99, 161–167. [CrossRef]
- Kokhmetova, A.M.; Ali, S.; Sapakhova, Z.; Atishova, M.N. Identification of Genotypes-Carriers of Resistance to Tan Spot Ptr ToxA and Ptr ToxB of Pyrenophora Tritici-Repentis in Common Wheat Collection. Vavilovskii Zhurnal Genet Selektsii 2018, 22, 978–986. [CrossRef]
- Madenova, A.; Kokhmetova, A.; Sapakhova, Z.; Galymbek, K.; Keishilov, Z.; Akan, K.; Yesserkenov, A. Effect of Common Bunt [Tilletia Caries (DC) Tul] Infection on Agronomic Traits and Resistance of Wheat Entries. Research on Crops 2020, 21, 791–797. [CrossRef]
- Taj, M.; Sajjad, M.; Li, M.; Yasmeen, A.; Mubarik, M.S.; Kaniganti, S.; He, C. Potential Targets for CRISPR/Cas Knockdowns to Enhance Genetic Resistance Against Some Diseases in Wheat (Triticum Aestivum L.). Front Genet 2022, 13. [CrossRef]
- Mu, K.; Ren, X.; Yang, H.; Zhang, T.; Yan, W.; Yuan, F.; Wu, J.; Kang, Z.; Han, D.; Deng, R.; et al. CRISPR-Cas12a-Based Diagnostics of Wheat Fungal Diseases. J Agric Food Chem 2022, 70, 7240–7247. [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A Deletion Mutation in TaHRC Confers Fhb1 Resistance to Fusarium Head Blight in Wheat. Nat Genet 2019, 51, 1099–1105. [CrossRef]
- Chen, H.; Su, Z.; Tian, B.; Liu, Y.; Pang, Y.; Kavetskyi, V.; Trick, H.N.; Bai, G. Development and Optimization of a Barley Stripe Mosaic Virus-Mediated Gene Editing System to Improve Fusarium Head Blight Resistance in Wheat. Plant Biotechnol J 2022, 20, 1018–1020. [CrossRef]
- Karmacharya, A.; Li, D.; Leng, Y.; Shi, G.; Liu, Z.; Yang, S.; Du, Y.; Dai, W.; Zhong, S. Targeting Disease Susceptibility Genes in Wheat Through Wide Hybridization with Maize Expressing Cas9 and Guide RNA. Molecular Plant-Microbe Interactions 2023, 36, 554–557. [CrossRef]
- Brauer, E.K.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Schernthaner, J.; Subramaniam, R.; Ouellet, T.; Genome Editing of a Deoxynivalenol-Induced Transcription Factor Confers Resistance to Fusarium graminearum in Wheat. Mol Plant Microbe Interact. 2020, 33, 553-560. [CrossRef]
- Jones, D.A.B.; Rybak, K.; Hossain, M.; Bertazzoni, S.; Williams, A.; Tan, K.C.; Phan, H.T.T.; Hane, J.K. Repeat-Induced Point Mutations Driving Parastagonospora Nodorum Genomic Diversity Are Balanced by Selection against Non-Synonymous Mutations. Commun Biol 2024, 7. [CrossRef]
- Poddar, S.; Tanaka, J.; Running, K.L.D.; Kariyawasam, G.K.; Faris, J.D.; Friesen, T.L.; Cho, M.J.; Cate, J.H.D.; Staskawicz, B. Optimization of Highly Efficient Exogenous-DNA-Free Cas9-Ribonucleoprotein Mediated Gene Editing in Disease Susceptibility Loci in Wheat (Triticum Aestivum L.). Front Plant Sci 2023, 13. [CrossRef]
- Khan, H.; McDonald, M.C.; Williams, S.J.; Solomon, P.S. Assessing the Efficacy of CRISPR/Cas9 Genome Editing in the Wheat Pathogen Parastagonspora Nodorum. Fungal Biol Biotechnol 2020, 7. [CrossRef]
- Zou, S.; Xu, Y.; Li, Q.; Wei, Y.; Zhang, Y.; Tang, D. Wheat Powdery Mildew Resistance: From Gene Identification to Immunity Deployment. Front Plant Sci 2023, 14.
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-Edited Powdery Mildew Resistance in Wheat without Growth Penalties. Nature 2022, 602, 455–460. [CrossRef]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous Modification of Three Homoeologs of TaEDR1 by Genome Editing Enhances Powdery Mildew Resistance in Wheat. Plant Journal 2017, 91, 714–724. [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat Biotechnol 2014, 32, 947–951. [CrossRef]
- Kokhmetova, A.; Rathan, N.D.; Sehgal, D.; Malysheva, A.; Kumarbayeva, M.; Nurzhuma, M.; Bolatbekova, A.; Krishnappa, G.; Gultyaeva, E.; Kokhmetova, A.; et al. QTL Mapping for Seedling and Adult Plant Resistance to Stripe and Leaf Rust in Two Winter Wheat Populations. Front Genet 2023, 14. [CrossRef]
- Ochilov, B.O.; Turakulov, K.S.; Meliev, S.K.; Melikuziev, F.A.; Aytenov, I.S.; Murodova, S.M.; Khalillaeva, G.O.; Chinikulov, B.K.; Azimova, L.A.; Urinov, A.M.; et al. Development of Yellow Rust-Resistant and High-Yielding Bread Wheat (Triticum Aestivum L.) Lines Using Marker-Assisted Backcrossing Strategies. Int J Mol Sci 2025, 26. [CrossRef]
- Malysheva, A.; Kokhmetova, A.; Urazaliev, R.; Kumarbayeva, M.; Keishilov, Z.; Nurzhuma, M.; Bolatbekova, A.; Kokhmetova, A. Phenotyping and Identification of Molecular Markers Associated with Leaf Rust Resistance in the Wheat Germplasm from Kazakhstan, CIMMYT and ICARDA. Plants 2023, 12. [CrossRef]
- Liu, S.; Zhang, F.; Su, J.; Fang, A.; Tian, B.; Yu, Y.; Bi, C.; Ma, D.; Xiao, S.; Yang, Y. CRISPR-Targeted Mutagenesis of Mitogen-Activated Protein Kinase Phosphatase 1 Improves Both Immunity and Yield in Wheat. Plant Biotechnol J 2024, 22, 1929–1941. [CrossRef]
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W.; et al. Inactivation of a Wheat Protein Kinase Gene Confers Broad-Spectrum Resistance to Rust Fungi. Cell 2022, 185, 2961-2974.e19. [CrossRef]
- Liu, S.; Liu, H.; Guo, M.; Pan, Y.; Hao, C.; Hou, J.; Yan, L.; Zhang, X.; Chen, X.; Li, T. Knockout of GRAIN WIDTH2 Has a Dual Effect on Enhancing Leaf Rust Resistance and Increasing Grain Weight in Wheat. Plant Biotechnol J 2024, 22, 2007–2009. [CrossRef]
- Conde, S.; Catarino, S.; Ferreira, S.; Temudo, M.P.; Monteiro, F. Rice Pests and Diseases Around the World: Literature-Based Assessment with Emphasis on Africa and Asia. Agriculture (Switzerland) 2025, 15.
- Foster, A.J.; Martin-Urdiroz, M.; Yan, X.; Wright, S.; Soanes, D.M.; Talbot, N.J. CRISPR-Cas9 Ribonucleoprotein-Mediated Co-Editing and Counterselection in the Rice Blast Fungus 2018.
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/ Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS One 2016, 11. [CrossRef]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z.; Sodmergen Disruption of OsSEC3A Increases the Content of Salicylic Acid and Induces Plant Defense Responses in Rice. J Exp Bot 2018, 69, 1051–1064. [CrossRef]
- Wang, R.J.; Zhao, J.; Bhadauria, V.; Peng, Y.L. Efficient Gene Editing with an Arg-TRNA Promoter-Driven CRISPR/Cas9 in the Rice Blast Fungus Pyricularia Oryzae. Phytopathology Research 2024, 6. [CrossRef]
- Wang, F-Q.; Fan, F-J.; Li, W-Q.; Zhu, J-Y.; Wang, J.; Zhong, W.-G.; Yang, J. Knock-out Efficiency Analysis of Pi21 Gene Using CRISPR/Cas9 in Rice. Chinese Journal ofF Rice Science 2016, 30, 469-478. [CrossRef]
- Xu, P.; Wang, H.; Tu, R.; Liu, Q.; Wu, W.; Fu, X.; Cao, L.; Shen, X. Orientation improvement of blast resistance in rice via CRISPR/Cas9 system. Chinese Journal of Rice Science 2019, 33, 313-322. [CrossRef]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing Disease-Resistant Thermosensitive Male Sterile Rice by Multiplex Gene Editing. J Integr Plant Biol 2019, 61, 1201–1205. [CrossRef]
- Yang, J.; Fang, Y.; Wu, H.; Zhao, N.; Guo, X.; Mackon, E.; Peng, H.; Huang, S.; He, Y.; Qin, B.; et al. Improvement of Resistance to Rice Blast and Bacterial Leaf Streak by CRISPR/Cas9-Mediated Mutagenesis of Pi21 and OsSULTR3;6 in Rice (Oryza Sativa L.). Front Plant Sci 2023, 14. [CrossRef]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of Rice Varieties with Enhanced Resistances to Both Blast and Bacterial Blight Diseases via CRISPR/Cas9. Plant Biotechnol J 2022, 20, 876–885. [CrossRef]
- Peng, Q.; Li, J.; Xu, H.; Zhang, X.; Zhang, D.; Zhu, S. Editing Pi21 Gene to Improve the Blast Resistance of Cultivated Rice dalixiang by Using CRISPR/Cas9 System. Molecular Plant Breeding 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20220221.1756.019.htm.
- Wu, X.; Li, J-L.; Ceng, Q.-H.; Zhang, D.-S.; Xu, H.-F.; Jiang, X.; Song, Li.; Peng, Q.; Zhu, S.-S. Improvement of Blast Resistance of Rice Variety Dalixiang by CRISPR/Cas9 Gene Editing Technology. Seed 2021, 40, 50-55. [CrossRef]
- Zhu, Z.; Yin, J.; Chern, M.; Zhu, X.; Yang, C.; He, K.; Liu, Y.; He, M.; Wang, J.; Song, L.; et al. New Insights into Bsr-D1-Mediated Broad-Spectrum Resistance to Rice Blast. Mol Plant Pathol 2020, 21, 951–960. [CrossRef]
- Zhang, Y.; Zheng, L.; Xie, K. CRISPR/DCas9-Mediated Gene Silencing in Two Plant Fungal Pathogens. mSphere 2023, 8. [CrossRef]
- Ren, B.; Yan, F.; Kuang, Y.; Li, N.; Zhang, D.; Zhou, X.; Lin, H.; Zhou, H. Improved Base Editor for Efficiently Inducing Genetic Variations in Rice with CRISPR/Cas9-Guided Hyperactive HAID Mutant. Mol Plant 2018, 11, 623–626.
- Martínez-Moreno, F.; Solís, I.; Igartua, E. Barley History and Breeding in Spain. Agriculture (Switzerland) 2024, 14.
- Çelik Oğuz, A.; Karakaya, A. Genetic Diversity of Barley Foliar Fungal Pathogens. Agronomy 2021, 11.
- Poursafar, A.; Leng, Y.; Zhong, S. Development of a CRISPR/Cas9-Mediated Gene Knockout Method for Functional Genomics of the Barley Spot Blotch Pathogen Bipolaris Sorokiniana . PhytoFrontiersTM 2024, 4, 682–689. [CrossRef]
- Ge, C.; Moolhuijzen, P.; Hickey, L.; Wentzel, E.; Deng, W.; Dinglasan, E.G.; Ellwood, S.R. Physiological Changes in Barley Mlo-11 Powdery Mildew Resistance Conditioned by Tandem Repeat Copy Number. Int J Mol Sci 2020, 21, 1–18. [CrossRef]
- Mishra, R.; Tripathi, M.K.; Sikarwar, R.S.; Singh, Y.; Tripathi, N. Soybean (Glycine Max L. Merrill): A Multipurpose Legume Shaping Our World. Plant Cell Biotechnol Mol Biol 2024, 25, 17–37. [CrossRef]
- McCaghey, M.; Willbur, J.; Smith, D.L.; Kabbage, M. The Complexity of the Sclerotinia Sclerotiorum Pathosystem in Soybean: Virulence Factors, Resistance Mechanisms, and Their Exploitation to Control Sclerotinia Stem Rot. Trop Plant Pathol 2019, 44, 12–22.
- Bui, T.P.; Le, H.; Ta, D.T.; Nguyen, C.X.; Le, N.T.; Tran, T.T.; Van Nguyen, P.; Stacey, G.; Stacey, M.G.; Pham, N.B.; et al. Enhancing Powdery Mildew Resistance in Soybean by Targeted Mutation of MLO Genes Using the CRISPR/Cas9 System. BMC Plant Biol 2023, 23. [CrossRef]
- Dou, D.; Kale, S.D.; Liu, T.; Tang, Q.; Wang, X.; Arredondo, F.D.; Basnayake, S.; Whisson, S.; Drenth, A.; Maclean, D.; et al. Different Domains of Phytophthora Sojae Effector Avr4/6 Are Recognized by Soybean Resistance Genes Rps4 and Rps6. / 425 MPMI 2010, 23, 425–435. [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient Disruption and Replacement of an Effector Gene in the Oomycete Phytophthora Sojae Using CRISPR/Cas9. Mol Plant Pathol 2016, 17, 127–139. [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of Susceptibility as a Novel Breeding Strategy for Durable and Broad-Spectrum Resistance. Molecular Breeding 2010, 25, 1–12.
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol Plant Pathol 2012, 13, 414–430.
- Bü, R.; Hollricher, K. The Barley Mlo Gene: A Novel Control Element of Plant Pathogen Resistance; 1997; Vol. 88;.
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical Mystery Tour: MLO Proteins in Plant Immunity and Beyond. Journal of Physiology 2014, 204, 273–281.
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci Rep 2017, 7. [CrossRef]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int J Mol Sci 2021, 22, 1–18. [CrossRef]
- Santillán Martínez, M.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y. CRISPR/Cas9-Targeted Mutagenesis of the Tomato Susceptibility Gene PMR4 for Resistance against Powdery Mildew. BMC Plant Biol 2020, 20. [CrossRef]
- Li, R.; Cui, L.; Martina, M.; Bracuto, V.; Meijer-Dekens, F.; Wolters, A.M.A.; Moglia, A.; Bai, Y.; Acquadro, A. Less Is More: CRISPR/Cas9-Based Mutations in DND1 Gene Enhance Tomato Resistance to Powdery Mildew with Low Fitness Costs. BMC Plant Biol 2024, 24. [CrossRef]
- Li, R.; Maioli, A.; Yan, Z.; Bai, Y.; Valentino, D.; Milani, A.M.; Pompili, V.; Comino, C.; Lanteri, S.; Moglia, A.; et al. CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars. Int J Mol Sci 2022, 23. [CrossRef]
- Hong, Y.; Meng, J.; He, X.; Zhang, Y.; Liu, Y.; Zhang, C.; Qi, H.; Luan, Y. Editing Mir482b and Mir482c Simultaneously by Crispr/Cas9 Enhanced Tomato Resistance to Phytophthora Infestans. Phytopathology 2021, 111, 1008–1016. [CrossRef]
- Paula de Toledo Thomazella, D.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 Mediated Mutagenesis of a DMR6 Ortholog in Tomato Confers Broad-Spectrum Disease Resistance 2016.
- Debbarma, J.; Saikia, B.; Singha, D.L.; Das, D.; Keot, A.K.; Maharana, J.; Velmurugan, N.; Arunkumar, K.P.; Reddy, P.S.; Chikkaputtaiah, C. CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum Lycopersicum L.). Genes (Basel) 2023, 14. [CrossRef]
- Ramirez Gaona, M.; van Tuinen, A.; Schipper, D.; Kano, A.; Wolters, P.J.; Visser, R.G.F.; van Kan, J.A.L.; Wolters, A.M.A.; Bai, Y. Mutation of PUB17 in Tomato Leads to Reduced Susceptibility to Necrotrophic Fungi. Plant Biotechnol J 2023, 21, 2157–2159. [CrossRef]
- Perk, E.A.; D’Ambrosio, J.M.; Cerrudo, I.; Robuschi, L.; Juárez, M.; Vélez, P.; Mary, V.; Theumer, M.; Segretin, M.E.; Laxalt, A.M. Phosphoinositide-Specific Phospholipase C 2 (SlPLC2) Facilitates Vesicle Formation and Modulates Immune Signaling in Tomato Phytophthora Infestans Interactions 2025.
- Ortega-Salazar, I.; Crum, D.; Sbodio, A.O.; Sugiyama, Y.; Adaskaveg, A.; Wang, D.; Seymour, G.B.; Li, X.; Wang, S.C.; Blanco-Ulate, B. Double CRISPR Knockout of Pectin Degrading Enzymes Improves Tomato Shelf-Life While Ensuring Fruit Quality. Plants People Planet 2024, 6, 330–340. [CrossRef]
- Hanika, K.; Schipper, D.; Chinnappa, S.; Oortwijn, M.; Schouten, H.J.; Thomma, B.P.H.J.; Bai, Y. Impairment of Tomato WAT1 Enhances Resistance to Vascular Wilt Fungi Despite Severe Growth Defects. Front Plant Sci 2021, 12. [CrossRef]
- Park, J.H.; Kim, H. Harnessing CRISPR/Cas9 for Enhanced Disease Resistance in Hot Peppers: A Comparative Study on CaMLO2-Gene-Editing Efficiency across Six Cultivars. Int J Mol Sci 2023, 24. [CrossRef]
- Kim, H.; Choi, J.; Won, K.H. A Stable DNA-Free Screening System for CRISPR/RNPs-Mediated Gene Editing in Hot and Sweet Cultivars of Capsicum Annuum. BMC Plant Biol 2020, 20. [CrossRef]
- Mishra, R.; Mohanty, J.N.; Mahanty, B.; Joshi, R.K. A Single Transcript CRISPR/Cas9 Mediated Mutagenesis of CaERF28 Confers Anthracnose Resistance in Chilli Pepper (Capsicum Annuum L.). Planta 2021, 254. [CrossRef]
- Ferrero, M.; Valentino, D.; Milani, A.M.; Comino, C.; Lanteri, S.; Acquadro, A.; Moglia, A. Enhancing Tolerance to Phytophthora Spp. in Eggplant through DMR6–1 CRISPR/Cas9 Knockout. Plant Stress 2024, 14. [CrossRef]
- Vogel, J.; Somerville, S. Isolation and Characterization of Powdery Mildew-Resistant Arabidopsis Mutants; PNAS, 1999; Vol. 97;.
- Sun, K.; Wolters, A.M.A.; Loonen, A.E.H.M.; Huibers, R.P.; van der Vlugt, R.; Goverse, A.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Down-Regulation of Arabidopsis DND1 Orthologs in Potato and Tomato Leads to Broad-Spectrum Resistance to Late Blight and Powdery Mildew. Transgenic Res 2016, 25, 123–138. [CrossRef]
- Zhu, Q.H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. MiR482 Regulation of NBS-LRR Defense Genes during Fungal Pathogen Infection in Cotton. PLoS One 2013, 8. [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR Proteins: Adaptable Guards. Genome Biol 2006, 7.
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van Den Ackerveken, G. Downy Mildew Resistant 6 and DMR6-like Oxygenase 1 Are Partially Redundant but Distinct Suppressors of Immunity in Arabidopsis. Plant Journal 2015, 81, 210–222. [CrossRef]
- Krasikov, V.; Dekker, H.L.; Rep, M.; Takken, F.L.W. The Tomato Xylem Sap Protein XSP10 Is Required for Full Susceptibility to Fusarium Wilt Disease. J Exp Bot 2011, 62, 963–973. [CrossRef]
- Koo, Y.J.; Kim, M.A.; Kim, E.H.; Song, J.T.; Jung, C.; Moon, J.-K.; Kim, J.-H.; Seo, H.S.; Song, S.I.; Kim, J.-K.; Lee, J.S.; Cheong, J.-J.; Choi, Y.D. Overexpression of Salicylic Acid Carboxyl Methyltransferase Reduces Salicylic Acid-Mediated Pathogen Resistance in Arabidopsis thaliana. Plant Mol Biol 2007, 64, 1–15. doi.org/10.1007/s11103-006-9123-x.
- Gordon, T.R. Fusarium Oxysporum and the Fusarium Wilt Syndrome. Annu Rev Phytopathol 2025, 15. [CrossRef]
- Joshi, R. A Review of Fusarium Oxysporum on Its Plant Interaction and Industrial Use. Journal of Medicinal Plants Studies 2018, 6, 112–115. [CrossRef]
- Ramírez Gaona, M.; van Tuinen, A.; Schipper, D.; Ramos Peregrina, Á.; Visser, R.G.F.; van Kan, J.A.L.; Bai, Y.; Wolters, A.-M.A. Mutation of PUB21 in Tomato Leads to Reduced Susceptibility to Necrotrophic Fungi. BMC Plant Biol 2025, 25, 1038. [CrossRef]
- Silva, C.J.; Van Den Abeele, C.; Ortega-Salazar, I.; Papin, V.; Adaskaveg, J.A.; Wang, D.; Casteel, C.L.; Seymour, G.B.; Blanco-Ulate, B. Host Susceptibility Factors Render Ripe Tomato Fruit Vulnerable to Fungal Disease despite Active Immune Responses. J Exp Bot 2021, 72, 2696–2709. [CrossRef]
- Denancé, N.; Ranocha, P.; Oria, N.; Barlet, X.; Rivière, M.P.; Yadeta, K.A.; Hoffmann, L.; Perreau, F.; Clément, G.; Maia-Grondard, A.; et al. Arabidopsis Wat1 (Walls Are Thin1)-Mediated Resistance to the Bacterial Vascular Pathogen, Ralstonia Solanacearum, Is Accompanied by Cross-Regulation of Salicylic Acid and Tryptophan Metabolism. Plant Journal 2013, 73, 225–239. [CrossRef]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratge-Faillie, C.; Novak, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 Is a Vacuolar Auxin Transport Facilitator Required for Auxin Homoeostasis. Nat Commun 2013, 4. [CrossRef]
- Cui, L.; van den Munckhof, M.C.; Bai, Y.; Voorrips, R.E. Resistance to Anthracnose Rot Disease in Capsicum. Agronomy 2023, 13. [CrossRef]
- Sanati, S.; Razavi, B.M.; Hosseinzadeh, H. A Review of the Effects of Capsicum Annuum L. And Its Constituent, Capsaicin, in Metabolic Syndrome. Iran J Basic Med Sci 2018, 21, 439–448.
- Mieslerová, B.; Cook, R.T.A.; Wheater, C.P.; Lebeda, A. Ecology of Powdery Mildews – Influence of Abiotic Factors on their Development and Epidemiology. Critical Reviews in Plant Sciences 2022, 41, 365–390. [CrossRef]
- Mishra, R.; Rout, E.; Joshi, R.K. Identification of Resistant Sources Against Anthracnose Disease Caused by Colletotrichum Truncatum and Colletotrichum Gloeosporioides in Capsicum Annuum L. Proceedings of the National Academy of Sciences India Section B - Biological Sciences 2019, 89, 517–524. [CrossRef]
- Oo, M.M.; Oh, S.-K. Chilli Anthracnose (Colletotrichum Spp.) Disease and Its Management Approach. Korean Journal of Agricultural Science 2016, 43, 153–162. [CrossRef]
- Taher, D.; Solberg, S.; Prohens, J.; Chou, Y.Y.; Rakha, M.; Wu, T.H. World Vegetable Center Eggplant Collection: Origin, Composition, Seed Dissemination and Utilization in Breeding. Front Plant Sci 2017, 8. [CrossRef]
- Saini, H.; Thakur, R.; Gill, R.; Tyagi, K.; Goswami, M. CRISPR/Cas9-Gene Editing Approaches in Plant Breeding. GM Crops Food 2023, 14, 1–17. [CrossRef]
- Kusch, S.; Panstruga, R. Mlo-Based Resistance: An Apparently Universal “Weapon” to Defeat Powdery Mildew Disease. Molecular Plant-Microbe Interactions 2017, 30, 179–189. [CrossRef]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of Different Types of CRISPR/Cas-Based Systems in Bacteria. Microb Cell Fact 2020, 19.
- Koonin, E. V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr Opin Microbiol 2017, 37, 67–78. [CrossRef]
- Huang, J.; Gu, L.; Zhang, Y.; Yan, T.; Kong, G.; Kong, L.; Guo, B.; Qiu, M.; Wang, Y.; Jing, M.; et al. An Oomycete Plant Pathogen Reprograms Host Pre-MRNA Splicing to Subvert Immunity. Nat Commun 2017, 8. [CrossRef]
- Rasul, I.; Zafar, F.; Ali, M.A.; Nadeem, H.; Siddique, M.H.; Shahid, M.; Ashfaq, U.A.; Azeem, F. Genetic Basis for Biotic Stress Resistance in Plants from Solanaceae Family: A Review. Int J Agric Biol 2019, 22, 178–194.
- Pottinger, S.E.; Innes, R.W. RPS5-Mediated Disease Resistance: Fundamental Insights and Translational Applications. Annu. Rev. Phytopathol. 2020, 58, 139–60. [CrossRef]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-Delivered CRISPR/Cas9 System for Wheat Genome Editing. Plant Biotechnol J 2019, 17, 1623–1635. [CrossRef]
- Gan, W.C.; Ling, A.P.K. CRISPR/Cas9 in Plant Biotechnology: Applications and Challenges. Biotechnologia 2022, 103, 81–93. [CrossRef]
| Pathogen | Target Gene | Delivery Method | Type of Editing | References |
| Tomato | ||||
| Oidium neolycopersici | SlMlo1 |
Agrobacterium- mediated transformation |
Knockout | [72] |
| Oidium neolycopersici | SlMlo1, SlPelo |
Agrobacterium- mediated transformation |
Knockout | [73] |
| Oidium neolycopersici | PMR4 |
Agrobacterium- mediated transformation |
Knockout | [74] |
| Oidium neolycopersici | SlDND1 |
Agrobacterium- mediated transformation |
Knockout | [75] |
| Phytophthora infestans | SlPMR4 |
Agrobacterium- mediated transformation |
Knockout | [76] |
| Phytophthora infestans | miR482b, miR482c |
Agrobacterium- mediated transformation |
Knockout | [77] |
| Phytophthora capsici | SlDMR6-1 |
Agrobacterium- mediated transformation |
Knockout | [78] |
| Fusarium oxysporum | XSP10, SlSAMT |
Agrobacterium- mediated transformation |
Knockout | [79] |
| Fusarium oxysporum | SlPUB21, SlPUB17/SlPUB21 |
Agrobacterium- mediated transformation |
Knockout | [80] |
| Botrytis cinerea | SlPLC2 |
Agrobacterium- mediated transformation |
Knockout | [81] |
| Botrytis cinerea | SlPG2a, SlPL |
Agrobacterium- mediated transformation |
Knockout | [82] |
| Verticillium dahliae | SlWAT |
Agrobacterium- mediated transformation |
Knockout | [83] |
| Pepper | ||||
| Leveillula taurica | CaMLO2 | RNP | Knockout | [84] |
| Leveillula taurica | CaMLO2 | RNP | Knockout | [85] |
| Colletotrichum truncatum | CaERF28 | Agrobacterium-mediated transformation | Knockout | [86] |
| Eggplant | ||||
| Phytophthora infestans, Phytophthora capsici | SmDMR6 | Agrobacterium-mediated transformation | Knockout | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





