Submitted:
17 October 2025
Posted:
20 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
3. Results
| # | DOI | Title | Reference | Year | Terms Found | Classes * |
|---|---|---|---|---|---|---|
| 1 | 10.1007/s12553-024-00895-y | Current trends and future perspectives in hadron therapy: radiobiology | [101] | 2024 | DNA repair | [1] |
| 2 | 10.3389/fonc.2021.687672 | The Time for Chronotherapy in Radiation Oncology | [102] | 2021 | DNA repair | [1] |
| 3 | 10.1667/RADE-24-00037.1 | What's changed in 75 years of RadRes? - An australian perspective on selected topics | [103] | 2024 | DNA repair | [1] |
| 4 | 10.3389/fnano.2025.1603334 | Radiobiological perspective on metrics for quantifying dose enhancement effects of High-Z nanoparticles | [104] | 2025 | DNA repair, precision oncology | [1, 2] |
| 5 | 10.1016/j.clon.2021.09.003 | Can Rational Combination of Ultra-high Dose Rate FLASH Radiotherapy with Immunotherapy Provide a Novel Approach to Cancer Treatment? | [105] | 2021 | flash radiotherapy | [5] |
| 6 | 10.1016/j.canlet.2025.217895 | Unraveling the dual nature of FLASH radiotherapy: From normal tissue sparing to tumor control | [15] | 2025 | flash radiotherapy, radiosensitivity | [5, 3] |
| 7 | 10.18632/oncotarget.10996 | Next generation multi-scale biophysical characterization of high precision cancer particle radiotherapy using clinical proton, helium-, carbon- and oxygen ion beams | [106] | 2016 | precision oncology | [2] |
| 8 | 10.3390/ijms26136375 | Radiation Therapy Personalization in Cancer Treatment: Strategies and Perspectives | [107] | 2025 | precision oncology | [2] |
| 9 | 10.1080/14737140.2018.1458615 | The effects of radiotherapy on the survival of patients with unresectable non-small cell lung cancer | [108] | 2018 | precision oncology | [2] |
| 10 | 10.1016/j.hoc.2019.07.001 | Toward a New Framework for Clinical Radiation Biology | [109] | 2019 | precision oncology | [2] |
| 11 | 10.1016/S0007-4551(18)30383-7 | Kidney cancer and radiotherapy: Radioresistance and beyond | [110] | 2018 | radioresistance | [4] |
| 12 | 10.1093/jrr/rrz048 | Carbon-ion irradiation overcomes HPV-integration/E2 gene-disruption induced radioresistance of cervical keratinocytes | [111] | 2019 | radioresistance, radiosensitivity | [4, 3] |
| 13 | 10.1016/j.radonc.2022.08.001 | Accurate prediction of long-term risk of biochemical failure after salvage radiotherapy including the impact of pelvic node irradiation | [112] | 2022 | radiosensitivity | [3] |
| 14 | 10.1186/s13014-024-02566-8 | Alpha/beta values in pediatric medulloblastoma: implications for tailored approaches in radiation oncology | [113] | 2025 | radiosensitivity | [3] |
| 15 | 10.32471/exp-oncology.2312-8852.vol-43-no-3.16554 | Can SARS-CoV-2 change individual radiation sensitivity of the patients recovered from COVID-19? (experimental and theoretical background). | [114] | 2021 | radiosensitivity | [3] |
| 16 | 10.3390/cells12030360 | Comparison of Radiation Response between 2D and 3D Cell Culture Models of Different Human Cancer Cell Lines | [115] | 2023 | radiosensitivity | [3] |
| 17 | 10.2298/SARH220131085N | Current aspects of radiobiology in modern radiotherapy – our clinical experience | [116] | 2022 | radiosensitivity | [3] |
| 18 | 10.1002/mp.13751 | Genomics models in radiotherapy: From mechanistic to machine learning | [117] | 2020 | radiosensitivity | [3] |
| 19 | 10.1016/j.rpor.2014.11.006 | Hypersensitivity to chemoradiation in FANCA carrier with cervical carcinoma-A case report and review of the literature. | [118] | 2015 | radiosensitivity | [3] |
| 20 | 10.3390/cancers8030028 | Paradigm Shift in Radiation Biology/Radiation Oncology Exploitation of the H2O2 Effect for Radiotherapy Using Low-LET (Linear Energy Transfer) Radiation such as X-rays and High-Energy Electrons | [119] | 2016 | radiosensitivity | [3] |
| 21 | 10.1259/bjr.20170949 | Radiation biology and oncology in the genomic era | [120] | 2018 | radiosensitivity | [3] |
| 22 | 10.1088/1361-6560/aac814 | Radiobiological parameters in a tumour control probability model for prostate cancer LDR brachytherapy | [121] | 2018 | radiosensitivity | [3] |
| 23 | 10.1136/bmjopen-2021-059345 | The COMPLETE trial: HolistiC early respOnse assessMent for oroPharyngeaL cancEr paTiEnts; Protocol for an observational study | [122] | 2022 | radiosensitivity | [3] |
| 24 | 10.1016/j.radonc.2023.109868 | Voxel-based analysis: Roadmap for clinical translation | [123] | 2023 | radiosensitivity | [3] |
| 25 | 10.1002/cnr2.1126 | Progressive breast fibrosis caused by extreme radiosensitivity: Oncocytogenetic diagnosis and treatment by reconstructive flap surgery. | [124] | 2019 | radiosensitivity, DNA repair | [3, 1] |
| 26 | 10.1002/mp.15177 | Radiobiological comparison between Cobalt-60 and Iridium-192 high-dose-rate brachytherapy sources: Part I—cervical cancer | [125] | 2021 | radiosensitivity, hypoxia-induced | [3, 5] |
| 27 | 10.1016/j.ijrobp.2015.11.013 | Influence of Nucleoshuttling of the ATM Protein in the Healthy Tissues Response to Radiation Therapy: Toward a Molecular Classification of Human Radiosensitivity | [126] | 2016 | radiosensitivity, radioresistance | [3, 4] |
| 28 | 10.3390/proteomes13020025 | Deciphering Radiotherapy Resistance: A Proteomic Perspective | [127] | 2025 | radiosensitivity, radioresistance, DNA repair, precision oncology | [3, 4, 1, 2] |
3.1. CASE STUDIES
3.2. IMPLEMENTATION PLAN IN PRECISION RADIOTHERAPY
3.3. Contributions, Practical Implications, Limitations, and Future Work
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Declaration of Generative AI and AI-Assisted Technologies in the Writing Process
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation / Term | Meaning | Context / Explanation |
| ≥3 Toxicity | Grade 3 or Higher Adverse Events | Represents severe clinical side effects per standardized toxicity grading. |
| ≥40 Gy/s | Dose-Rate Threshold for FLASH-RT | Represents ultrafast radiation rate preserving normal tissue integrity. |
| 0.291–0.669 (~0.378 span) | Range of Observed Cosine Scores | Captures the minimum-to-maximum similarity observed between LLM outputs. |
| 0.5235 ± 0.0271 (95% CI 0.4964–0.5506) | Aggregate Similarity Statistics | Demonstrates moderate overall alignment and high reproducibility of language/semantic structure between models. |
| 15 Standardized Terms | Controlled Vocabulary Set | Used for harmonizing semantic meaning across Scopus, PubMed, and Web of Science records. |
| 2014 / 2015–2024 windows | Temporal limits of corpus | Ensure focus on contemporary, translational radiotherapy. |
| 2014–2025 Analytic Window | Defined Temporal Scope of Corpus | Restriction ensuring semantic stability and consistent term usage across databases. |
| 2D / 3D | Two-dimensional / Three-dimensional | Cell-culture models compared for radiation response. |
| 3D Spheroids | Three-Dimensional Cell Cultures | In-vitro tumor analogues reproducing microenvironmental gradients and resistance. |
| 60Co / ¹⁹²Ir | Cobalt-60 / Iridium-192 Isotopes | Common brachytherapy sources; compared for efficacy, RBE, and safety. |
| a | Abstracts Corpus | The suffix “a” in Topic_0a–Topic_9a refers to topic models generated from abstracts rather than titles. |
| Adaptive Clinical Protocols | Dynamically Updated Treatment Guidelines | Clinical pathways that adjust based on biomarker or imaging feedback. |
| Adaptive Radiotherapy Planning | Treatment Optimization Based on Patient Response | Future goal using AI-derived biomarkers for closed-loop, real-time dose adjustment. |
| Adjacency definition | Mathematical rule for linking nodes | Directed adjacency alters numeric scales of network metrics. |
| Advanced Radiotherapy Technologies | Emerging Modalities (FLASH-RT, Brachytherapy, Particle Therapy) | High-precision and/or ultrafast radiation technologies providing enhanced therapeutic index. |
| Advanced Technologies Cluster | Thematic Category in LDA Model | Identified area with limited samples causing wider uncertainty in validation. |
| AGENT | Therapeutic Agent | Denotes a drug, molecule, or compound used in combination with radiation therapy. |
| AI | Artificial Intelligence | Central methodological pillar of the study; refers to machine-learning and computational modeling approaches. |
| AI Models (Validated) | Certified Predictive Algorithms | Used to plan or adjust treatments, ensuring regulatory and scientific reliability. |
| AI synthesis | Artificial Intelligence synthesis | Refers to AI-based interpretation and summarization phase of the pipeline. |
| AI-assisted | Artificial Intelligence–Assisted | Denotes the use of AI in processing, interpreting, and modeling scientific literature. |
| AI-based | Artificial Intelligence-based | Refers to the study’s computational modeling methods. |
| AI-based methods | Artificial-intelligence analytical techniques | Include topic modeling, text mining, and co-occurrence network analysis. |
| AI-based Modelling | Artificial Intelligence–Based Modeling | Framework for harmonizing heterogeneous datasets into unified evidence models. |
| AI-derived Biomarkers | Machine-Learned Predictive Molecular Indicators; Artificial Intelligence–Generated Predictive Features | Proposed for real-time, adaptive radiotherapy planning linked to patient-specific biological response; Used for adaptive treatment and real-time learning protocols. |
| Alpha/Beta (α/β) | LQ model tissue parameters | Reported in pediatric medulloblastoma for tailoring dose/fractionation. |
| ATM | Ataxia-Telangiectasia Mutated | DNA-damage response kinase; nucleoshuttling linked to tissue radiosensitivity. |
| ATR | Ataxia Telangiectasia and Rad3-Related Protein | Parallel kinase to ATM; monitors replication stress and coordinates DNA repair. |
| Average Degree = 9.5 | Mean number of links per node | Quantifies average lexical connectivity. |
| Average Weighted Degree = 1962.2 | Mean sum of edge weights | Captures total co-occurrence frequency per term. |
| BER | Base Excision Repair | DNA repair pathway correcting small, non-helix-distorting base lesions caused by ionizing radiation. |
| BIOLOGICAL | Biological Effect or Endpoint | Used in Topic_2a for biophysical modeling linking energy deposition to cellular effect. |
| Biomarker signature | Composite molecular indicator | Selection criterion for case-study inclusion and stratification. |
| Biomedical Stopwords | Domain-specific uninformative terms | Filtered to improve semantic precision. |
| Biomedical subject areas | Domain filter in Scopus | Restricts search to clinically relevant literature. |
| Blue–Red Color Bar | Visual encoding of co-occurrence strength | Blue = weak association; red = strong association. |
| BMAL1, CLOCK, PER, CRY | Core Circadian Genes | Regulate cellular timing of DNA repair; targeted in chronoradiotherapy to reduce toxicity. |
| Brachytherapy | Internal radiotherapy using implanted sources | Another advanced modality considered in the framework. |
| Brachytherapy dose escalation | Targeted increase of internal RT dose | Inclusion term; linked to voxel-level and TCP modeling. |
| Carcinoma | Cancer Type Originating from Epithelial Cells | Frequently used term in radiation oncology literature (breast, esophageal, prostate). |
| CD8⁺ Lymphocytes | Cytotoxic T Cells | Immune effectors preserved after FLASH-RT, supporting anti-tumor immunity. |
| Cell-Fit-HD | High-Density Cell Microbeam Technology | Experimental system quantifying DNA-damage kinetics and repair precision. |
| Central node | Highest-connectivity vertex | “Cancer” identified as the core of the radiotherapy supergraph. |
| cGAS–STING | Cyclic GMP–AMP Synthase / Stimulator of Interferon Genes | Cytosolic DNA-sensing pathway linking radiation damage to immune activation. |
| ChatGPT | Generative Pretrained Transformer (GPT-5 model, OpenAI) | AI-based language model used under human supervision to improve clarity, grammar, and precision during manuscript preparation. |
| Chronoradiotherapy | Time-Synchronized Radiation Delivery | Aligns irradiation with circadian repair peaks to enhance efficacy and minimize side effects. |
| CI | Confidence Interval | Statistical interval representing uncertainty around the mean cosine-similarity estimates (95% CI: 0.4964–0.5506). |
| CiteSpace | Citation Space Mapping Tool | Bibliometric software for co-authorship and trend analysis. |
| Classe 1 – DNA Repair and Molecular Response | Class 1 Category | Focused on DNA damage signaling, repair pathways (HR, NHEJ, NER, BER), and checkpoint activation; aligns molecular biology with therapeutic personalization. |
| Classe 2 – Precision Oncology and Genomic Modeling | Class 2 Category | Integrates genomic profiling, machine-learning models, and patient stratification to personalize radiation therapy. |
| Classe 3 – Individual Radiosensitivity and Clinical Risk | Class 3 Category | Addresses variability in patient radiation response, genetic predisposition, and predictive biomarkers. |
| Classe 4 – Radioresistance and Associated Mechanisms | Class 4 Category | Explores resistance mechanisms (hypoxia, metabolism, stemness, viral integration) and corresponding therapeutic interventions. |
| Classe 5 – Advanced Technologies and Innovative Radiotherapy | Class 5 Category | Encompasses next-generation technologies including FLASH-RT, hadron therapy, voxel-based analysis, and high-Z nanoparticle radiosensitization. |
| CLINICAL / ONCOLOGY / RADIOBIOLOGY / MEDICAL / DEVELOPMENT / ELSEVIER / RIGHT / RESERVED | Publishing or Institutional Terms | Topic_8a includes frequent non-scientific tokens derived from editorial metadata, likely due to text-parsing of abstracts from publishers (Elsevier, etc.). |
| Clinical trial phase | Trial stage indicator | Used to filter translational maturity of studies. |
| Clinical–Anatomical Axis | Axis describing patient- and site-specific features; Dimension grouping disease sites, therapies, and outcomes | Encompasses disease sites, therapy types, outcomes; Derived from topic modeling to describe clinical themes. |
| Clustering coefficient | Likelihood that connected nodes form closed triangles | High value shows local semantic cohesion. |
| Co-Occurrence Supergraph | Graph of term co-appearances; Combined network of all significant co-occurring terms | Reveals structural relationships among concepts in the literature; Integrates molecular, clinical, and technological vocabularies into one semantic structure. |
| Combined modality | Multimodal treatment (e.g., RT + IO) | Inclusion term bridging lab and clinic. |
| COMPLETE (trial) | HolistiC early respOnse assessMent for oroPharyngeaL cancEr paTiEnts | Protocol integrating imaging and early-response assessment in oropharyngeal cancer. |
| COMPLETE Protocol | Combined Multiparametric Imaging, Radiomics and Machine-Learning Framework | Integrates imaging and omics data for personalized radiotherapy planning. |
| Computational Oncology | Field Integrating AI and Cancer Research | The disciplinary frame under which the study’s integrative modeling approach is positioned. |
| COPPE | Instituto Alberto Luiz Coimbra de Pós-Graduação e Pesquisa de Engenharia | UFRJ’s graduate engineering institute; associated with the Nanotechnology Engineering Program. |
| Cosine Similarity | Vector-Space Similarity Measure | Quantifies the semantic and lexical overlap between two text representations (range 0–1). Used here to assess alignment between outputs of different LLMs. |
| Cost per Controlled Case | Economic Indicator | Total treatment cost divided by number of patients with complete or partial sustained response; measures cost-effectiveness. |
| COVID-19 | Coronavirus Disease 2019 | Systemic factor influencing radiosensitivity and patient management. |
| Cross-Model Consistency | Comparative Validation Concept | Indicates methodological reliability by reproducing semantically equivalent results across distinct LLM architectures. |
| CSV | Comma-Separated Values | Log format for timestamped model inferences and auditing. |
| CTCAE | Common Terminology Criteria for Adverse Events | Global clinical-trial standard for grading radiotherapy-related adverse events. |
| Data Repositories | Centralized Databases for Clinical and Molecular Information | National archives enabling interoperability and machine-learning-based outcome prediction. |
| DAY | Treatment Day / Fraction Day | Represents time variable in radiotherapy fractionation (Topic_7a). |
| Decision-Ready Framework | Actionable Computational Infrastructure | Describes the output: a structured knowledge base usable for policy, funding, and clinical translation. |
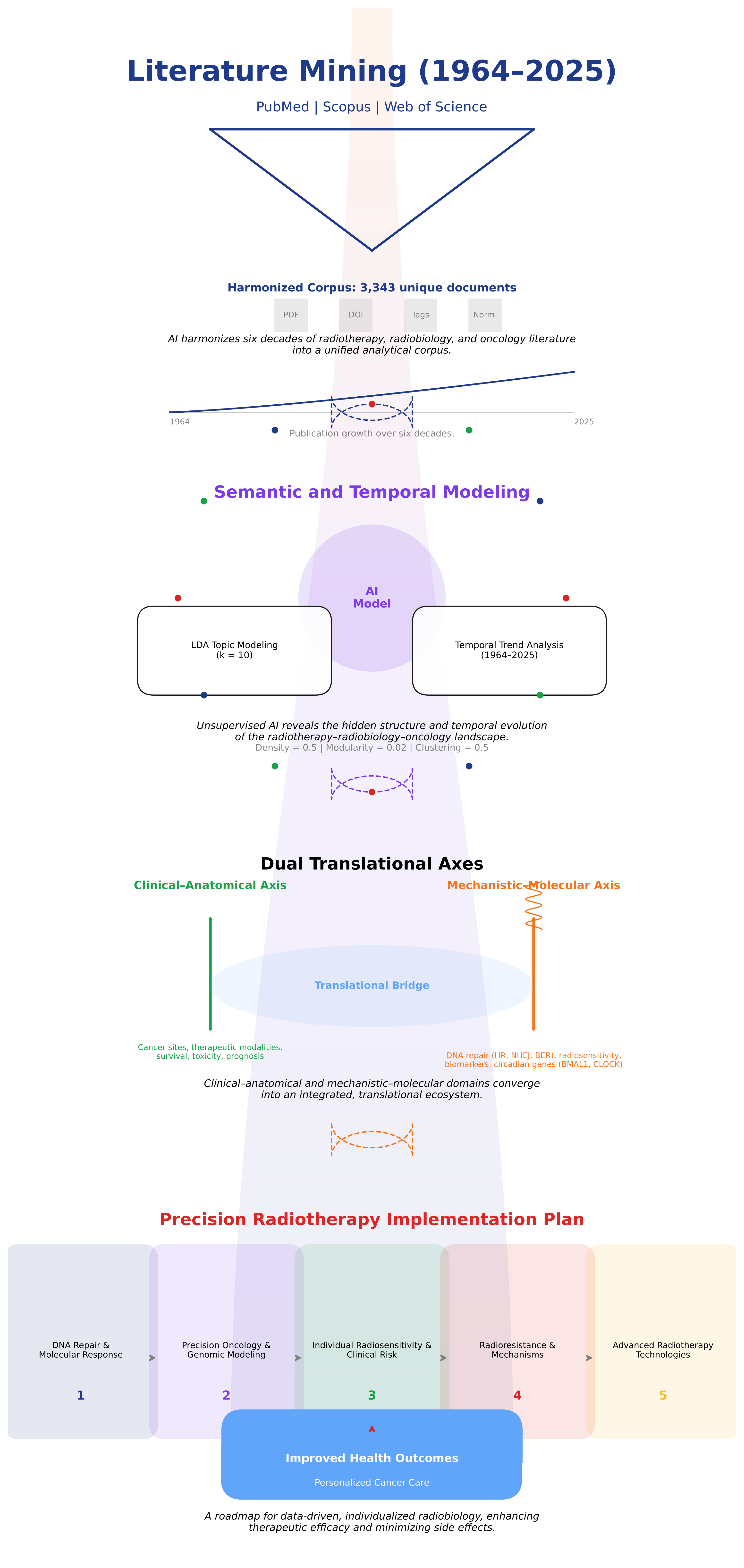
| Deduplication | Removal of duplicate entries | 594 records removed by ID and 25 by title → 3 343 unique publications. |
| Density | Ratio of existing to possible edges; Ratio of actual to possible edges | 0.5 in the directed setup, signifying a tightly connected lexical field; Indicates a dense and cohesive co-occurrence network. |
| DER / SER / RBE | Dose Enhancement Ratio / Sensitization Enhancement Ratio / Relative Biological Effectiveness | Quantitative radiobiological indices comparing biological responses between radiation modalities. |
| Diameter | Longest shortest path in the network; Longest shortest-path length | Here equals 1 due to directed adjacency; reflects condensed connectivity; Reflects immediate connectivity among principal terms in a directed adjacency setup. |
| Directed / Weighted Graph | Graph with edge orientation and numerical weights | Models term co-occurrence frequency and directionality in the semantic network. |
| DNA | Deoxyribonucleic Acid | Central mechanistic concept related to damage, repair, and radiosensitivity. |
| DNA Repair | Deoxyribonucleic Acid Repair | Core biological process for maintaining genomic integrity after radiation exposure; Major research theme representing molecular response to radiation. |
| DOI | Digital Object Identifier | Persistent identifier ensuring record uniqueness and traceability. |
| Dose–Response Model | Quantitative relation between dose and effect; Mathematical link between dose and biological effect | Appears among inclusion keywords; supports mechanistic theme; Underpins LDA topics and clinical correlation analysis. |
| E2 | HPV Regulatory Protein | Loss disrupts viral genome control; restoration of E2-associated checkpoints observed with carbon-ion irradiation. |
| Edge Thickness | Graph-visualization parameter | Increases proportionally with co-occurrence weight. |
| Editorial Workflow (“Llama → GPT-4o”) | Sequential Model Use | Denotes the pipeline where Llama3 generated drafts and GPT-4o refined phrasing, without altering data or analytic interpretations. |
| Effect | Biological or Clinical Outcome | Used to represent both therapeutic effects and adverse effects of radiation exposure. |
| ENERGY | Beam Energy | Physical parameter determining penetration depth and RBE of particle beams (Topic_2a). |
| EORTC | European Organisation for Research and Treatment of Cancer | European body with harmonized toxicity and clinical-trial reporting frameworks. |
| Eq. | Equation | Used to identify model equations from Eq. 11 to Eq. 20. |
| Equations 1–20 (Supplementary Information) | Numerical LDA Output Tables | Provide word probabilities and model parameters for transparency. |
| Equity / Access Metrics | Health Policy Indicators | Includes survival, toxicity, cost per controlled case, and hospitalizations for public health alignment. |
| EXPRESSION / GENE / PROTEIN / BLOOD | Biomarker-related Terms | In Topic_6a, these describe molecular and clinical biomarkers used for radiosensitivity prediction. |
| Factor | Biological or Statistical Factor | Represents either biological modifiers (e.g., hypoxia) or regression factors in predictive models. |
| FANCA | Fanconi Anemia Complementation Group A | Germline carrier state linked to hypersensitivity to chemoradiation. |
| Feasibility / Translational Potential | Applied Assessment Metrics | Used to evaluate how physical models and clinical data can be realistically implemented within current technological and clinical constraints. |
| Feature Space | Multidimensional Representation of Texts | Conceptual domain in which cosine similarity evaluates orientation and semantic overlap between document vectors. |
| FIU | Florida International University | U.S. collaborator institution for computational and AI analysis. |
| FLASH | Ultrafast Radiotherapy (FLASH-RT); Ultrafast radiotherapy at ultra-high dose rates; Ultrafast (> 40 Gy s⁻¹) radiation-delivery technique; Ultrafast (≥ 40 Gy s⁻¹) radiotherapy | High-dose-rate radiation mode producing reduced normal-tissue toxicity; investigated for normal-tissue sparing and tumor control; one of the advanced technologies highlighted in Class 5; modern modality delivering high-dose-rate pulses with reduced normal-tissue damage. |
| FLASH Radiotherapy and Brachytherapy | Advanced Techniques | Explored for synergistic effects in tissue protection and immune modulation. |
| FRACTION | Dose Fraction | In Topic_9a, refers to division of total dose into multiple treatment sessions—key variable in radiobiological modeling. |
| Fraction / Fractionation | Division of Total Radiation Dose into Multiple Sessions | Appears among frequent title/abstract terms; optimization variable flagged by abstracts analysis. |
| Frequency Trimming | Vocabulary Cutoff Procedure | Eliminates extremely rare or overly common tokens. |
| G ≥ 3 | Grade 3 or Higher Toxicity | Severe adverse-event category per CTCAE or RTOG/EORTC scales; measures safety profile. |
| GARD | Genomic Adjusted Radiation Dose | Integrates RSI and gene-expression data to personalize dose prescription. |
| Genomic / Proteomic Modeling | Molecular Data Integration Approaches | Computational modeling of genomic (gene-level) and proteomic (protein-level) datasets to predict radiosensitivity and outcomes. |
| Genomic Classifier | Model Assigning Molecular Subtypes | Used for precision oncology decisions. |
| Genomic Modeling | Predictive Modeling Based on Genetic Data; Mechanistic to Machine-Learning Frameworks | Used for individualized therapy planning in precision oncology; used to personalize radiotherapy (predict response/toxicity). |
| Gephi | Open-source Network-Analysis Software | Used for calculating density, diameter, clustering, and modularity of the co-occurrence network. |
| GPT-4o | OpenAI’s Multimodal Large Language Model (Omni Version) | Used to refine language, maintain consistency, and ensure terminological precision; served only in editorial and validation steps. |
| GPT-5 | Generative Pretrained Transformer, Version 5 | Model used within ChatGPT for generative text refinement and consistency checking. |
| Grammarly (v.2025) | AI-Powered English Language Writing Assistant | Utilized for grammatical and stylistic correction to ensure readability for international readers. |
| Gy | Gray | SI unit of absorbed radiation dose (1 Gy = 1 J/kg); frequent in topics 4a, 7a, 9a related to dosimetry and toxicity. |
| Gy / Gy s⁻¹ | Gray / Gray per second | SI units of absorbed dose and dose rate. |
| H₂O₂ | Hydrogen Peroxide | Central to radiosensitization concepts exploiting oxidative effects (low-LET contexts). |
| Hadron Therapy | Particle-Beam Radiotherapy (Protons, Carbon Ions); Particle-Based Radiotherapy (Using Protons or Heavy Ions) | Emerging high-precision radiation technique; advanced technology referenced in the framework as part of “Innovative Radiotherapy Technologies.” |
| Head and Neck / Breast / Prostate Cancer | Disease-Site Descriptors | Define clinical–anatomical axes in Topic_0t and related clusters. |
| Heavy Charged Particles | Proton/Helium/Carbon/Oxygen Ion Beams | “Next-generation multi-scale” characterization and high-LET effects. |
| HIF | Hypoxia-Inducible Factor | Transcription factor activated by VHL loss; mediates hypoxia-related radioresistance. |
| High-End Modalities | Advanced Treatment Technologies | Includes FLASH-RT, hadron therapy, and other precision radiotherapy innovations. |
| High-LET | High Linear Energy Transfer | Radiation that deposits dense energy tracks (e.g., heavy ions); generates complex DNA lesions. |
| High-LET Modalities | Heavy-Ion or Particle Therapies | Used in Class 4 to reverse resistant phenotypes via dense-ionization damage. |
| High-LET Radiation | High Linear Energy Transfer Radiation | Mentioned indirectly in prior results, relevant here as part of the “system-level dynamic modeling” refinement. |
| High-Z | High Atomic-Number Elements | Nanoparticles used to enhance dose deposition and radiosensitization. |
| High-Z Nanoparticles | High Atomic-Number Nanomaterials | Act as radiosensitizers by amplifying local dose and reactive-species generation. |
| High-Z Nanotechnologies | High Atomic Number Nanomaterials | Proposed future pathway to couple with ultrafast radiotherapy for dose amplification and radiosensitization. |
| High-Z Platforms | High Atomic Number Platforms | Refers to nanomaterials designed to enhance radiotherapy through dose amplification. |
| Hospitalization Rate | Frequency of Radiotherapy-Related Admissions | Indicator of adverse-event management effectiveness. |
| HPV | Human Papillomavirus | Oncogenic virus associated with cervical-cancer radioresistance; carbon-ion therapy shown to overcome it. |
| HPV / E2 | Human Papillomavirus / Regulatory Gene | Integration/E2 disruption associated with cervical radioresistance; carbon ions reported to overcome it. |
| HR | Homologous Recombination | Accurate double-strand-break repair pathway; its inhibition increases radiosensitization. |
| Hybrid Modeling / Model Fusion | Combined Output Strategy | Not used in this study; models were compared for agreement, not merged to produce shared outputs. |
| Hypotheses 1–4 | Structural Inferences Derived from Topology | Define conceptual axes: (1) cancer + radiotherapy centrality; (2) personalization trends; (3) biomarker emphasis; (4) transversal field integration. |
| ID Numbers | Unique Database Identifiers (e.g., PMID, DOI, EID) | Used for automated deduplication across sources. |
| IIPPR | Integrated Implementation Plan in Precision Radiotherapy | The overarching operational and translational framework structuring radiotherapy research into five classes with progressive integration into health policy. |
| IMA | Instituto de Macromoléculas Professora Eloisa Mano | UFRJ institute specializing in polymers and macromolecular science. |
| Imaging | Medical Imaging | Used for diagnosis, planning, and verification in radiotherapy (e.g., MRI, CT, PET). |
| Immunoradiotherapy | Radiotherapy Combined with Immune Modulation | Emergent interdisciplinary theme. |
| Immunotherapy | Immune-Based Cancer Therapy | Appears in Topic_5a; integration of radiation with immunotherapy strategies. |
| In vitro | Laboratory-Based Experiments | Denotes cellular studies that feed mechanistic insight. |
| Individualized Radiosensitivity Assessment | Patient-Specific Radiation Response Evaluation | Quantifies how individual genetic and biological profiles modify treatment tolerance and efficacy. |
| Integrated Implementation Plan in Precision Radiotherapy (IIPPR) | Translational Framework | Unifies molecular, clinical, and technological insights into five interdependent operational blocks for precision oncology and public-health integration. |
| Interdisciplinary Maturity | Cross-Disciplinary Integration | Refers to the field’s evolution into a coherent system combining biology, physics, and clinical practice. |
| Ion | Ion Beam | Refers to charged particle beams (proton, helium, carbon, oxygen) used in particle radiotherapy. |
| ION / PARTICLE / PROTON / CARBON / PHOTON | Ion-Beam and Particle Radiation Terms | Terms appearing together in Topic_2a; describe physical particles used in hadron therapy. |
| Irradiation | Exposure to Ionizing Radiation | Common in Topics 6a and 7a; refers to experimental or clinical exposure processes. |
| k = 10 Topics | Number of LDA Topics | Defines model granularity; ten topics selected for interpretability and stability. |
| Kidney Cancer VHL/HIF Axis | von Hippel–Lindau / Hypoxia-Inducible Factor | Mechanistic route to pseudohypoxia and resistance. |
| KORTUC II | Kochi Oxidative Radiotherapy for Unresectable Carcinomas Type II | Technique using hydrogen peroxide (H₂O₂) to increase tumor oxygenation and radiosensitivity. |
| LaBioS | Laboratório de Biopolímeros e Sustentabilidade | Laboratory supporting the experimental and organizational phases of the study. |
| LabOPTIMA | Laboratory for Optimization, Data Analytics, and Artificial Intelligence in Materials Science | UFRJ-based lab responsible for data-centric analysis and computational modeling; mentioned as a contributor to data processing and validation. |
| LDA | Latent Dirichlet Allocation | Unsupervised topic-modeling algorithm used for semantic extraction of dominant research axes. |
| LDR | Low-Dose-Rate | Brachytherapy context (e.g., prostate cancer TCP modeling). |
| LDR Brachytherapy Sources ⁶⁰Co / ¹⁹²Ir | Cobalt-60 / Iridium-192 | Compared radiobiologically (e.g., cervical cancer HDR; RBE differences). |
| Lemmatization | Word-Form Normalization | Reduces inflectional variants to base forms for stable modeling. |
| LET | Linear Energy Transfer | Energy deposition per track length; key in radiobiology and particle therapy. |
| LEVEL | Expression Level / Dosimetric Level | Indicates quantitative measurement of gene, protein, or dose levels (Topic_6a). |
| LINE | Cell Line | Experimental model system used for in-vitro radiation studies (Topic_1a). |
| Linear | Linear Model (Linear-Quadratic Model) | Refers to the linear component of the LQ model used to describe cell survival curves in radiotherapy modeling. |
| Llama-3 (8B) | Large Language Model (8 Billion Parameters); Open-Source Large Language Model (8 Billion Parameters) | Referenced as a semantic-processing model used during analysis, highlighting technical limitations and multilingual challenges; used for initial draft generation and offline semantic analysis; suitable for secure, local computation. |
| LLM | Large Language Model | AI model trained on massive text corpora; in this study, Llama-3 (8B) and GPT-4o were compared. |
| LLMs | Large Language Models | Used for semantic synthesis under fixed prompts; outputs logged for auditability; models (like GPT-type) used in PaperProcessor for semantic synthesis and summarization. |
| Local Control / Systemic Control | On-Site vs Distant Tumor Suppression | Metrics evaluating therapeutic success at tumor site and metastasis prevention. |
| Machine Learning | AI Technique for Pattern Recognition and Prediction | Supports risk stratification, biomarker identification, and outcome modeling. |
| Mean (μ) | Arithmetic Average | Represents central tendency of similarity values (~ 0.52 overall). |
| Mean Clustering Coefficient = 0.5 | Local Cohesion Metric | Reveals that many terms form tightly knit clusters (≈ 1140 triangles). |
| Mechanism / Molecular | Mechanistic and Molecular Basis | Indicates interest in the cellular and sub-cellular mechanisms of radiation injury and repair. |
| Mechanistic–Molecular Axis | Axis Describing Biological Mechanisms; Dimension Encompassing DNA Repair and Biomarkers | Encompasses cellular responses, DNA repair, and biomarkers; describes mechanistic research focus across decades. |
| Median Cohort | Summary Statistic in Cohort Analyses | Appears in abstract-level signals of methodological layer. |
| MeSH | Medical Subject Headings | Controlled vocabulary in PubMed employed for precise field-restricted searching. |
| Metadata | Supplementary Bibliographic Descriptors | Retained for reproducibility and cross-referencing (authors, year, source title, etc.). |
| MicroRNAs / Cytokines | Small Regulatory RNAs / Signaling Proteins | Linked to radiation toxicity and inflammatory responses in prostate cancer. |
| MMRd | Mismatch Repair Deficiency | Genetic defect leading to hypermutation; associated with increased immunogenicity and favorable prognosis. |
| Model | Computational or Predictive Model | Refers to mathematical frameworks (e.g., dose-response, LQ, biological modeling). |
| Modularity | Measure of Community Segregation; Community-Segregation Index | Low modularity indicates overlapping thematic communities; very low value (≈ 0.02) shows themes overlap strongly with one weakly connected component. |
| MOUSE | Animal Model | Indicates in-vivo preclinical testing in mice (Topic_7a). |
| Multi-Omic Biomarkers | Combined Genomic, Transcriptomic, and Proteomic Indicators | Used to stratify patients and personalize treatment strategies. |
| Multilingual Biomedical Corpora | Multi-Language Scientific Data Sets | Describes the heterogeneity of textual sources posing semantic-alignment challenges. |
| n | Sample Size | Number of text pairs analyzed per class (Class 1: n = 6; Class 2: n = 6; Class 3: n = 18; Class 4: n = 4; Class 5: n = 3). |
| NER | Nucleotide Excision Repair | DNA repair process that removes bulky helix-distorting lesions; important for radiation and oxidative stress responses. |
| NER / SER / DER Metrics | Nucleotide Excision Repair / Sensitization Enhancement Ratio / Dose Enhancement Ratio | Quantitative parameters for evaluating biological amplification of radiation effects. |
| NHEJ | Non-Homologous End Joining | Fast, error-prone DNA repair mechanism; hyperactivation linked to radioresistance. |
| no_below = 2–5 / no_above = 0.5 | Frequency-Trimming Parameters | Filter words appearing in fewer than 2–5 documents or in > 50 % of texts, ensuring vocabulary stability. |
| Node / Edge | Vertex / Connection | Represent keywords and their co-occurrence relations. |
| Normal | Normal Tissue | Opposed to tumor tissue; represents non-targeted biological matter whose protection is critical in treatment planning. |
| Normalization / Standardization | Data-Cleaning Processes | Harmonized author, title, and metadata formats across databases before merging. |
| NSCLC | Non-Small Cell Lung Cancer | Disease context in studies on radiotherapy benefits. |
| NSMP | No Specific Molecular Profile | Endometrial cancer subtype lacking defined genomic markers; used for risk-based treatment calibration. |
| O | Overall (e.g., Overall Survival – OS) | Shortened reference to clinical endpoints (Topic_3a); likely from expressions such as “overall survival.” |
| Offline Analyses | Local Computation Without Internet Connectivity | Performed using Llama-3 for data security, sovereignty, and reproducibility. |
| Omics Data | Integrated Genomic, Transcriptomic, Proteomic Datasets | Used across classes for multi-scale modeling of tumor behavior. |
| Oncology | Medical Field of Cancer Diagnosis and Treatment | Provides the clinical component in the interdisciplinary framework. |
| OpenAI | Artificial Intelligence Research Organization | Developer of ChatGPT; referenced for model attribution. |
| OS | Overall Survival | Proportion of patients alive at specified follow-up times (1 / 3 / 5 years) after radiotherapy. |
| P | p-value (Statistical Significance) | Appears in Topic_3a and 4a; denotes statistical significance in clinical or survival analyses. |
| PaperProcessor | LLM-Guided Semantic Extraction Script | Pipeline component for document-level summarization and tagging. |
| Parameter / Radiobiological | Model Coefficients / Domain Qualifier | Signal the quantitative calibration layer in abstracts. |
| Particle Therapy | Proton or Heavy-Ion Radiation Treatment | High-precision, high-LET modality for deep or radioresistant tumors. |
| passes = 10 | Number of LDA Training Iterations | Controls convergence during topic-model optimization. |
| Pathway | Biological Pathway | Indicates molecular signaling cascades affected by radiation (e.g., DNA damage-repair pathways). |
| Pearson Correlation Coefficient | Statistical Measure (r); Statistical Correlation Index | Indicates robustness of network stability across runs (r > 0.9); confirms stability of node/community ordering between directed and undirected graphs. |
| PENt | Programa de Engenharia da Nanotecnologia (Nanotechnology Engineering Program, COPPE/UFRJ) | Academic home of the AI and semantic-modeling pipeline described. |
| Peripheral Nodes | Lower-Degree Vertices at Network Edges | Terms like proton, stereotactic, imaging anchor technology subfields. |
| Personalized / Protocol | Individualized Plans / Standardized Procedures | Bridge terms in co-occurrence graphs linking themes. |
| PFS | Progression-Free Survival | Interval during which a patient remains free from tumor progression or recurrence. |
| PLAN / DOS | Treatment Planning and Dosimetry | In Topic_7a, “plan” and “dos” (truncated for dose or dosimetry) refer to planning and dose-distribution modeling. |
| PNG | Portable Network Graphics | Raster format for graphical visualization outputs. |
| POLE | DNA Polymerase Epsilon (Mutated Subtype) | Mutation defining a favorable genomic subtype in endometrial cancer; used in precision oncology stratification. |
| PORTEC-3 / PORTEC-4 | Post-operative Radiation Therapy for Endometrial Carcinoma Trials | Landmark clinical studies incorporating molecular subtyping into radiotherapy decision frameworks. |
| Precision Oncology | Data-Driven Personalized Cancer Care; Data-Driven Personalized Cancer Treatment | Cross-cutting theme linking biomarkers to clinical protocols; integrates molecular profiling and computational modeling to guide clinical decisions. |
| Precision Radiotherapy | Data-Guided, Patient-Specific Treatment Framework | Conceptual endpoint of the integrated ecosystem. |
| Precision Radiotherapy Corpus | Consolidated Dataset | Final ≈ 45 deduplicated records informing thematic and network analyses. |
| Precision Radiotherapy Implementation Plan (PRIP) | Operational Framework; AI-Derived Operational Framework | Practical output of the study – translates semantic and molecular insights into five actionable clinical classes; translates computational and molecular insights into clinical strategies. |
| Preclinical | Preclinical Study; Experimental (Non-Clinical) Phase | Experimental phase prior to human clinical trials; linked to animal or in-vitro testing (Topic_5a); denotes laboratory or animal studies preceding human trials. |
| Prediction / Response / Normal | High-Centrality Nodes | “Semantic hinges” connecting mechanistic and clinical themes in the supergraph. |
| Prediction Models | Computational or Statistical Outcome-Forecasting Tools | Represent the data-driven dimension emerging after 2010. |
| Predictive Systems | AI-Based Forecasting Models | Refer to evolving models that anticipate treatment response and guide dynamic planning. |
| Progression-Free Interval | Time to Recurrence | Quantitative measure for disease stability post-therapy. |
| Proteomics | Large-Scale Protein Analysis | Identifies functional markers of radioresistance and therapeutic targets. |
| Proton / Ion Beams | Particle-Beam Radiotherapy Modalities | Core of hadron-therapy and dose-delivery-physics studies. |
| Proton Therapy | Particle-Based Radiotherapy; Proton Therapy; Charged-Particle Radiation Modality | Advanced technique using proton beams for accurate dose delivery with minimal collateral damage; part of the technological wave correlated with publication surge. |
| Public Health Integration | Application of Precision Frameworks to Health-System Policies | Links AI-guided radiotherapy models to equity and access. |
| PubMed | U.S. National Library of Medicine Database (MEDLINE) | Primary biomedical source queried with MeSH and free-text terms; provided ≈ 66 % of final records. |
| PubMed, Scopus, Web of Science (WoS) | Scientific Databases | Used for bibliometric data collection and harmonization. |
| Quantitative Metrics | Numerical Parameters or Indicators | Include dose, LET, RBE, similarity scores, or statistical validation values supporting model precision. |
| RAD51 / BRCA1 | DNA Repair Genes | Over-expression correlates with radioresistance; potential biomarkers for therapeutic targeting. |
| RAD51, PARP1, CHK1, MAPK15 | Key DNA Repair / Stress Response Proteins | Biomarkers and therapeutic targets integrated into proteomic and genomic modeling layers. |
| Radiat / Cell / Dose / Tumor / Patient / Cancer / Radiobiolog | High-Frequency Lexical Stems | Most recurrent words in the processed corpus; define thematic backbone. |
| Radiobiological | Related to Radiation Biology | Describes models or parameters linking radiation dose to biological effects. |
| Radiobiological Integration | Coupling Biological Models and Clinical Protocols | Incorporates biological parameters (e.g., radiosensitivity indices) into planning. |
| Radiobiological Modeling | Quantitative Biological Modeling of Dose–Response | Used to predict tissue effects and optimize treatment. |
| Radiobiology | Study of Biological Effects of Ionizing Radiation | Core discipline analyzed alongside radiotherapy and oncology. |
| Radiomics | Quantitative Image-Feature Extraction | Supports predictive modeling and risk assessment integrated into adaptive radiotherapy. |
| Radioresistance Mechanisms | Molecular and Cellular Resistance Pathways | Refers to signaling and repair mechanisms that make tumor cells less responsive to radiation. |
| Radiotherapy (RT) | Use of Ionizing Radiation to Treat Cancer | Central disciplinary axis in all queries. |
| RadRes | Radiation Research (Journal) | Appears in a title summarizing 75 years of the field. |
| random_state = 42 | Random-Seed Setting | Ensures reproducible topic-model results. |
| Rate (Dose-Rate) | Dose Per Unit Time | Critical radiobiological variable, particularly relevant for FLASH-RT and LDR contexts. |
| RBE | Relative Biological Effectiveness | Quantifies biological potency of one radiation type relative to another. |
| Repair | DNA or Cellular Repair | Biological process restoring damaged DNA or cellular integrity after irradiation. |
| Reproducibility | Methodological Consistency; Methodological Standards | Refers to achieving consistent quantitative and terminological results across independent AI systems; ensures that pre-processing and modeling steps can be independently verified. |
| RESPONSE | Biological or Clinical Response | Describes radiation-induced effects, gene-expression responses, or treatment outcomes. |
| Risk / Volume | Dosimetric and Anatomical Covariates | Terms indicating planning and outcome modeling foci. |
| RSI | Radiosensitivity Index | Genomic predictor of individual radiosensitivity used for dose calibration. |
| RT | Radiotherapy | Appears in several topics and class definitions. |
| RTOG | Radiation Therapy Oncology Group | U.S. co-operative group providing standardized toxicity and outcome reporting criteria. |
| SARS-CoV-2 / COVID-19 | Virus / Disease | Examined for potential changes in individual radiation sensitivity. |
| SBRT | Stereotactic Body Radiotherapy | Highly conformal, high-precision modality for small or lung tumors; advanced form of external beam radiotherapy delivering high-precision doses to small tumor volumes (directly referenced in Topic_4t). |
| Scopus | Elsevier’s Multidisciplinary Abstract and Citation Database | Used for automated retrieval of bibliographic records on radiotherapy, radiobiology, and oncology. |
| Scopus / PubMed / Web of Science (WoS) | Major Bibliographic Databases | Primary data sources for corpus construction (1964–2025); used for refining, validating, and harmonizing corpus selection. |
| SD / ± | Standard Deviation | Expresses the dispersion of cosine-similarity values within each class. |
| Semantic Supergraph Analysis | Advanced Network Modeling Approach | Suggested future technique for identifying underexplored or emerging clusters within the scientific corpus. |
| Stereotactic Body Radiotherapy (SBRT) | High-Precision, Image-Guided Radiation Technique | Marks transition to modern radiotherapy (post-2010 growth phase). |
| Stopword Filtering | Removal of High-Frequency Function Words | Biomedical-specific list applied before modeling. |
| Supergraph | Global Integrated Co-Occurrence Network | Encompasses molecular, clinical, and technological domains. |
| Supergraph Inset | Central Visual in Fig. 1 | Depicts term relationships with colored weighted edges (blue = weak, red = strong). |
| SURVIVAL | Cellular or Patient Survival | Appears in Topics 1a and 3a, referring to both cell-survival assays and patient outcome metrics. |
| SUS / Unified Health System (SUS) | Sistema Único de Saúde (Brazil’s Unified Health System) | Framework for pilot implementation, ensuring scalability, equity, and cost-effectiveness; Brazil’s public-health system alignment target. |
| SVG | Scalable Vector Graphics | Visualization format for network maps. |
| t | Titles Corpus | The suffix “t” in Topic_0t–Topic_9t designates topics derived from article titles rather than abstracts. |
| Table 2 / Figure 2 | Plan Visualization Elements | Table summarizes indicators; figure visualizes network linking plan classes, health metrics, and strategic goals. |
| TARGET | Radiation Target Volume | Anatomical or biological region receiving prescribed radiation dose (Topic_7a). |
| TARGETED (THERAPY) | Molecularly Targeted Therapy | Used in Topic_5a; describes agents designed to act on specific molecular pathways. |
| TCGA | The Cancer Genome Atlas | Genomic classification system defining molecular subtypes across cancers; applied for adaptive radiotherapy. |
| TCP / NTCP | Tumor Control Probability / Normal Tissue Complication Probability | Radiobiological models estimating treatment success vs. toxicity. |
| Temporal Evolution Curve | Publication-Frequency Over Time | Depicts output growth and term-usage expansion (sharp post-2010 increase). |
| Term-Frequency Curve | Temporal Plot of Keyword Occurrence | Used to link bibliometric evolution to network topology. |
| TF-IDF | Term Frequency–Inverse Document Frequency | Weighting scheme representing documents numerically for cosine-similarity computation; emphasizes discriminative words. |
| Therap / Radiotherapi (Radiotherapy Lemmatized Form) | Lemmatized Word Stems | Generated during text preprocessing before word-cloud and LDA analysis. |
| Therapeutic Window | Efficacy–Toxicity Balance | Keyword guiding selection of translationally relevant articles. |
| Tissue / Normal | Biological Material (Tumor vs. Normal) | Distinction between targeted tumor tissue and healthy tissue, relevant to toxicity models (Topic_9a). |
| TITLE-ABS-KEY | Title–Abstract–Keyword Query Field in Scopus | Syntax specifying that Boolean terms be searched in all three metadata fields. |
| Topic_0a–Topic_9a | Abstract-Based LDA Topics | Capture methodological and biological dimensions (e.g., FLASH-RT, RBE, immunotherapy). |
| Topic_0t–Topic_9t | Title-Based LDA Topics | Identify clusters such as tumor sites, radiation types, or molecular mechanisms; each topic represents a statistically derived keyword cluster from titles. |
| Toxicity | Measure of Treatment-Induced Adverse Effects | One of the outcome indicators for therapy optimization. |
| TP53 / p53abn | Tumor Protein 53 / Abnormal TP53 Subtype | Gene regulating cell cycle and apoptosis; mutation indicates poor prognosis in endometrial and cervical cancers. |
| Translational Continuity | Bridging Experimental and Clinical Domains | Describes how findings progress from bench to bedside within a single analytical cycle. |
| Translational Publication Types / Translational Trials | Research Bridging Lab and Clinic / Studies Bridging Lab Findings and Clinical Implementation | Filters applied in PubMed to maximize relevance; reflected by terms “clinic,” “trial,” “meta,” “evalu.” |
| Transparency | Disclosure of AI Tool Use | Explicitly described to comply with academic-integrity and reproducibility standards. |
| Two-Dimensional Thematic Landscape | Dual-Axis Conceptual Map | Represents integration of clinical and molecular radiotherapy research. |
| UFRJ | Universidade Federal do Rio de Janeiro (Federal University of Rio de Janeiro) | Institutional affiliation of multiple authors. |
| Undirected, Thresholded Version | Simplified Graph with Bidirectional Edges and Minimum-Weight Cutoff | Used to test robustness of results. |
| VHL | von Hippel–Lindau Gene | Mutation induces pseudohypoxia and pro-survival signaling in renal carcinoma. |
| Vitro (from in vitro) | In-Vitro Studies | Laboratory studies conducted outside living organisms, often in cell cultures, used to study radiation-response mechanisms. |
| VOSviewer | Visualization of Similarities Viewer | Software for bibliometric and co-occurrence network visualization. |
| Voxel / Voxel-Based Analysis / Voxel-Based Mapping / Voxel-Level Analytics | Volumetric Pixel and Spatial Dose–Response Modeling Methods | Voxel = smallest unit in 3D medical imaging and dosimetry; used for dose/response mapping and adaptive planning; links radiobiological effects to 3-D anatomical regions for spatially resolved optimization. |
| Weakly Connected Component | Subgraph with at Least One Directional Path Between All Nodes | Indicates overall semantic unity of the dataset. |
| Web of Science (WoS) | Clarivate’s Multidisciplinary Citation Index | Used with direct keyword search to complement PubMed and Scopus coverage. |
| Weighted Degree | Sum of Edge Weights for a Node | Counts total co-occurrence frequency rather than binary presence. |
| Word Cloud | Frequency-Scaled Visual Representation of Keywords | Summarizes lexical prominence from titles and abstracts. |
| α-Parameter | Linear Component of the LQ Model | Represents cell-killing probability per unit dose in radiobiological modeling. |
| α/β Values | Linear–Quadratic Model Parameters | Describe tissue-specific radiation response; used in pediatric and comparative modeling. |
| γH2AX | Phosphorylated Histone H2AX | Biomarker of DNA double-strand breaks; persistence indicates inefficient repair. |
Appendix A
Appendix A.1. Topics from Titles (t)
Appendix A.2. Topics from Abstracts (a)
References
- Song, Z.; Hwang, G.-Y.; Zhang, X.; Huang, S.; Park, B.-K. A Scientific-Article Key-Insight Extraction System Based on Multi-Actor of Fine-Tuned Open-Source Large Language Models. Sci Rep 2025, 15, 1608. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Exploring the Intersection of Machine Learning and Big Data: A Survey. Machine Learning and Knowledge Extraction 2025, 7, 13. [Google Scholar] [CrossRef]
- Elahi, M.M.A.; Hossain, Md.M.; Karim, M.R.; Zain, M.F.M.; Shearer, C. A Review on Alkali-Activated Binders: Materials Composition and Fresh Properties of Concrete. Construction and Building Materials 2020, 260, 119788–119788. [Google Scholar] [CrossRef]
- Shi, F.; Evans, J. Surprising Combinations of Research Contents and Contexts Are Related to Impact and Emerge with Scientific Outsiders from Distant Disciplines. Nat Commun 2023, 14, 1641. [Google Scholar] [CrossRef]
- Masseroli, M.; Mons, B.; Bongcam-Rudloff, E.; Ceri, S.; Kel, A.; Rechenmann, F.; Lisacek, F.; Romano, P. Integrated Bio-Search: Challenges and Trends for the Integration, Search and Comprehensive Processing of Biological Information. BMC Bioinformatics 2014, 15, S2. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ward, L.; Chard, K.; Blaiszik, B.; Foster, I. Challenges and Advances in Information Extraction from Scientific Literature: A Review. JOM 2021, 73, 3383–3400. [Google Scholar] [CrossRef]
- Vuković, D.B.; Dekpo-Adza, S.; Matović, S. AI Integration in Financial Services: A Systematic Review of Trends and Regulatory Challenges. Humanit Soc Sci Commun 2025, 12, 562. [Google Scholar] [CrossRef]
- Giamellaro, M.; Buxton, C.; Taylor, J.; Ayotte-Beaudet, J.-P.; L’Heureux, K.; Beaudry, M.-C. The Landscape of Research on Contextualized Science Learning: A Bibliometric Network Review. Science Education 2025, 109, 851–875. [Google Scholar] [CrossRef]
- Neumann, E.; Thomas, J. Knowledge Assembly for the Life Sciences. Drug Discovery Today 2002, 7, s160–s162. [Google Scholar] [CrossRef]
- Zheng, X.; Peng, P.; Wang, Y.; Bian, L.; Zhao, K.; Shi, A.; Jiang, Z.; Zhao, L.; Jiang, J.; Zhang, S. The Impact of Exercise during Radiotherapy on Treatment-Related Side Effects in Breast Cancer Patients: A Systematic Review and Meta-Analysis. International Journal of Nursing Studies 2025, 163, 104990. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Louie, A.V.; Kotecha, R.; Saxena, A.; Zhang, Y.; Guckenberger, M.; Kim, M.-S.; Scorsetti, M.; Slotman, B.J.; Lo, S.S.; et al. Stereotactic Body Radiotherapy for Non-Spine Bone Metastases: A Meta-Analysis and International Stereotactic Radiosurgery Society (ISRS) Clinical Practice Guidelines. Radiotherapy and Oncology 2025, 205, 110717. [Google Scholar] [CrossRef] [PubMed]
- Kishan, A.U.; Sun, Y.; Tree, A.C.; Hall, E.; Dearnaley, D.; Catton, C.N.; Lukka, H.R.; Pond, G.; Lee, W.R.; Sandler, H.M.; et al. Hypofractionated Radiotherapy for Prostate Cancer (HYDRA): An Individual Patient Data Meta-Analysis of Randomised Trials in the MARCAP Consortium. The Lancet Oncology 2025, 26, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Javadnia, P.; Bahadori, A.R.; Ghanaatpisheh, A.; Dahaghin, S.; Rajabi, M.; Davari, A.; Sheikhvatan, M.; Ranji, S.; Shafiee, S.; Tafakhori, A. The Safety and Efficacy of Robotic Radiosurgery and Radiotherapy in the Management of Skull Base Tumors: A Systematic Review and Meta-Analysis. Neurosurg Rev 2025, 48, 39. [Google Scholar] [CrossRef]
- Huang, R.S.; Chow, R.; Benour, A.; Chen, D.; Boldt, G.; Wallis, C.J.D.; Swaminath, A.; Simone, C.B.; Lock, M.; Raman, S. Comparative Efficacy and Safety of Ablative Therapies in the Management of Primary Localised Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. The Lancet Oncology 2025, 26, 387–398. [Google Scholar] [CrossRef]
- Guo, Y.; Hao, S.; Huang, Q.; Di, C.; Gan, L.; Xie, Y.; Li, Q.; Si, J. Unraveling the Dual Nature of FLASH Radiotherapy: From Normal Tissue Sparing to Tumor Control. Cancer Letters 2025, 630, 217895. [Google Scholar] [CrossRef]
- Tan, V.S.; Padayachee, J.; Rodrigues, G.B.; Navarro, I.; Shah, P.S.; Palma, D.A.; Barry, A.; Fazelzad, R.; Raphael, J.; Helou, J. Stereotactic Ablative Radiotherapy for Oligoprogressive Solid Tumours: A Systematic Review and Meta-Analysis. Radiotherapy and Oncology 2024, 200, 110505. [Google Scholar] [CrossRef]
- Rodrigues, A.C.L.F.; Tos, S.M.; Shaaban, A.; Mantziaris, G.; Trifiletti, D.M.; Sheehan, J. Proton Beam and Carbon Ion Radiotherapy in Skull Base Chordoma: A Systematic Review, Meta-Analysis and Meta-Regression with Trial Sequential Analysis. Neurosurg Rev 2024, 47, 893. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Zhao, X.; Liu, X.; Zhang, Y.; Tian, J.; Li, M. Treatment Options for Unilateral Vestibular Schwannoma: A Network Meta-Analysis. BMC Cancer 2024, 24, 1490. [Google Scholar] [CrossRef]
- Duckett, K.A.; Kassir, M.F.; Nguyen, S.A.; Brennan, E.A.; Chera, B.S.; Sterba, K.R.; Halbert, C.H.; Hill, E.G.; McCay, J.; Puram, S.V.; et al. Factors Associated with Head and Neck Cancer Postoperative Radiotherapy Delays: A Systematic Review and Meta-analysis. Otolaryngol.--head neck surg. 2024, 171, 1265–1282. [Google Scholar] [CrossRef]
- Daloiso, A.; Cazzador, D.; Concheri, S.; Tealdo, G.; Zanoletti, E. Long-Term Hearing Outcome For Vestibular Schwannomas After Microsurgery And Radiotherapy: A Systematic Review and Meta-Analysis. Otolaryngol.--head neck surg. 2024, 171, 1670–1681. [Google Scholar] [CrossRef]
- Boterberg, T.; Dunlea, C.; Harrabi, S.; Janssens, G.; Laprie, A.; Whitfield, G.; Gaze, M. Contemporary Paediatric Radiation Oncology. Arch Dis Child 2023, 108, 332–337. [Google Scholar] [CrossRef]
- Waqar, M.; Trifiletti, D.M.; McBain, C.; O’Connor, J.; Coope, D.J.; Akkari, L.; Quinones-Hinojosa, A.; Borst, G.R. Early Therapeutic Interventions for Newly Diagnosed Glioblastoma: Rationale and Review of the Literature. Curr Oncol Rep 2022, 24, 311–324. [Google Scholar] [CrossRef]
- Kempson, I. Nanoparticle-Based Radiosensitization. IJMS 2020, 21, 2879. [Google Scholar] [CrossRef]
- Sidibe, I.; Biau, J.; Graff, P. Carcinomes épidermoïdes de la cavité buccale et du pharyngo-larynx. Quels volumes ganglionnaires traiter en radiothérapie externe ? Cancer/Radiothérapie 2019, 23, 696–700. [Google Scholar] [CrossRef]
- O’Regan, T.; Robinson, L.; Newton-Hughes, A.; Strudwick, R. A Review of Visual Ethnography: Radiography Viewed through a Different Lens. Radiography 2019, 25, S9–S13. [Google Scholar] [CrossRef] [PubMed]
- Bleiker, J.; Morgan-Trimmer, S.; Knapp, K.; Hopkins, S. Navigating the Maze: Qualitative Research Methodologies and Their Philosophical Foundations. Radiography 2019, 25, S4–S8. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Clementel, E.; Eaton, D.J.; Greer, P.B.; Haworth, A.; Ishikura, S.; Kry, S.F.; Lehmann, J.; Lye, J.; Monti, A.F.; et al. A Virtual Dosimetry Audit – Towards Transferability of Gamma Index Analysis between Clinical Trial QA Groups. Radiotherapy and Oncology 2017, 125, 398–404. [Google Scholar] [CrossRef]
- Cosset, J.-M. L’aube de la radiothérapie, entre coups de génie, drames et controverses. Cancer/Radiothérapie 2016, 20, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Cai, D.; Zhong, Q.; Dou, W.; Yuan, B. Impact of Breast Size on Dosimetry and Radiobiology of VMAT Left-sided Breast-conserving Conventional Fractionation Radiotherapy under Setup Errors. J Applied Clin Med Phys 2025, 26, e70151. [Google Scholar] [CrossRef]
- Zhang, B.; Zhong, R.; Shen, G.; Tuo, C.; Dong, Y.; Wang, W.; Zhang, M.; Tong, G.; Zhang, H.; Yuan, B.; et al. The Space Radiobiological Exposure Facility on the China Space Station. Astrobiology 2025, ast.2024.0027. [Google Scholar] [CrossRef]
- Xu, T.; Liu, F.; He, J.; Xu, P.; Qu, J.; Wang, H.; Yue, J.; Yang, Q.; Wu, W.; Zeng, G.; et al. Leveraging Zebrafish Models for Advancing Radiobiology: Mechanisms, Applications, and Future Prospects in Radiation Exposure Research. Environmental Research 2025, 266, 120504. [Google Scholar] [CrossRef]
- Taliaferro, L.P.; Brenner, D.J.; Amundson, S.A.; Garty, G.; Zhang, Y.; Davies, E.W.; Carrier, F.; Ross, J.R.; Cline, J.M.; Chao, N.J. Centers for Medical Countermeasures against Radiation Consortium: Past, Present, and Beyond. Radiation Research 2025, 204. [Google Scholar] [CrossRef]
- Rovituso, M.; Groenendijk, C.F.; Van Der Wal, E.; Van Burik, W.; Ibrahimi, A.; Rituerto Prieto, H.; Brown, J.M.C.; Weber, U.; Simeonov, Y.; Fontana, M.; et al. Characterisation of the HollandPTC R&D Proton Beamline for Physics and Radiobiology Studies. Physica Medica 2025, 130, 104883. [Google Scholar] [CrossRef]
- Pouget, J.-P.; Gabina, P.M.; Herrmann, K.; Deandreis, D.; Konijnenberg, M.; Taieb, D.; Van Leeuwen, F.W.B.; Kurth, J.; Eberlein, U.; Lassmann, M.; et al. EANM Expert Opinion: How Can Lessons from Radiobiology Be Applied to the Design of Clinical Trials? Part I: Back to the Basics of Absorbed Dose–Response and Threshold Absorbed Doses. Eur J Nucl Med Mol Imaging 2025, 52, 1210–1222. [Google Scholar] [CrossRef]
- Oliveira Dias, J.; Sampaio Fagundes, I.; Bisio, M.D.C.; Da Silva Barboza, V.; Jacinto, A.A.; Altei, W.F. Extracellular Vesicles as the Common Denominator among the 7 Rs of Radiobiology: From the Cellular Level to Clinical Practice. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2025, 1880, 189315. [Google Scholar] [CrossRef]
- Mendes, F.; Terry, S.Y.A.; Spiegelberg, D.; Cornelissen, B.; Bolcaen, J.; Nonnekens, J. ; On behalf of the European Working group of Radiobiology of Molecular Radionuclide Therapy Recommendations for Reporting Preclinical Radiobiological Studies in Targeted Radionuclide Therapy. Eur J Nucl Med Mol Imaging 2025, 52, 3066–3070. [Google Scholar] [CrossRef] [PubMed]
- Kazmierska-Grebowska, P.; Jankowski, M.M.; Obrador, E.; Kolodziejczyk-Czepas, J.; Litwinienko, G.; Grebowski, J. Nanotechnology Meets Radiobiology: Fullerenols and Metallofullerenols as Nano-Shields in Radiotherapy. Biomedicine & Pharmacotherapy 2025, 184, 117915. [Google Scholar] [CrossRef]
- Hill, M.A.; Silvestre Patallo, I.; Aitkenhead, A.H.; Bazalova-Carter, M.; Carter, R.; Nill, S.; Nisbet, A.; Ghita-Pettigrew, M.; Poirier, Y.; Prise, K.M.; et al. The Importance of Standardization and Challenges of Dosimetry in Conventional Preclinical Radiation Biology Research. British Journal of Radiology 2025, 98, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Shang, Y.; Zhang, S.; Fan, S. The Interplay between RNA m6A Modification and Radiation Biology of Cancerous and Non-Cancerous Tissues: A Narrative Review. Cancer Biol Med 2025, 21, 1120–1140. [Google Scholar] [CrossRef] [PubMed]
- Charalampopoulou, A.; Barcellini, A.; Magro, G.; Bellini, A.; Borgna, S.S.; Fulgini, G.; Ivaldi, G.B.; Mereghetti, A.; Orlandi, E.; Pullia, M.G.; et al. Advancing Radiobiology: Investigating the Effects of Photon, Proton, and Carbon-Ion Irradiation on PANC-1 Cells in 2D and 3D Tumor Models. Current Oncology 2025, 32, 49. [Google Scholar] [CrossRef]
- Azevedo, T.A.; Abrantes, A.M.; Carvalho, J. Radiobiological Modeling with Monte Carlo Tools – Simulating Cellular Responses to Ionizing Radiation. Technol Cancer Res Treat 2025, 24, 15330338251350909. [Google Scholar] [CrossRef]
- Akolawala, Q.; Accardo, A. Engineered Cell Microenvironments: A Benchmark Tool for Radiobiology. ACS Appl. Mater. Interfaces 2025, 17, 5563–5577. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, F.; Ghorbani, M.; Seif, F.; Deevband, M.R.; Pursamimi, M.; Babai, S.; Tavakoli, M. Comparison Study between Different Treatment Planning Techniques for Medulloblastoma in Terms of Dosimetry, Radiobiology, and Secondary Cancer Risk. Journal of Cancer Research and Therapeutics 2025, 21, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Punshon, L.D.; Fabbrizi, M.R.; Phoenix, B.; Green, S.; Parsons, J.L. Current Insights into the Radiobiology of Boron Neutron Capture Therapy and the Potential for Further Improving Biological Effectiveness. Cells 2024, 13, 2065. [Google Scholar] [CrossRef] [PubMed]
- Gardner, L.L.; Thompson, S.J.; O’Connor, J.D.; McMahon, S.J. Modelling Radiobiology. Phys. Med. Biol. 2024, 69, 18TR01. [Google Scholar] [CrossRef]
- Zhou, Z.; Guan, B.; Xia, H.; Zheng, R.; Xu, B. Particle Radiotherapy in the Era of Radioimmunotherapy. Cancer Letters 2023, 567, 216268. [Google Scholar] [CrossRef]
- Tuleasca, C.; Tripathi, M.; Starnoni, D.; Daniel, R.T.; Reyns, N.; Levivier, M. Radiobiology of Radiosurgery for Neurosurgeons. Neurology India 2023, 71, S14–S20. [Google Scholar] [CrossRef]
- Ma, C.M.C. Pulsed Low Dose-Rate Radiotherapy: Radiobiology and Dosimetry. Phys. Med. Biol. 2022, 67, 03TR01. [Google Scholar] [CrossRef]
- Barcellos-Hoff, M.H. The Radiobiology of TGFβ. Seminars in Cancer Biology 2022, 86, 857–867. [Google Scholar] [CrossRef]
- Morris, Z.S.; Wang, A.Z.; Knox, S.J. The Radiobiology of Radiopharmaceuticals. Seminars in Radiation Oncology 2021, 31, 20–27. [Google Scholar] [CrossRef]
- Sgouros, G.; Hobbs, R.; Josefsson, A. Dosimetry and Radiobiology of Alpha-Particle Emitting Radionuclides. CRP 2018, 11, 209–214. [Google Scholar] [CrossRef]
- Thiagarajan, A.; Yamada, Y. Radiobiology and Radiotherapy of Brain Metastases. Clin Exp Metastasis 2017, 34, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Valenza, C.; Saldanha, E.F.; Gong, Y.; De Placido, P.; Gritsch, D.; Ortiz, H.; Trapani, D.; Conforti, F.; Cremolini, C.; Peters, S.; et al. Circulating Tumor DNA Clearance as a Predictive Biomarker of Pathologic Complete Response in Patients with Solid Tumors Treated with Neoadjuvant Immune Checkpoint Inhibitors: A Systematic Review and Meta-Analysis. Annals of Oncology 2025, 36, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Soong, R.Y.; Low, C.E.; Ong, V.; Sim, I.; Lee, C.; Lee, F.; Chew, L.; Yau, C.E.; Lee, A.R.Y.B.; Chen, M.Z. Exercise Interventions for Depression, Anxiety, and Quality of Life in Older Adults With Cancer: A Systematic Review and Meta-Analysis. JAMA Netw Open 2025, 8, e2457859. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Han, H.; Li, H.; Wei, L.; Wu, H.; Li, C.; Xie, X.; Zhang, B.; Li, Z.; Chen, X.; et al. Cancer Risk Subsequent to Cardiovascular Disease: A Prospective Population-Based Study and Meta-Analysis. BMC Med 2025, 23, 192. [Google Scholar] [CrossRef]
- Rischin, D.; Porceddu, S.; Day, F.; Brungs, D.P.; Christie, H.; Jackson, J.E.; Stein, B.N.; Su, Y.B.; Ladwa, R.; Adams, G.; et al. Adjuvant Cemiplimab or Placebo in High-Risk Cutaneous Squamous-Cell Carcinoma. N Engl J Med 2025, 393, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Mamounas, E.P.; Bandos, H.; White, J.R.; Julian, T.B.; Khan, A.J.; Shaitelman, S.F.; Torres, M.A.; Vicini, F.A.; Ganz, P.A.; McCloskey, S.A.; et al. Omitting Regional Nodal Irradiation after Response to Neoadjuvant Chemotherapy. N Engl J Med 2025, 392, 2113–2124. [Google Scholar] [CrossRef]
- Kuerer, H.M.; Valero, V.; Smith, B.D.; Krishnamurthy, S.; Diego, E.J.; Johnson, H.M.; Lin, H.; Shen, Y.; Lucci, A.; Shaitelman, S.F.; et al. Selective Elimination of Breast Surgery for Invasive Breast Cancer: A Nonrandomized Clinical Trial. JAMA Oncol 2025, 11, 529. [Google Scholar] [CrossRef]
- Hwang, E.S.; Hyslop, T.; Lynch, T.; Ryser, M.D.; Weiss, A.; Wolf, A.; Norris, K.; Witten, M.; Grimm, L.; Schnitt, S.; et al. Active Monitoring With or Without Endocrine Therapy for Low-Risk Ductal Carcinoma In Situ: The COMET Randomized Clinical Trial. JAMA 2025, 333, 972. [Google Scholar] [CrossRef]
- Haddad, R.; Fayette, J.; Teixeira, M.; Prabhash, K.; Mesia, R.; Kawecki, A.; Dechaphunkul, A.; Dinis, J.; Guo, Y.; Masuda, M.; et al. Atezolizumab in High-Risk Locally Advanced Squamous Cell Carcinoma of the Head and Neck: A Randomized Clinical Trial. JAMA 2025, 333, 1599. [Google Scholar] [CrossRef]
- Galsky, M.D.; Witjes, J.A.; Gschwend, J.E.; Milowsky, M.I.; Schenker, M.; Valderrama, B.P.; Tomita, Y.; Bamias, A.; Lebret, T.; Shariat, S.F.; et al. Adjuvant Nivolumab in High-Risk Muscle-Invasive Urothelial Carcinoma: Expanded Efficacy From CheckMate 274. JCO 2025, 43, 15–21. [Google Scholar] [CrossRef]
- Fekrvand, S.; Abolhassani, H.; Esfahani, Z.H.; Fard, N.N.G.; Amiri, M.; Salehi, H.; Almasi-Hashiani, A.; Saeedi-Boroujeni, A.; Fathi, N.; Mohtashami, M.; et al. Cancer Trends in Inborn Errors of Immunity: A Systematic Review and Meta-Analysis. J Clin Immunol 2025, 45, 34. [Google Scholar] [CrossRef]
- Courneya, K.S.; Vardy, J.L.; O’Callaghan, C.J.; Gill, S.; Friedenreich, C.M.; Wong, R.K.S.; Dhillon, H.M.; Coyle, V.; Chua, N.S.; Jonker, D.J.; et al. Structured Exercise after Adjuvant Chemotherapy for Colon Cancer. N Engl J Med 2025, 393, 13–25. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, L.; Pang, Y.; Shi, J.; Gao, H.; Sun, Y.; Chen, J.; Fu, H.; Cai, J.; Yu, L.; et al. 68Ga-MY6349 PET/CT Imaging to Assess Trop2 Expression in Multiple Types of Cancer. Journal of Clinical Investigation 2025, 135, e185408. [Google Scholar] [CrossRef] [PubMed]
- Bettariga, F.; Galvao, D.A.; Taaffe, D.R.; Bishop, C.; Lopez, P.; Maestroni, L.; Quinto, G.; Crainich, U.; Verdini, E.; Bandini, E.; et al. Association of Muscle Strength and Cardiorespiratory Fitness with All-Cause and Cancer-Specific Mortality in Patients Diagnosed with Cancer: A Systematic Review with Meta-Analysis. Br J Sports Med 2025, 59, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Huang, Y.; Zeng, L.; Li, N.; Hao, L.; Yue, J.; Li, S.; Deng, J.; Yu, F.; Hu, X. The Genetically Predicted Causal Associations between Circulating 3-Hydroxybutyrate Levels and Malignant Neoplasms: A Pan-Cancer Mendelian Randomization Study. Clinical Nutrition 2024, 43, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhu, C.; Li, L.; Li, Y.; Chen, Y.; Hu, X. The Effect of Targeted Palliative Care Interventions on Depression, Quality of Life and Caregiver Burden in Informal Caregivers of Advanced Cancer Patients: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. International Journal of Nursing Studies 2024, 160, 104895. [Google Scholar] [CrossRef]
- Quan, L.; Liu, J.; Wang, Y.; Yang, F.; Yang, Z.; Ju, J.; Shuai, Y.; Wei, T.; Yue, J.; Wang, X.; et al. Exploring Risk Factors for Endocrine-Related Immune-Related Adverse Events: Insights from Meta-Analysis and Mendelian Randomization. Human Vaccines & Immunotherapeutics 2024, 20, 2410557. [Google Scholar] [CrossRef]
- Nichols, A.C.; Theurer, J.; Prisman, E.; Read, N.; Berthelet, E.; Tran, E.; Fung, K.; De Almeida, J.R.; Bayley, A.; Goldstein, D.P.; et al. Radiotherapy Versus Transoral Robotic Surgery for Oropharyngeal Squamous Cell Carcinoma: Final Results of the ORATOR Randomized Trial. JCO 2024, 42, 4023–4028. [Google Scholar] [CrossRef]
- Ludmir, E.B.; Sherry, A.D.; Fellman, B.M.; Liu, S.; Bathala, T.; Haymaker, C.; Medina-Rosales, M.N.; Reuben, A.; Holliday, E.B.; Smith, G.L.; et al. Addition of Metastasis-Directed Therapy to Systemic Therapy for Oligometastatic Pancreatic Ductal Adenocarcinoma (EXTEND): A Multicenter, Randomized Phase II Trial. JCO 2024, 42, 3795–3805. [Google Scholar] [CrossRef]
- Long, Y.; Zhou, Z.; Zhou, S.; Zhang, G. The Effectiveness of Different Non-Pharmacological Therapies on Cancer-Related Fatigue in Cancer Patients:A Network Meta-Analysis. International Journal of Nursing Studies 2024, 160, 104904. [Google Scholar] [CrossRef]
- Baker, J.; Noguchi, N.; Marinovich, M.L.; Sprague, B.L.; Salisbury, E.; Houssami, N. Atypical Ductal or Lobular Hyperplasia, Lobular Carcinoma in-Situ, Flat Epithelial Atypia, and Future Risk of Developing Breast Cancer: Systematic Review and Meta-Analysis. The Breast 2024, 78, 103807. [Google Scholar] [CrossRef]
- Pelosi, D.; Cacciagrano, D.; Piangerelli, M. Explainability and Interpretability in Concept and Data Drift: A Systematic Literature Review. Algorithms 2025, 18, 443. [Google Scholar] [CrossRef]
- Bibri, S.E.; Huang, J.; Jagatheesaperumal, S.K.; Krogstie, J. The Synergistic Interplay of Artificial Intelligence and Digital Twin in Environmentally Planning Sustainable Smart Cities: A Comprehensive Systematic Review. Environmental Science and Ecotechnology 2024, 20, 100433. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, S.; Ouyang, C.; Chen, M.; Liu, C.; Zhang, J.; Yu, L.; Wang, F.; Xie, Y.; Li, J.; et al. Artificial Intelligence for Geoscience: Progress, Challenges, and Perspectives. Innovation (Camb) 2024, 5, 100691. [Google Scholar] [CrossRef]
- Gomes Souza, F.; Bhansali, S.; Pal, K.; Silveira Maranhão, F. da; Santos Oliveira, M.; Valladão, V.S.; Brandão e Silva, D.S.; Silva, G.B. A 30-Year Review on Nanocomposites: Comprehensive Bibliometric Insights into Microstructural, Electrical, and Mechanical Properties Assisted by Artificial Intelligence. Materials 2024, 17, 1088. [Google Scholar] [CrossRef]
- Valladão, V.S.; da Silva, E.O.; Souza Jr., F. G. A Data Mining Study of Zinc Oxide Nanostructures Utilization in Drug Delivery Nanosystems. Macromolecular Symposia 2024, 413, 2300141. [Google Scholar] [CrossRef]
- Gomes, F.; Bhansali, S.; Valladão, V.; Brandão, D.; Silva, G.; Maranhão, F.; Pal, K.; Thiré, R.; Araújo, J.; Batista, A.; et al. Advancing Dye-Sensitized Solar Cells: Synergistic Effects of Polyaniline, Graphene Oxide, and Carbon Nanotubes for Enhanced Efficiency and Sustainability Developments. 2025. [CrossRef]
- Gomes Souza, F.; Pal, K.; Maranhão, F.; Zanoni, C.; Brandão, D.; Colão, M.; Silva, G.; Ampah, J.; Velasco, K. Advancing Hybrid Nanocatalyst Research: A Python-basedVisualization of Similarity Analysis for Interdisciplinary andSustainable Development. CNANO 2024, 20, 830–856. [Google Scholar] [CrossRef]
- Almeida, T.M.D.; Souza, F.G. de; Pal, K. Bibliometric Analysis of the Hot Theme “Phytosynthesized Nanoparticles.”. ABEB 2020, 4, 1–5. [Google Scholar] [CrossRef]
- Gomes Souza, F.; Pal, K.; Ampah, J.D.; Dantas, M.C.; Araújo, A.; Maranhão, F.; Domingues, P. Biofuels and Nanocatalysts: Python Boosting Visualization of Similarities. Materials 2023, 16, 1175. [Google Scholar] [CrossRef]
- Santos, J.F.; Del Rocío Silva-Calpa, L.; De Souza, F.G.; Pal, K. Central Countries’ and Brazil’s Contributions to Nanotechnology. CNM 2024, 9, 109–147. [Google Scholar] [CrossRef]
- Daher, E.; Junior, F.G. de S.; Carelo, J.; Brandão, V. Drug Delivery Polymers: An Analysis Based on Literature Text Mining. Brazilian Journal of Experimental Design, Data Analysis and Inferential Statistics 2021, 1, 40–55. [Google Scholar] [CrossRef]
- Gomes S., F.; Velasco, K.; Cunha, S.; Santos, J.; Aboelkheir, M.G.; Sumini, M.; Thiré, R.; Duarte, P.C.; Andrade, A.J.P.; Díaz-Martín, R.D.; et al. Leveraging Large Language Models for Accelerated Learning and Innovation in Biogenic Tissue-Engineered Vascular Grafts. Journal of Drug Delivery Science and Technology 2025, 108, 106935. [Google Scholar] [CrossRef]
- Junior, F.G. de S.; Barradas, T.N.; Caetano, V.F.; Becerra, A. Nanoparticles Improving Polyaniline Electrical Conductivity: A Meta-Analysis Study. Brazilian Journal of Experimental Design, Data Analysis and Inferential Statistics 2022, 2, 25–58. [Google Scholar] [CrossRef]
- Costa, V.C.; Junior, F.G. de S.; Thomas, S.; Filho, R.D.T.; Sousa, L.D.C.; Filho, S.T.; Carvalho, F.V.D.; Maranhão, F.D.S.; Aboelkheir, M.G.; Lima, N.R.B. de; et al. Nanotechnology in Concrete: A Bibliometric Review. Brazilian Journal of Experimental Design, Data Analysis and Inferential Statistics 2021, 1, 100–113. [Google Scholar] [CrossRef]
- Daher, E.; Junior, F.G. de S.; Brandão, V.; Carelo, J. Poly(Lactic Acid)-PLA: An Analysis Based on Literature Text Mining. Brazilian Journal of Experimental Design, Data Analysis and Inferential Statistics 2021, 1, 56–68. [Google Scholar] [CrossRef]
- Valladão, V.S.; Souza Júnior, F.G. de; Thode Filho, S.; Maranhão, F. da S.; Ribeiro, L. de S.; Carneiro, M.E. da S.; Santos, R.K. da S. dos Sentiment Mapping of Microplastic Awareness in Educational Environments. Polímeros 2025, 35, e20250017. [Google Scholar] [CrossRef]
- Gomes, F.; Bhansali, S.; Valladão, V.; Maranhão, F.; Brandão, D.; Delfino, C.; Asthana, N. Sustainable Catalysts: Advances in Geopolymer-Catalyzed Reactions and Their Applications. Journal of Molecular Structure 2025, 142017. [Google Scholar] [CrossRef]
- Delfino, C.; Perez Cordovés, A.I.; F.G., S.J. The Use of Biosensor as a New Trend in Cancer: Bibliometric Analysis from 2007 to 2017. RDMS 2018, 7. [CrossRef]
- Parvin, N.; Joo, S.W.; Jung, J.H.; Mandal, T.K. Unlocking the Future: Carbon Nanotubes as Pioneers in Sensing Technologies. Chemosensors 2025, 13, 225. [Google Scholar] [CrossRef]
- Santos, F.D.D.P.B. dos; Junior, F.G. de S. Up-and-Coming Oil-Sorbing Green Fibers: A Text Mining Study. Brazilian Journal of Experimental Design, Data Analysis and Inferential Statistics 2021, 1, 114–129. [Google Scholar] [CrossRef]
- Gomes, F.; Bhansali, S.; Maranhão, F.; Valladão, V.; Velasco, K. What Are the Future Directions for Microplastics Characterization? A Regex-Llama Data Mining Approach for Identifying Emerging Trends. An. Acad. Bras. Ciênc. 2025, 97, e20241345. [Google Scholar] [CrossRef] [PubMed]
- Yafen Wang; Yuhan Ying; Fan Wang Radiomics in Head and Neck Cancer: A Web of Science-Based Bibliometric and Visualized Analysis. medsc 2025, 6. [CrossRef]
- Andrei, V.; Arandjelović, O. Complex Temporal Topic Evolution Modelling Using the Kullback-Leibler Divergence and the Bhattacharyya Distance. J Bioinform Sys Biology 2016, 2016, 16. [Google Scholar] [CrossRef]
- Tabibi, S.; Ali, L.; Jyothi, H.; Asasfeh, E.; Abdelgani, W. Integration of Artificial Intelligence in Predicting Radiotherapy Outcomes for Glioblastoma Multiforme (GBM). IJFMR 2025, 7, 51688. [Google Scholar] [CrossRef]
- Lastrucci, A.; Wandael, Y.; Ricci, R.; Maccioni, G.; Giansanti, D. The Integration of Deep Learning in Radiotherapy: Exploring Challenges, Opportunities, and Future Directions through an Umbrella Review. Diagnostics 2024, 14, 939. [Google Scholar] [CrossRef]
- Momin, S.; Fu, Y.; Lei, Y.; Roper, J.; Bradley, J.D.; Curran, W.J.; Liu, T.; Yang, X. Knowledge-Based Radiation Treatment Planning: A Data-Driven Method Survey 2020.
- Trifiletti, D.M.; Showalter, T.N. Big Data and Comparative Effectiveness Research in Radiation Oncology: Synergy and Accelerated Discovery. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef]
- F.G. Souza Jr. PaperProcessor - Text Extraction, NLP, and Topic Modeling for Academic Reports 2024.
- Story, M.D.; Davis, A.J.; Sishc, B.J. Current Trends and Future Perspectives in Hadron Therapy: Radiobiology. Health Technol. 2024, 14, 867–872. [Google Scholar] [CrossRef]
- Bermúdez-Guzmán, L.; Blanco-Saborío, A.; Ramírez-Zamora, J.; Lovo, E. The Time for Chronotherapy in Radiation Oncology. Front. Oncol. 2021, 11, 687672. [Google Scholar] [CrossRef]
- Martin, O.A.; Sykes, P.J.; Lavin, M.; Engels, E.; Martin, R.F. What’s Changed in 75 Years of RadRes? – An Australian Perspective on Selected Topics. Radiation Research 2024, 202. [Google Scholar] [CrossRef]
- Luo, Y. Radiobiological Perspective on Metrics for Quantifying Dose Enhancement Effects of High-Z Nanoparticles. Front. Nanotechnol. 2025, 7, 1603334. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, Z.; Perentesis, J.P.; Khuntia, D.; Pfister, S.X.; Sharma, R.A. Can Rational Combination of Ultra-High Dose Rate FLASH Radiotherapy with Immunotherapy Provide a Novel Approach to Cancer Treatment? Clinical Oncology 2021, 33, 713–722. [Google Scholar] [CrossRef]
- Dokic, I.; Mairani, A.; Niklas, M.; Zimmermann, F.; Chaudhri, N.; Krunic, D.; Tessonnier, T.; Ferrari, A.; Parodi, K.; Jäkel, O.; et al. Next Generation Multi-Scale Biophysical Characterization of High Precision Cancer Particle Radiotherapy Using Clinical Proton, Helium-, Carbon- and Oxygen Ion Beams. Oncotarget 2016, 7, 56676–56689. [Google Scholar] [CrossRef] [PubMed]
- Calvaruso, M.; Pucci, G.; Alberghina, C.; Minafra, L. Radiation Therapy Personalization in Cancer Treatment: Strategies and Perspectives. IJMS 2025, 26, 6375. [Google Scholar] [CrossRef] [PubMed]
- Tini, P.; Nardone, V.; Pastina, P.; Pirtoli, L.; Correale, P.; Giordano, A. The Effects of Radiotherapy on the Survival of Patients with Unresectable Non-Small Cell Lung Cancer. Expert Review of Anticancer Therapy 2018, 18, 593–602. [Google Scholar] [CrossRef]
- Willers, H.; Keane, F.K.; Kamran, S.C. Toward a New Framework for Clinical Radiation Biology. Hematology/Oncology Clinics of North America 2019, 33, 929–945. [Google Scholar] [CrossRef]
- Mery, B.; Rancoule, C.; Rowinski, E.; Bosacki, C.; Vallard, A.; Guy, J.-B.; Magné, N. Cancer du rein et radiothérapie: radiorésistance et au-delà. Bulletin du Cancer 2018, 105, S280–S285. [Google Scholar] [CrossRef]
- Arians, N.; Nicolay, N.H.; Brons, S.; Koerber, S.A.; Jaschke, C.; Vercruysse, M.; Daffinger, S.; Rühle, A.; Debus, J.; Lindel, K. Carbon-Ion Irradiation Overcomes HPV-Integration/E2 Gene-Disruption Induced Radioresistance of Cervical Keratinocytes. Journal of Radiation Research 2019, 60, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Cozzarini, C.; Olivieri, M.; Magli, A.; Cante, D.; Noris Chiorda, B.; Munoz, F.; Faiella, A.; Olivetta, E.; Deantoni, C.; Fodor, A.; et al. Accurate Prediction of Long-Term Risk of Biochemical Failure after Salvage Radiotherapy Including the Impact of Pelvic Node Irradiation. Radiotherapy and Oncology 2022, 175, 26–32. [Google Scholar] [CrossRef]
- Jazmati, D.; Sohn, D.; Hörner-Rieber, J.; Qin, N.; Bölke, E.; Haussmann, J.; Schwarz, R.; Niggemeier, N.D.; Borkhardt, A.; Babor, F.; et al. Alpha/Beta Values in Pediatric Medulloblastoma: Implications for Tailored Approaches in Radiation Oncology. Radiat Oncol 2025, 20, 17. [Google Scholar] [CrossRef]
- Chekhun, V.F.; Domina, E.A. Can SARS-CoV-2 Change Individual Radiation Sensitivity of the Patients Recovered from COVID-19? (Experimental and Theoretical Background). Experimental Oncology 2021, 43, 277–280. [Google Scholar] [CrossRef]
- Raitanen, J.; Barta, B.; Hacker, M.; Georg, D.; Balber, T.; Mitterhauser, M. Comparison of Radiation Response between 2D and 3D Cell Culture Models of Different Human Cancer Cell Lines. Cells 2023, 12, 360. [Google Scholar] [CrossRef]
- Nikitovic, M.; Stanojkovic, T. Current Aspects of Radiobiology in Modern Radiotherapy - Our Clinical Experience. Srp Arh Celok Lek 2022, 150, 732–736. [Google Scholar] [CrossRef]
- Kang, J.; Coates, J.T.; Strawderman, R.L.; Rosenstein, B.S.; Kerns, S.L. Genomics Models in Radiotherapy: From Mechanistic to Machine Learning. Medical Physics 2020, 47. [Google Scholar] [CrossRef] [PubMed]
- Sirák, I.; Šinkorová, Z.; Šenkeříková, M.; Špaček, J.; Laco, J.; Vošmiková, H.; John, S.; Petera, J. Hypersensitivity to Chemoradiation in FANCA Carrier with Cervical Carcinoma—A Case Report and Review of the Literature. Reports of Practical Oncology & Radiotherapy 2015, 20, 309–315. [Google Scholar] [CrossRef]
- Ogawa, Y. Paradigm Shift in Radiation Biology/Radiation Oncology—Exploitation of the “H2O2 Effect” for Radiotherapy Using Low-LET (Linear Energy Transfer) Radiation Such as X-Rays and High-Energy Electrons. Cancers 2016, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Kerns, S.L.; Chuang, K.-H.; Hall, W.; Werner, Z.; Chen, Y.; Ostrer, H.; West, C.; Rosenstein, B. Radiation Biology and Oncology in the Genomic Era. The British Journal of Radiology 2018, 91, 20170949. [Google Scholar] [CrossRef]
- Her, E.J.; Reynolds, H.M.; Mears, C.; Williams, S.; Moorehouse, C.; Millar, J.L.; Ebert, M.A.; Haworth, A. Radiobiological Parameters in a Tumour Control Probability Model for Prostate Cancer LDR Brachytherapy. Phys. Med. Biol. 2018, 63, 135011. [Google Scholar] [CrossRef]
- Verduijn, G.M.; Capala, M.E.; Sijtsema, N.D.; Lauwers, I.; Hernandez Tamames, J.A.; Heemsbergen, W.D.; Sewnaik, A.; Hardillo, J.A.; Mast, H.; Van Norden, Y.; et al. The COMPLETE Trial: HolistiC Early respOnse assessMent for oroPharyngeaL cancEr paTiEnts; Protocol for an Observational Study. BMJ Open 2022, 12, e059345. [Google Scholar] [CrossRef]
- McWilliam, A.; Palma, G.; Abravan, A.; Acosta, O.; Appelt, A.; Aznar, M.; Monti, S.; Onjukka, E.; Panettieri, V.; Placidi, L.; et al. Voxel-Based Analysis: Roadmap for Clinical Translation. Radiotherapy and Oncology 2023, 188, 109868. [Google Scholar] [CrossRef]
- Mészáros, N.; Farkas, G.; Székely, G.; Kocsis, Z.S.; Kelemen, P.B.; Fodor, J.; Polgár, C.; Jurányi, Z. Progressive Breast Fibrosis Caused by Extreme Radiosensitivity: Oncocytogenetic Diagnosis and Treatment by Reconstructive Flap Surgery. Cancer Reports 2019, 2, e1126. [Google Scholar] [CrossRef] [PubMed]
- Dayyani, M.; Hoseinian-Azghadi, E.; Miri-Hakimabad, H.; Rafat-Motavalli, L.; Abdollahi, S.; Mohammadi, N. Radiobiological Comparison between Cobalt-60 and Iridium-192 High-dose-rate Brachytherapy Sources: Part I—Cervical Cancer. Medical Physics 2021, 48, 6213–6225. [Google Scholar] [CrossRef]
- Granzotto, A.; Benadjaoud, M.A.; Vogin, G.; Devic, C.; Ferlazzo, M.L.; Bodgi, L.; Pereira, S.; Sonzogni, L.; Forcheron, F.; Viau, M.; et al. Influence of Nucleoshuttling of the ATM Protein in the Healthy Tissues Response to Radiation Therapy: Toward a Molecular Classification of Human Radiosensitivity. International Journal of Radiation Oncology*Biology*Physics 2016, 94, 450–460. [Google Scholar] [CrossRef]
- Perico, D.; Mauri, P. Deciphering Radiotherapy Resistance: A Proteomic Perspective. Proteomes 2025, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A Clinically Applicable Molecular-Based Classification for Endometrial Cancers. Br J Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Van Den Heerik, A.S.V.M.; Horeweg, N.; De Boer, S.M.; Bosse, T.; Creutzberg, C.L. Adjuvant Therapy for Endometrial Cancer in the Era of Molecular Classification: Radiotherapy, Chemoradiation and Novel Targets for Therapy. International Journal of Gynecological Cancer 2021, 31, 594–604. [Google Scholar] [CrossRef] [PubMed]
- León-Castillo, A.; Gilvazquez, E.; Nout, R.; Smit, V.T.; McAlpine, J.N.; McConechy, M.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; et al. Clinicopathological and Molecular Characterisation of ‘Multiple-classifier’ Endometrial Carcinomas. The Journal of Pathology 2020, 250, 312–322. [Google Scholar] [CrossRef]
- Vermij, L.; Smit, V.; Nout, R.; Bosse, T. Incorporation of Molecular Characteristics into Endometrial Cancer Management. Histopathology 2020, 76, 52–63. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Levine, D.A. Integrated Genomic Characterization of Endometrial Carcinoma. Nature 2013, 497, 67–73. [CrossRef]
- Yen, T.-T.; Wang, T.-L.; Fader, A.N.; Shih, I.-M.; Gaillard, S. Molecular Classification and Emerging Targeted Therapy in Endometrial Cancer. International Journal of Gynecological Pathology 2020, 39, 26–35. [Google Scholar] [CrossRef]
- Goulder, A.; Gaillard, S.L. Molecular Classification of Endometrial Cancer: Entering an Era of Precision Medicine. J Gynecol Oncol 2022, 33, e47. [Google Scholar] [CrossRef]
- Bosse, T.; Nout, R.A.; McAlpine, J.N.; McConechy, M.K.; Britton, H.; Hussein, Y.R.; Gonzalez, C.; Ganesan, R.; Steele, J.C.; Harrison, B.T.; et al. Molecular Classification of Grade 3 Endometrioid Endometrial Cancers Identifies Distinct Prognostic Subgroups. American Journal of Surgical Pathology 2018, 42, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Puechl, A.M.; Spinosa, D.; Berchuck, A.; Secord, A.A.; Drury, K.E.; Broadwater, G.; Wong, J.; Whitaker, R.; Devos, N.; Corcoran, D.L.; et al. Molecular Classification to Prognosticate Response in Medically Managed Endometrial Cancers and Endometrial Intraepithelial Neoplasia. Cancers 2021, 13, 2847. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, A.; Bosse, T.; McAlpine, J.N. The Emerging Role of Molecular Pathology in Directing the Systemic Treatment of Endometrial Cancer. Ther Adv Med Oncol 2021, 13, 17588359211035959. [Google Scholar] [CrossRef] [PubMed]
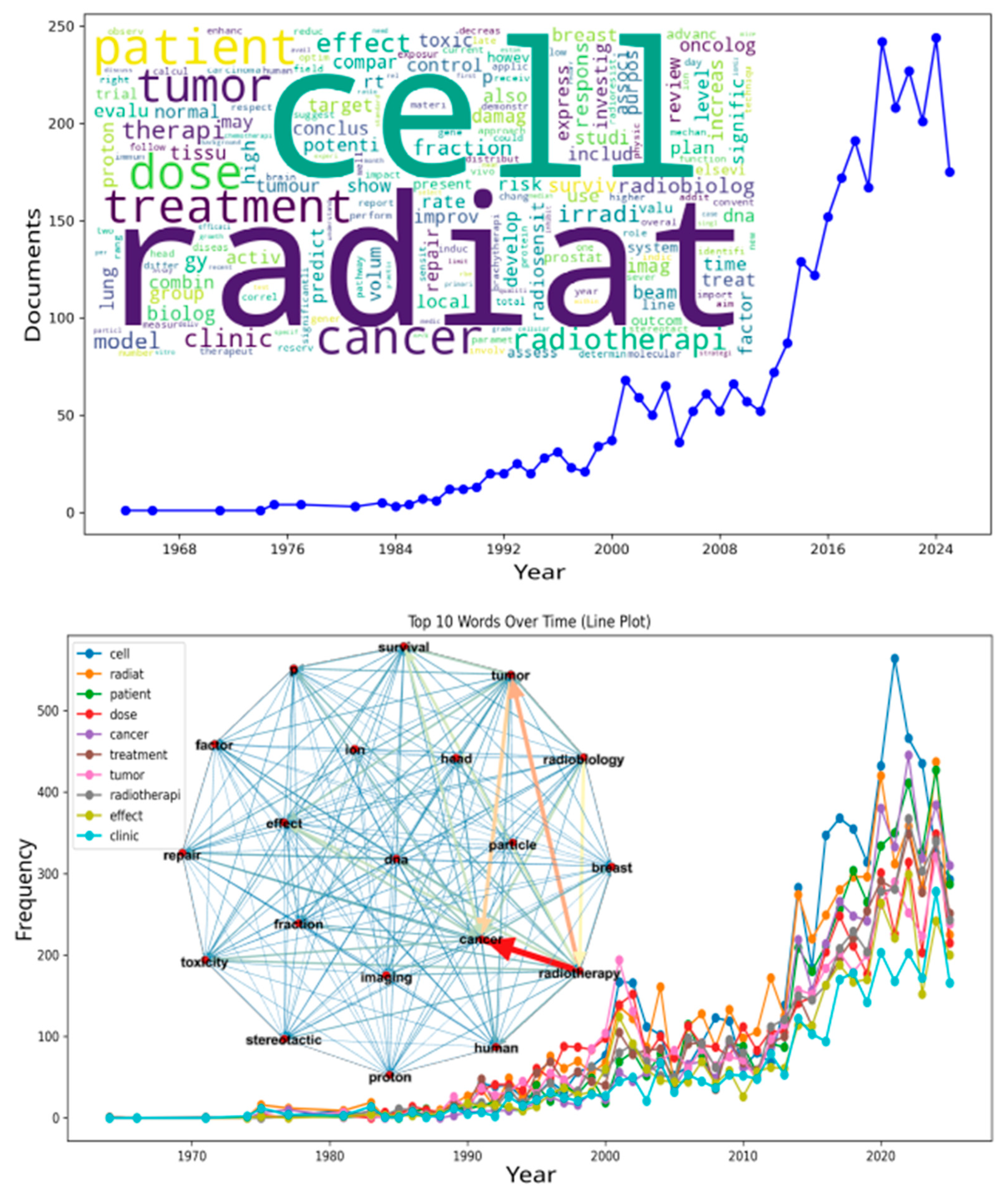
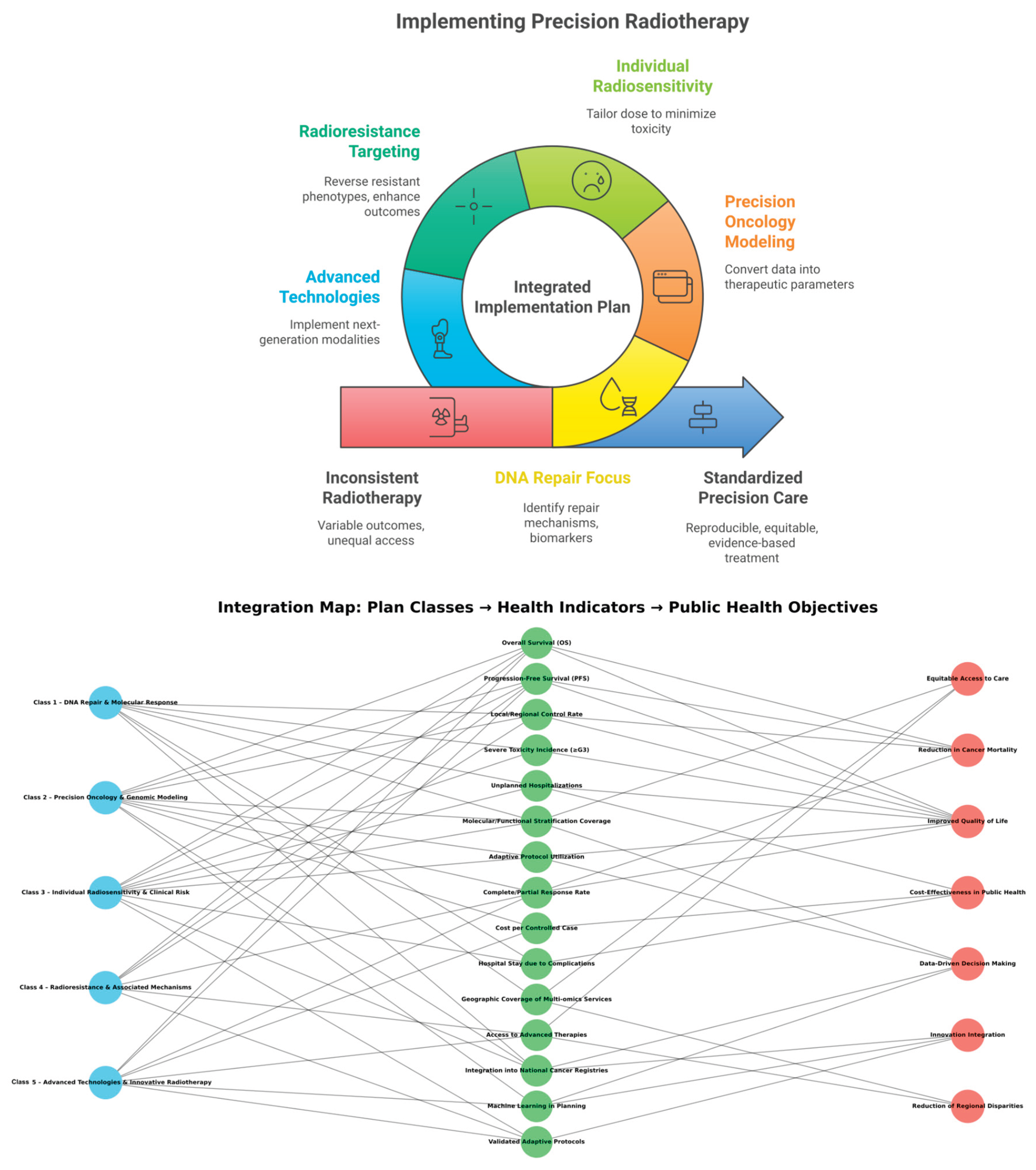
| Indicator | Related Plan Class(es) | Description and Measurement Method | Expected Public Health Impact |
|---|---|---|---|
| Overall survival (OS) at 1, 3, and 5 years | 2, 3, 4, 5 | Percentage of patients alive 1, 3, and 5 years after radiotherapy, stratified by tumor type and treatment protocol. | Improved longevity and reduction in cancer-specific mortality, particularly in high-risk tumors. |
| Progression-free survival (PFS) | 2, 3, 4, 5 | Mean time to recurrence or disease progression after personalized radiotherapy. | Greater disease control, reduced retreatment, and lower hospital costs. |
| Local and regional control ratel | 1, 2, 3, 4 | Proportion of patients without recurrence in the tumor bed or regional lymph nodes after 12 and 24 months. | Increased therapeutic efficacy and improved population prognosis. |
| Incidence of acute and late toxicities (grade ≥3) | 1, 2, 3 | Standardized reporting (RTOG/EORTC or CTCAE) of severe adverse events related to radiotherapy. | Reduced disabling side effects and improved post-treatment quality of life. |
| Rate of unplanned hospitalizations related to treatment | 1, 3 | Percentage of patients requiring emergency hospitalization due to radiotherapy complications. | Savings in hospital resources and reduced burden on inpatient units. |
| Percentage of patients undergoing molecular and functional stratification prior to treatment | 1, 2, 3 | Proportion of patients who underwent genomic, proteomic, or functional testing before radiotherapy initiation. | Greater precision in therapeutic planning and optimization of resource allocation. |
| Proportion of treatments with adaptive protocols | 2, 3, 5 | Percentage of patients whose radiotherapy plan was adjusted during treatment based on clinical response and/or biomarkers. | Reduced toxicity, improved efficacy, and more rational use of technology. |
| Complete and partial response rate | 2, 4, 5 | Percentage of patients with complete disappearance or significant reduction of tumor after treatment. | Direct indicator of personalized protocol effectiveness. |
| Average cost per patient with tumor control achieved | 2, 5 | Total treatment cost divided by the number of patients with sustained complete or partial response. | Increased economic efficiency of the public health system. |
| Mean hospital stay due to treatment complications | 1, 3 | Average number of days of hospitalization caused by radiotherapy-related adverse events.. | Reduced hospital bed usage and expanded treatment capacity. |
| Geographic coverage of services with multi-omic and radiobiological capacity | 1, 2, 3 | Percentage of health regions with infrastructure for multi-omic testing and radiobiological integration. | Reduction of regional inequalities in access to advanced treatments. |
| Percentage of patients with access to advanced therapies in the public health system (SUS) | 4, 5 | Proportion of patients receiving modalities such as FLASH-RT, optimized brachytherapy, or particle therapy. | Equity in access to innovative technologies across the national territory. |
| Proportion of cases integrated into national registries with molecular and radiomic data | 1, 2, 3, 4, 5 | Percentage of patients with clinical, molecular, and imaging data stored in national repositories. | Strengthened epidemiological surveillance and translational research. |
| Utilization rate of machine learning in clinical planning | 2, 3, 5 | Percentage of cases planned or adjusted using validated AI models. | Standardization of evidence-based practices and increased outcome predictability. |
| Number of adaptive protocols validated and incorporated into national guidelines | 3, 4, 5 | Number of adaptive clinical protocols formally approved and implemented in the public health system | Continuous improvement of care quality and systematic technological updating. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





