Submitted:
14 October 2025
Posted:
15 October 2025
You are already at the latest version
Abstract
Keywords:
Essentials
- Deep learning-based auto segmentation models achieve high accuracy and substantially reduced inter-observer variability in glioblastoma radiotherapy planning (pp. 6-8)
- Biologically informed mathematical modeling integrates tumor growth dynamics with imaging, enabling personalized radiotherapy dose mapping strategies (pp. 8-11)
- Radiogenomic models integrating imaging and molecular data predict status of key biomarkers, supporting non-invasive tumor subtyping and personalized therapy (pp. 11-13)
- Multi-institutional datasets, model interpretability, and standardized validation protocols remain critical barriers to clinical adoption of artificial intelligence-guided radiotherapy (pp. 13-15)
- Recent advances including adaptive radiotherapy, multimodal integration, and foundation models enable personalization and real-time adaptability in glioblastoma radiotherapy (pp.15-17)
Summary Statement
Introduction
- Treatment preparation encompasses the delineation of target volumes and organs at risk, the adoption of dose prescriptions, and the determination of treatment plan through simulation.
- Treatment delivery involves the fractionation of the simulated plan into multiple sessions and the systematic administration of radiation according to the established plan.
- Treatment adaptation entails the continuous monitoring of treatment execution and the modification of the plan when anatomical or physiological changes compromise the ability of the initial plan to satisfy predefined dosimetric and clinical constraints.
Current Challenges in Glioblastoma Management and Treatment
Tumor Delineation and Auto Segmentation
Personalized and Biologically Informed Tumor-Progression Radiotherapy
Modification of Treatment, Patient Response Prediction, and Triage During Therapy
Radiogenomics and Non-Invasive Biomarker Integration
Interpretability and Explainability in Deep Learning Models
AI-Driven Solutions and Current Trends in Technology
Concluding Remarks
Conflict of Interest
Abbreviations
References
- Kanderi T, Munakomi S, Gupta V. Glioblastoma Multiforme. StatPearls. Treasure Island (FL): StatPearls Publishing; 2025.
- McKinnon C, Nandhabalan M, Murray SA, Plaha P. Glioblastoma: clinical presentation, diagnosis, and management. Bmj. 2021;374:n1560. Epub 20210714. PubMed PMID: 34261630. [CrossRef]
- Brown TJ, Brennan MC, Li M, Church EW, Brandmeir NJ, Rakszawski KL, Patel AS, Rizk EB, Suki D, Sawaya R, Glantz M. Association of the Extent of Resection With Survival in Glioblastoma: A Systematic Review and Meta-analysis. JAMA Oncol. 2016;2(11):1460–9. PubMed PMID: 27310651; PMCID: PMC6438173. [CrossRef]
- Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, Bendszus M, Balana C, Chinot O, Dirven L, French P, Hegi ME, Jakola AS, Platten M, Roth P, Rudà R, Short S, Smits M, Taphoorn MJB, von Deimling A, Westphal M, Soffietti R, Reifenberger G, Wick W. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18(3):170–86. Epub 20201208. PubMed PMID: 33293629; PMCID: PMC7904519. [CrossRef]
- Peeken JC, Molina-Romero M, Diehl C, Menze BH, Straube C, Meyer B, Zimmer C, Wiestler B, Combs SE. Deep learning derived tumor infiltration maps for personalized target definition in Glioblastoma radiotherapy. Radiotherapy and Oncology. 2019;138:166–72. [CrossRef]
- Rončević A, Koruga N, Soldo Koruga A, Rončević R, Rotim T, Šimundić T, Kretić D, Perić M, Turk T, Štimac D. Personalized Treatment of Glioblastoma: Current State and Future Perspective. Biomedicines. 2023;11(6):1579. [CrossRef]
- Erices JI, Bizama C, Niechi I, Uribe D, Rosales A, Fabres K, Navarro-Martínez G, Torres Á, San Martín R, Roa JC, Quezada-Monrás C. Glioblastoma Microenvironment and Invasiveness: New Insights and Therapeutic Targets. Int J Mol Sci. 2023;24(8). Epub 20230411. PubMed PMID: 37108208; PMCID: PMC10139189. [CrossRef]
- Fathi Kazerooni A, Nabil M, Zeinali Zadeh M, Firouznia K, Azmoudeh-Ardalan F, Frangi AF, Davatzikos C, Saligheh Rad H. Characterization of active and infiltrative tumorous subregions from normal tissue in brain gliomas using multiparametric MRI. J Magn Reson Imaging. 2018;48(4):938–50. PubMed PMID: 29412496; PMCID: PMC6081259. Epub 20180207. [CrossRef]
- Kruser TJ, Bosch WR, Badiyan SN, Bovi JA, Ghia AJ, Kim MM, Solanki AA, Sachdev S, Tsien C, Wang TJC, Mehta MP, McMullen KP. NRG brain tumor specialists consensus guidelines for glioblastoma contouring. J Neurooncol. 2019;143(1):157–66. [CrossRef]
- Poel R, Rüfenacht E, Ermis E, Müller M, Fix MK, Aebersold DM, Manser P, Reyes M. Impact of random outliers in auto-segmented targets on radiotherapy treatment plans for glioblastoma. Radiat Oncol. 2022;17(1):170. [CrossRef]
- Sidibe I, Tensaouti F, Gilhodes J, Cabarrou B, Filleron T, Desmoulin F, Ken S, Noël G, Truc G, Sunyach MP, Charissoux M, Magné N, Lotterie JA, Roques M, Péran P, Cohen-Jonathan Moyal E, Laprie A. Pseudoprogression in GBM versus true progression in patients with glioblastoma: A multiapproach analysis. Radiother Oncol. 2023;181:109486. Epub 20230124. PubMed PMID: 36706959. [CrossRef]
- Cramer CK, Cummings TL, Andrews RN, Strowd R, Rapp SR, Shaw EG, Chan MD, Lesser GJ. Treatment of Radiation-Induced Cognitive Decline in Adult Brain Tumor Patients. Curr Treat Options Oncol. 2019;20(5):42. Epub 20190408. PubMed PMID: 30963289; PMCID: PMC6594685. [CrossRef]
- Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, Harting I, Sahm F, Prager M, Nowosielski M, Wick A, Nolden M, Radbruch A, Debus J, Schlemmer H-P, Heiland S, Platten M, Von Deimling A, Van Den Bent MJ, Gorlia T, Wick W, Bendszus M, Maier-Hein KH. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. The Lancet Oncology. 2019;20(5):728–40. [CrossRef]
- Mansoorian S, Schmidt M, Weissmann T, Delev D, Heiland DH, Coras R, Stritzelberger J, Saake M, Höfler D, Schubert P, Schmitter C, Lettmaier S, Filimonova I, Frey B, Gaipl US, Distel LV, Semrau S, Bert C, Eze C, Schönecker S, Belka C, Blümcke I, Uder M, Schnell O, Dörfler A, Fietkau R, Putz F. Reirradiation for recurrent glioblastoma: the significance of the residual tumor volume. J Neurooncol. 2025. [CrossRef]
- Shaver M, Kohanteb P, Chiou C, Bardis M, Chantaduly C, Bota D, Filippi C, Weinberg B, Grinband J, Chow D, Chang P. Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging. Cancers. 2019;11(6):829. [CrossRef]
- Lu S-L, Xiao F-R, Cheng JC-H, Yang W-C, Cheng Y-H, Chang Y-C, Lin J-Y, Liang C-H, Lu J-T, Chen Y-F, Hsu F-M. Randomized multi-reader evaluation of automated detection and segmentation of brain tumors in stereotactic radiosurgery with deep neural networks. Neuro-Oncology. 2021;23(9):1560–8. [CrossRef]
- Doolan PJ, Charalambous S, Roussakis Y, Leczynski A, Peratikou M, Benjamin M, Ferentinos K, Strouthos I, Zamboglou C, Karagiannis E. A clinical evaluation of the performance of five commercial artificial intelligence contouring systems for radiotherapy. Front Oncol. 2023;13:1213068. Epub 20230804. PubMed PMID: 37601695; PMCID: PMC10436522. [CrossRef]
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber M-A, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin H-C, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. [CrossRef]
- Marey A, Arjmand P, Alerab ADS, Eslami MJ, Saad AM, Sanchez N, Umair M. Explainability, transparency and black box challenges of AI in radiology: impact on patient care in cardiovascular radiology. Egypt J Radiol Nucl Med. 2024;55(1):183. [CrossRef]
- Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M, Prastawa M, Alberts E, Lipkova J, Freymann J, Kirby J, Bilello M, Fathallah-Shaykh H, Wiest R, Kirschke J, Wiestler B, Colen R, Kotrotsou A, Lamontagne P, Marcus D, Milchenko M, Nazeri A, Weber M-A, Mahajan A, Baid U, Gerstner E, Kwon D, Acharya G, Agarwal M, Alam M, Albiol A, Albiol A, Albiol FJ, Alex V, Allinson N, Amorim PHA, Amrutkar A, Anand G, Andermatt S, Arbel T, Arbelaez P, Avery A, Azmat M, Pranjal B, Bai W, Banerjee S, Barth B, Batchelder T, Batmanghelich K, Battistella E, Beers A, Belyaev M, Bendszus M, Benson E, Bernal J, Bharath HN, Biros G, Bisdas S, Brown J, Cabezas M, Cao S, Cardoso JM, Carver EN, Casamitjana A, Castillo LS, Catà M, Cattin P, Cerigues A, Chagas VS, Chandra S, Chang Y-J, Chang S, Chang K, Chazalon J, Chen S, Chen W, Chen JW, Chen Z, Cheng K, Choudhury AR, Chylla R, Clérigues A, Colleman S, Colmeiro RGR, Combalia M, Costa A, Cui X, Dai Z, Dai L, Daza LA, Deutsch E, Ding C, Dong C, Dong S, Dudzik W, Eaton-Rosen Z, Egan G, Escudero G, Estienne T, Everson R, Fabrizio J, Fan Y, Fang L, Feng X, Ferrante E, Fidon L, Fischer M, French AP, Fridman N, Fu H, Fuentes D, Gao Y, Gates E, Gering D, Gholami A, Gierke W, Glocker B, Gong M, González-Villá S, Grosges T, Guan Y, Guo S, Gupta S, Han W-S, Han IS, Harmuth K, He H, Hernández-Sabaté A, Herrmann E, Himthani N, Hsu W, Hsu C, Hu X, Hu X, Hu Y, Hu Y, Hua R, Huang T-Y, Huang W, Huffel SV, Huo Q, Vivek HV, Iftekharuddin KM, Isensee F, Islam M, Jackson AS, Jambawalikar SR, Jesson A, Jian W, Jin P, Jose VJM, Jungo A, Kainz B, Kamnitsas K, Kao P-Y, Karnawat A, Kellermeier T, Kermi A, Keutzer K, Khadir MT, Khened M, Kickingereder P, Kim G, King N, Knapp H, Knecht U, Kohli L, Kong D, Kong X, Koppers S, Kori A, Krishnamurthi G, Krivov E, Kumar P, Kushibar K, Lachinov D, Lambrou T, Lee J, Lee C, Lee Y, Lee M, Lefkovits S, Lefkovits L, Levitt J, Li T, Li H, Li W, Li H, Li X, Li Y, Li H, Li Z, Li X, Li Z, Li X, Lin Z-S, Lin F, Lio P, Liu C, Liu B, Liu X, Liu M, Liu J, Liu L, Llado X, Lopez MM, Lorenzo PR, Lu Z, Luo L, Luo Z, Ma J, Ma K, Mackie T, Madabushi A, Mahmoudi I, Maier-Hein KH, Maji P, Mammen CP, Mang A, Manjunath BS, Marcinkiewicz M, McDonagh S, McKenna S, McKinley R, Mehl M, Mehta S, Mehta R, Meier R, Meinel C, Merhof D, Meyer C, Miller R, Mitra S, Moiyadi A, Molina-Garcia D, Monteiro MAB, Mrukwa G, Myronenko A, Nalepa J, Ngo T, Nie D, Ning H, Niu C, Nuechterlein NK, Oermann E, Oliveira A, Oliveira DDC, Oliver A, Osman AFI, Ou Y-N, Ourselin S, Paragios N, Park MS, Paschke B, Pauloski JG, Pawar K, Pawlowski N, Pei L, Peng S, Pereira SM, Perez-Beteta J, Perez-Garcia VM, Pezold S, Pham B, Phophalia A, Piella G, Pillai GN, Piraud M, Pisov M, Popli A, Pound MP, Pourreza R, Prasanna P, Prkovska V, Pridmore TP, Puch S, Puybareau É, Qian B, Qiao X, Rajchl M, Rane S, Rebsamen M, Ren H, Ren X, Revanuru K, Rezaei M, Rippel O, Rivera LC, Robert C, Rosen B, Rueckert D, Safwan M, Salem M, Salvi J, Sanchez I, Sánchez I, Santos HM, Sartor E, Schellingerhout D, Scheufele K, Scott MR, Scussel AA, Sedlar S, Serrano-Rubio JP, Shah NJ, Shah N, Shaikh M, Shankar BU, Shboul Z, Shen H, Shen D, Shen L, Shen H, Shenoy V, Shi F, Shin HE, Shu H, Sima D, Sinclair M, Smedby O, Snyder JM, Soltaninejad M, Song G, Soni M, Stawiaski J, Subramanian S, Sun L, Sun R, Sun J, Sun K, Sun Y, Sun G, Sun S, Suter YR, Szilagyi L, Talbar S, Tao D, Teng Z, Thakur S, Thakur MH, Tharakan S, Tiwari P, Tochon G, Tran T, Tsai YM, Tseng K-L, Tuan TA, Turlapov V, Tustison N, Vakalopoulou M, Valverde S, Vanguri R, Vasiliev E, Ventura J, Vera L, Vercauteren T, Verrastro CA, Vidyaratne L, Vilaplana V, Vivekanandan A, Wang G, Wang Q, Wang CJ, Wang W, Wang D, Wang R, Wang Y, Wang C, Wen N, Wen X, Weninger L, Wick W, Wu S, Wu Q, Wu Y, Xia Y, Xu Y, Xu X, Xu P, Yang T-L, Yang X, Yang H-Y, Yang J, Yang H, Yang G, Yao H, Ye X, Yin C, Young-Moxon B, Yu J, Yue X, Zhang S, Zhang A, Zhang K, Zhang X, Zhang L, Zhang X, Zhang Y, Zhang L, Zhang J, Zhang X, Zhang T, Zhao S, Zhao Y, Zhao X, Zhao L, Zheng Y, Zhong L, Zhou C, Zhou X, Zhou F, Zhu H, Zhu J, Zhuge Y, Zong W, Kalpathy-Cramer J, Farahani K, Davatzikos C, Leemput KV, Menze B. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. Apollo—University of Cambridge Repository; 2018.
- Dora L, Agrawal S, Panda R, Abraham A. State-of-the-Art Methods for Brain Tissue Segmentation: A Review. IEEE Rev Biomed Eng. 2017;10:235–49. [CrossRef]
- Işın A, Direkoğlu C, Şah M. Review of MRI-based Brain Tumor Image Segmentation Using Deep Learning Methods. Procedia Computer Science. 2016;102:317–24. [CrossRef]
- Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, Farahani K, Kalpathy-Cramer J, Kitamura FC, Pati S, Prevedello L, Rudie J, Sako C, Shinohara R, Bergquist T, Chai R, Eddy J, Elliott J, Reade W, Schaffter T, Yu T, Zheng J, Davatzikos C, Mongan J, Hess C, Cha S, Villanueva-Meyer J, Freymann JB, Kirby JS, Wiestler B, Crivellaro P, Colen RR, Kotrotsou A, Marcus D, Milchenko M, Nazeri A, Fathallah-Shaykh H, Wiest R, Jakab A, Weber M-A, Mahajan A, Menze B, Flanders AE, Bakas S. RSNA-ASNR-MICCAI-BraTS-2021. The Cancer Imaging Archive; 2023.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–11. [CrossRef]
- Zeineldin RA, Karar ME, Burgert O, Mathis-Ullrich F. Multimodal CNN Networks for Brain Tumor Segmentation in MRI: A BraTS 2022 Challenge Solution. In: Bakas S, Crimi A, Baid U, Malec S, Pytlarz M, Baheti B, Zenk M, Dorent R, editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer Nature Switzerland; 2023. p. 127–37.
- Ferreira A, Solak N, Li J, Dammann P, Kleesiek J, Alves V, Egger J. Enhanced Data Augmentation Using Synthetic Data for Brain Tumour Segmentation. In: Baid U, Dorent R, Malec S, Pytlarz M, Su R, Wijethilake N, Bakas S, Crimi A, editors. Brain Tumor Segmentation, and Cross-Modality Domain Adaptation for Medical Image Segmentation. Cham: Springer Nature Switzerland; 2024. p. 79–93.
- Moradi N, Ferreira A, Puladi B, Kleesiek J, Fatemizadeh E, Luijten G, Alves V, Egger J. Comparative Analysis of nnUNet and MedNeXt for Head and Neck Tumor Segmentation in MRI-Guided Radiotherapy. In: Wahid KA, Dede C, Naser MA, Fuller CD, editors. Head and Neck Tumor Segmentation for MR-Guided Applications. Cham: Springer Nature Switzerland; 2025. p. 136–53.
- Xue J, Wang B, Ming Y, Liu X, Jiang Z, Wang C, Liu X, Chen L, Qu J, Xu S, Tang X, Mao Y, Liu Y, Li D. Deep learning–based detection and segmentation-assisted management of brain metastases. Neuro-Oncology. 2020;22(4):505–14. [CrossRef]
- Naceur MB, Saouli R, Akil M, Kachouri R. Fully Automatic Brain Tumor Segmentation using End-To-End Incremental Deep Neural Networks in MRI images. Computer Methods and Programs in Biomedicine. 2018;166:39–49. [CrossRef]
- Chang K, Beers AL, Bai HX, Brown JM, Ly KI, Li X, Senders JT, Kavouridis VK, Boaro A, Su C, Bi WL, Rapalino O, Liao W, Shen Q, Zhou H, Xiao B, Wang Y, Zhang PJ, Pinho MC, Wen PY, Batchelor TT, Boxerman JL, Arnaout O, Rosen BR, Gerstner ER, Yang L, Huang RY, Kalpathy-Cramer J. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro-Oncology. 2019;21(11):1412–22. [CrossRef]
- Ranjbarzadeh R, Bagherian Kasgari A, Jafarzadeh Ghoushchi S, Anari S, Naseri M, Bendechache M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci Rep. 2021;11(1):10930. [CrossRef]
- Deng W, Shi Q, Luo K, Yang Y, Ning N. Brain Tumor Segmentation Based on Improved Convolutional Neural Network in Combination with Non-quantifiable Local Texture Feature. J Med Syst. 2019;43(6):152. [CrossRef]
- Zhuge Y, Krauze AV, Ning H, Cheng JY, Arora BC, Camphausen K, Miller RW. Brain tumor segmentation using holistically nested neural networks in MRI images. Medical Physics. 2017;44(10):5234–43. [CrossRef]
- Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2018. p. 287–97.
- Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging. 2016;35(5):1240–51. [CrossRef]
- Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H. Brain tumor segmentation with Deep Neural Networks. Medical Image Analysis. 2017;35:18–31. [CrossRef]
- Soltaninejad M, Zhang L, Lambrou T, Allinson N, Ye X. Multimodal MRI brain tumor segmentation using random forests with features learned from fully convolutional neural network. arXiv; 2017.
- Hussain S, Anwar SM, Majid M, editors. Brain tumor segmentation using cascaded deep convolutional neural network. 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2017 2017/07//. Seogwipo: IEEE.
- Tian S, Liu Y, Mao X, Xu X, He S, Jia L, Zhang W, Peng P, Wang J. A multicenter study on deep learning for glioblastoma auto-segmentation with prior knowledge in multimodal imaging. Cancer Science. 2024;115(10):3415–25. [CrossRef]
- Bibault J-E, Giraud P. Deep learning for automated segmentation in radiotherapy: a narrative review. British Journal of Radiology. 2024;97(1153):13–20. [CrossRef]
- Unkelbach J, Bortfeld T, Cardenas CE, Gregoire V, Hager W, Heijmen B, Jeraj R, Korreman SS, Ludwig R, Pouymayou B, Shusharina N, Söderberg J, Toma-Dasu I, Troost EGC, Vasquez Osorio E. The role of computational methods for automating and improving clinical target volume definition. Radiotherapy and Oncology. 2020;153:15–25. [CrossRef]
- Metz M-C, Ezhov I, Peeken JC, Buchner JA, Lipkova J, Kofler F, Waldmannstetter D, Delbridge C, Diehl C, Bernhardt D, Schmidt-Graf F, Gempt J, Combs SE, Zimmer C, Menze B, Wiestler B. Toward image-based personalization of glioblastoma therapy: A clinical and biological validation study of a novel, deep learning-driven tumor growth model. Neurooncol Adv. 2024;6(1):vdad171. [CrossRef]
- Lipkova J, Angelikopoulos P, Wu S, Alberts E, Wiestler B, Diehl C, Preibisch C, Pyka T, Combs SE, Hadjidoukas P, Van Leemput K, Koumoutsakos P, Lowengrub J, Menze B. Personalized Radiotherapy Design for Glioblastoma: Integrating Mathematical Tumor Models, Multimodal Scans, and Bayesian Inference. IEEE Trans Med Imaging. 2019;38(8):1875–84. [CrossRef]
- Dextraze K, Saha A, Kim D, Narang S, Lehrer M, Rao A, Narang S, Rao D, Ahmed S, Madhugiri V, Fuller CD, Kim MM, Krishnan S, Rao G, Rao A. Spatial habitats from multiparametric MR imaging are associated with signaling pathway activities and survival in glioblastoma. Oncotarget. 2017;8(68):112992–3001. [CrossRef]
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discovery. 2022;12(1):31–46. [CrossRef]
- Kwak S, Akbari H, Garcia JA, Mohan S, Dicker Y, Sako C, Matsumoto Y, Nasrallah MP, Shalaby M, O’Rourke DM, Shinohara RT, Liu F, Badve C, Barnholtz-Sloan JS, Sloan AE, Lee M, Jain R, Cepeda S, Chakravarti A, Palmer JD, Dicker AP, Shukla G, Flanders AE, Shi W, Woodworth GF, Davatzikos C. Predicting peritumoral glioblastoma infiltration and subsequent recurrence using deep-learning–based analysis of multi-parametric magnetic resonance imaging. J Med Imag. 2024;11(05). [CrossRef]
- Hong JC, Eclov NCW, Dalal NH, Thomas SM, Stephens SJ, Malicki M, Shields S, Cobb A, Mowery YM, Niedzwiecki D, Tenenbaum JD, Palta M. System for High-Intensity Evaluation During Radiation Therapy (SHIELD-RT): A Prospective Randomized Study of Machine Learning–Directed Clinical Evaluations During Radiation and Chemoradiation. JCO. 2020;38(31):3652–61. [CrossRef]
- Gutsche R, Lohmann P, Hoevels M, Ruess D, Galldiks N, Visser-Vandewalle V, Treuer H, Ruge M, Kocher M. Radiomics outperforms semantic features for prediction of response to stereotactic radiosurgery in brain metastases. Radiotherapy and Oncology. 2022;166:37–43. [CrossRef]
- Yang Z, Zamarud A, Marianayagam NJ, Park DJ, Yener U, Soltys SG, Chang SD, Meola A, Jiang H, Lu W, Gu X. Deep learning-based overall survival prediction in patients with glioblastoma: An automatic end-to-end workflow using pre-resection basic structural multiparametric MRIs. Computers in Biology and Medicine. 2025;185:109436. [CrossRef]
- Tsang DS, Tsui G, McIntosh C, Purdie T, Bauman G, Dama H, Laperriere N, Millar B-A, Shultz DB, Ahmed S, Khandwala M, Hodgson DC. A pilot study of machine-learning based automated planning for primary brain tumours. Radiat Oncol. 2022;17(1):3. [CrossRef]
- Di Nunno V, Fordellone M, Minniti G, Asioli S, Conti A, Mazzatenta D, Balestrini D, Chiodini P, Agati R, Tonon C, Tosoni A, Gatto L, Bartolini S, Lodi R, Franceschi E. Machine learning in neuro-oncology: toward novel development fields. J Neurooncol. 2022;159(2):333–46. [CrossRef]
- Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis VK, Senders JT, Boaro A, Beers A, Zhang B, Capellini A, Liao W, Shen Q, Li X, Xiao B, Cryan J, Ramkissoon S, Ramkissoon L, Ligon K, Wen PY, Bindra RS, Woo J, Arnaout O, Gerstner ER, Zhang PJ, Rosen BR, Yang L, Huang RY, Kalpathy-Cramer J. Residual Convolutional Neural Network for the Determination of <i>IDH</i> Status in Low- and High-Grade Gliomas from MR Imaging. Clinical Cancer Research. 2018;24(5):1073–81. [CrossRef]
- Wong QH-W, Li KK-W, Wang W-W, Malta TM, Noushmehr H, Grabovska Y, Jones C, Chan AK-Y, Kwan JS-H, Huang QJ-Q, Wong GC-H, Li W-C, Liu X-Z, Chen H, Chan DT-M, Mao Y, Zhang Z-Y, Shi Z-F, Ng H-K. Molecular landscape of IDH-mutant primary astrocytoma Grade IV/glioblastomas. Modern Pathology. 2021;34(7):1245–60. [CrossRef]
- Ding J, Zhao R, Qiu Q, Chen J, Duan J, Cao X, Yin Y. Developing and validating a deep learning and radiomic model for glioma grading using multiplanar reconstructed magnetic resonance contrast-enhanced T1-weighted imaging: a robust, multi-institutional study. Quant Imaging Med Surg. 2022;12(2):1517–28. [CrossRef]
- Zhang X, Yan L-F, Hu Y-C, Li G, Yang Y, Han Y, Sun Y-Z, Liu Z-C, Tian Q, Han Z-Y, Liu L-D, Hu B-Q, Qiu Z-Y, Wang W, Cui G-B. Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget. 2017;8(29):47816–30. [CrossRef]
- Fathi Kazerooni A, Akbari H, Hu X, Bommineni V, Grigoriadis D, Toorens E, Sako C, Mamourian E, Ballinger D, Sussman R, Singh A, Verginadis II, Dahmane N, Koumenis C, Binder ZA, Bagley SJ, Mohan S, Hatzigeorgiou A, O’Rourke DM, Ganguly T, De S, Bakas S, Nasrallah MP, Davatzikos C. The radiogenomic and spatiogenomic landscapes of glioblastoma and their relationship to oncogenic drivers. Commun Med. 2025;5(1):55. [CrossRef]
- Lu J, Zhang Z-Y, Zhong S, Deng D, Yang W-Z, Wu S-W, Cheng Y, Bai Y, Mou Y-G. Evaluating the Diagnostic and Prognostic Value of Peripheral Immune Markers in Glioma Patients: A Prospective Multi-Institutional Cohort Study of 1282 Patients. JIR. 2025;Volume 18:7477–92. [CrossRef]
- Aman RA, Pratama MG, Satriawan RR, Ardiansyah IR, Suanjaya IKA. Diagnostic and Prognostic Values of miRNAs in High-Grade Gliomas: A Systematic Review. F1000Res. 2025;13:796. [CrossRef]
- Hasani F, Masrour M, Jazi K, Ahmadi P, Hosseini SS, Lu VM, Alborzi A. MicroRNA as a potential diagnostic and prognostic biomarker in brain gliomas: a systematic review and meta-analysis. Front Neurol. 2024;15:1357321. [CrossRef]
- Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, Lzicarova E, Svoboda M, Dolezelova H, Smrcka M, Vyzula R, Michalek J, Hajduch M, Slaby O. MiR-195, miR-196b, miR-181c, miR-21 expression levels and <i>O</i> -6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Science. 2011;102(12):2186–90. [CrossRef]
- Lan F, Yue X, Xia T. Exosomal microRNA-210 is a potentially non-invasive biomarker for the diagnosis and prognosis of glioma. Oncol Lett. 2020. [CrossRef]
- Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Science. 2018;109(9):2651–9. [CrossRef]
- Velu U, Singh A, Nittala R, Yang J, Vijayakumar S, Cherukuri C, Vance GR, Salvemini JD, Hathaway BF, Grady C, Roux JA, Lewis S. Precision Population Cancer Medicine in Brain Tumors: A Potential Roadmap to Improve Outcomes and Strategize the Steps to Bring Interdisciplinary Interventions. Cureus. 2024. [CrossRef]
- Silva PJ, Silva PA, Ramos KS. Genomic and Health Data as Fuel to Advance a Health Data Economy for Artificial Intelligence. BioMed Research International. 2025;2025(1):6565955. [CrossRef]
- Silva PJ, Rahimzadeh V, Powell R, Husain J, Grossman S, Hansen A, Hinkel J, Rosengarten R, Ory MG, Ramos KS. Health equity innovation in precision medicine: data stewardship and agency to expand representation in clinicogenomics. Health Research Policy and Systems. 2024;22(1):170. [CrossRef]
- Silva P, Janjan N, Ramos KS, Udeani G, Zhong L, Ory MG, Smith ML. External control arms: COVID-19 reveals the merits of using real world evidence in real-time for clinical and public health investigations. Front Med (Lausanne). 2023;10:1198088. Epub 20230706. PubMed PMID: 37484840; PMCID: PMC10359981. [CrossRef]
- d’Este SH, Nielsen MB, Hansen AE. Visualizing Glioma Infiltration by the Combination of Multimodality Imaging and Artificial Intelligence, a Systematic Review of the Literature. Diagnostics. 2021;11(4):592. [CrossRef]
- Holzinger A, Langs G, Denk H, Zatloukal K, Müller H. Causability and explainability of artificial intelligence in medicine. WIREs Data Min & Knowl. 2019;9(4):e1312. [CrossRef]
- AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Medical Physics. 2018;45(3):1150–8. [CrossRef]
- Bologna G, Hayashi Y. Characterization of Symbolic Rules Embedded in Deep DIMLP Networks: A Challenge to Transparency of Deep Learning. Journal of Artificial Intelligence and Soft Computing Research. 2017;7(4):265–86. [CrossRef]
- Cui S, Traverso A, Niraula D, Zou J, Luo Y, Owen D, El Naqa I, Wei L. Interpretable artificial intelligence in radiology and radiation oncology. The British Journal of Radiology. 2023;96(1150):20230142. [CrossRef]
- Sangwan H. QUANTIFYING EXPLAINABLE AI METHODS IN MEDICAL DIAGNOSIS: A STUDY IN SKIN CANCER. Health Informatics; 2024.
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int J Comput Vis. 2020;128(2):336–59. [CrossRef]
- Lundberg S, Lee S-I. A Unified Approach to Interpreting Model Predictions. arXiv; 2017.
- Sahlsten J, Jaskari J, Wahid KA, Ahmed S, Glerean E, He R, Kann BH, Mäkitie A, Fuller CD, Naser MA, Kaski K. Application of simultaneous uncertainty quantification and segmentation for oropharyngeal cancer use-case with Bayesian deep learning. Commun Med. 2024;4(1):110. [CrossRef]
- Alruily M, Mahmoud AA, Allahem H, Mostafa AM, Shabana H, Ezz M. Enhancing Breast Cancer Detection in Ultrasound Images: An Innovative Approach Using Progressive Fine-Tuning of Vision Transformer Models. International Journal of Intelligent Systems. 2024;2024(1):6528752. [CrossRef]
- Marin T, Zhuo Y, Lahoud RM, Tian F, Ma X, Xing F, Moteabbed M, Liu X, Grogg K, Shusharina N, Woo J, Lim R, Ma C, Chen YE, El Fakhri G. Deep learning-based GTV contouring modeling inter- and intra- observer variability in sarcomas. Radiother Oncol. 2022;167:269–76. Epub 20211119. PubMed PMID: 34808228; PMCID: PMC8934266. [CrossRef]
- Abbasi S, Lan H, Choupan J, Sheikh-Bahaei N, Pandey G, Varghese B. Deep learning for the harmonization of structural MRI scans: a survey. Biomed Eng Online. 2024;23(1):90. Epub 20240831. PubMed PMID: 39217355; PMCID: PMC11365220. [CrossRef]
- Ensenyat-Mendez M, Íñiguez-Muñoz S, Sesé B, Marzese DM. iGlioSub: an integrative transcriptomic and epigenomic classifier for glioblastoma molecular subtypes. BioData Mining. 2021;14(1):42. [CrossRef]
- Dona Lemus OM, Cao M, Cai B, Cummings M, Zheng D. Adaptive Radiotherapy: Next-Generation Radiotherapy. Cancers. 2024;16(6):1206. [CrossRef]
- Weykamp F, Meixner E, Arians N, Hoegen-Saßmannshausen P, Kim J-Y, Tawk B, Knoll M, Huber P, König L, Sander A, Mokry T, Meinzer C, Schlemmer H-P, Jäkel O, Debus J, Hörner-Rieber J. Daily AI-Based Treatment Adaptation under Weekly Offline MR Guidance in Chemoradiotherapy for Cervical Cancer 1: The AIM-C1 Trial. JCM. 2024;13(4):957. [CrossRef]
- Vuong W, Gupta S, Weight C, Almassi N, Nikolaev A, Tendulkar RD, Scott JG, Chan TA, Mian OY. Trial in Progress: Adaptive RADiation Therapy with Concurrent Sacituzumab Govitecan (SG) for Bladder Preservation in Patients with MIBC (RAD-SG). International Journal of Radiation Oncology*Biology*Physics. 2023;117(2):e447–e8. [CrossRef]
- Guevara B, Cullison K, Maziero D, Azzam GA, De La Fuente MI, Brown K, Valderrama A, Meshman J, Breto A, Ford JC, Mellon EA. Simulated Adaptive Radiotherapy for Shrinking Glioblastoma Resection Cavities on a Hybrid MRI-Linear Accelerator. Cancers (Basel). 2023;15(5). Epub 20230302. PubMed PMID: 36900346; PMCID: PMC10000839. [CrossRef]
- Paschali M, Chen Z, Blankemeier L, Varma M, Youssef A, Bluethgen C, Langlotz C, Gatidis S, Chaudhari A. Foundation Models in Radiology: What, How, Why, and Why Not. Radiology. 2025;314(2):e240597. PubMed PMID: 39903075; PMCID: PMC11868850. [CrossRef]
- Putz F, Beirami S, Schmidt MA, May MS, Grigo J, Weissmann T, Schubert P, Höfler D, Gomaa A, Hassen BT, Lettmaier S, Frey B, Gaipl US, Distel LV, Semrau S, Bert C, Fietkau R, Huang Y. The Segment Anything foundation model achieves favorable brain tumor auto-segmentation accuracy in MRI to support radiotherapy treatment planning. Strahlenther Onkol. 2025;201(3):255–65. Epub 20241106. PubMed PMID: 39503868; PMCID: PMC11839838. [CrossRef]
- Kebaili A, Lapuyade-Lahorgue J, Vera P, Ruan S. Multi-modal MRI synthesis with conditional latent diffusion models for data augmentation in tumor segmentation. Comput Med Imaging Graph. 2025;123:102532. Epub 20250321. PubMed PMID: 40121926. [CrossRef]
- Baroudi H, Brock KK, Cao W, Chen X, Chung C, Court LE, El Basha MD, Farhat M, Gay S, Gronberg MP, Gupta AC, Hernandez S, Huang K, Jaffray DA, Lim R, Marquez B, Nealon K, Netherton TJ, Nguyen CM, Reber B, Rhee DJ, Salazar RM, Shanker MD, Sjogreen C, Woodland M, Yang J, Yu C, Zhao Y. Automated Contouring and Planning in Radiation Therapy: What Is ‘Clinically Acceptable’? Diagnostics. 2023;13(4):667. [CrossRef]
- Hussain S, Anwar SM, Majid M. Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing. 2018;282:248–61. [CrossRef]
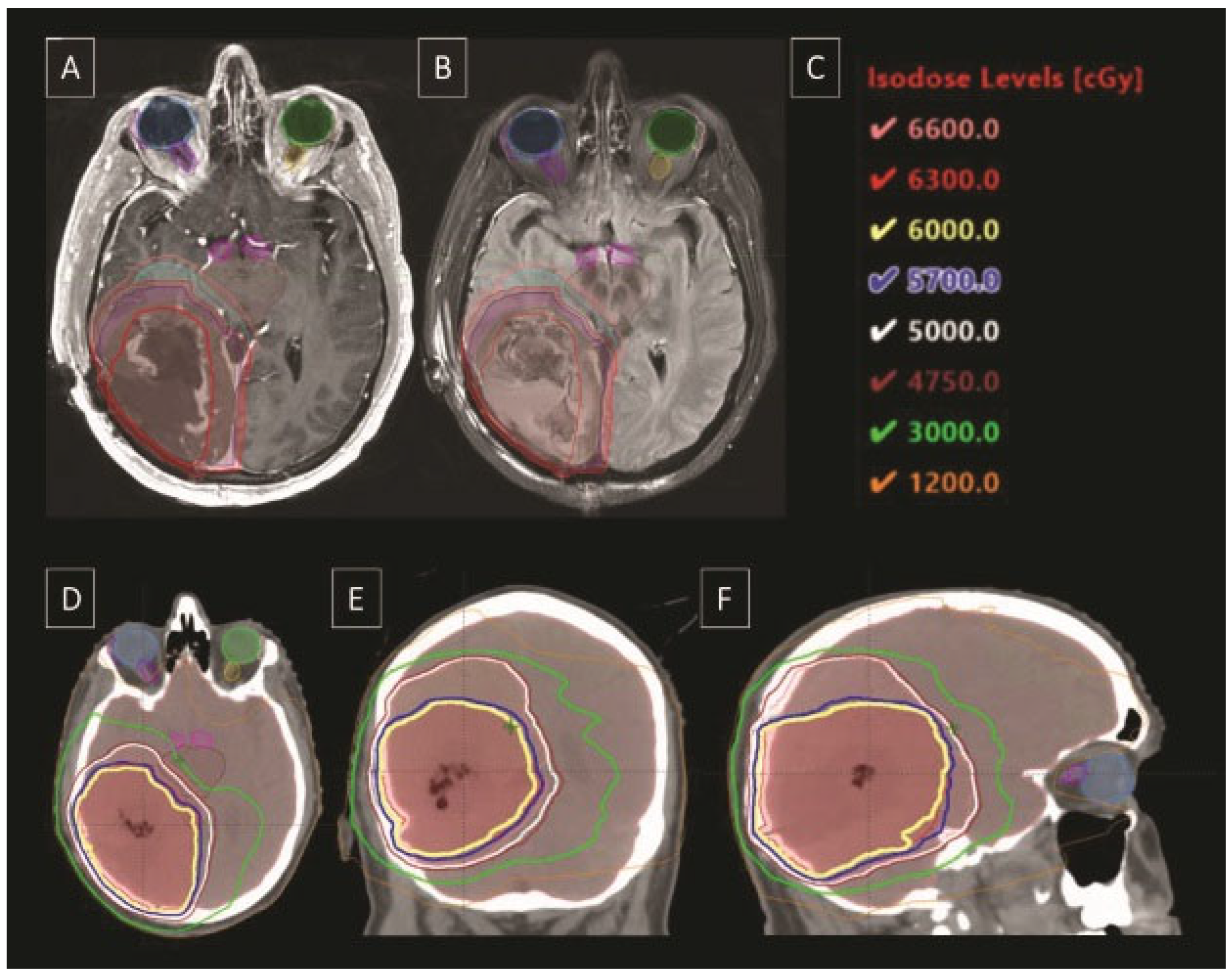
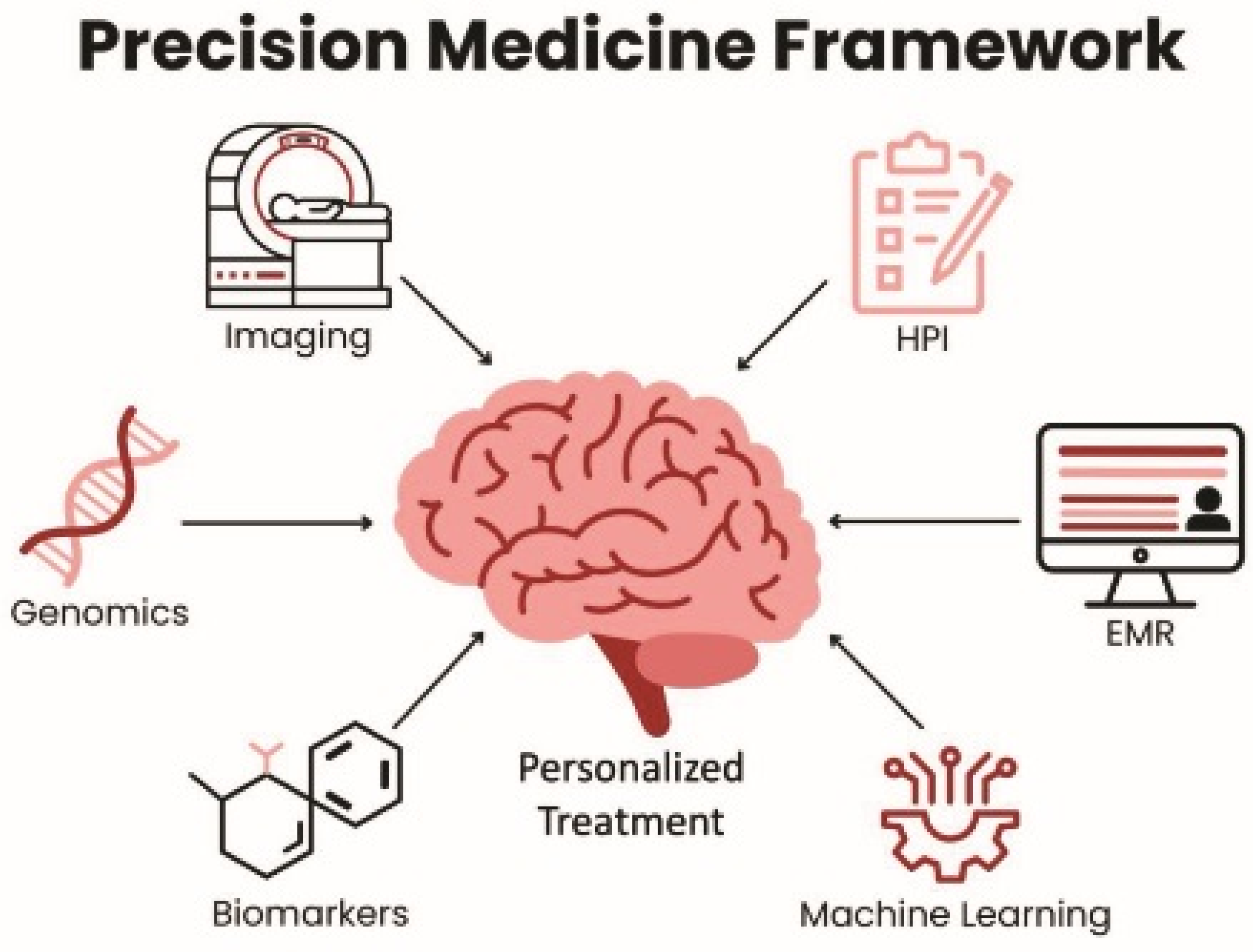
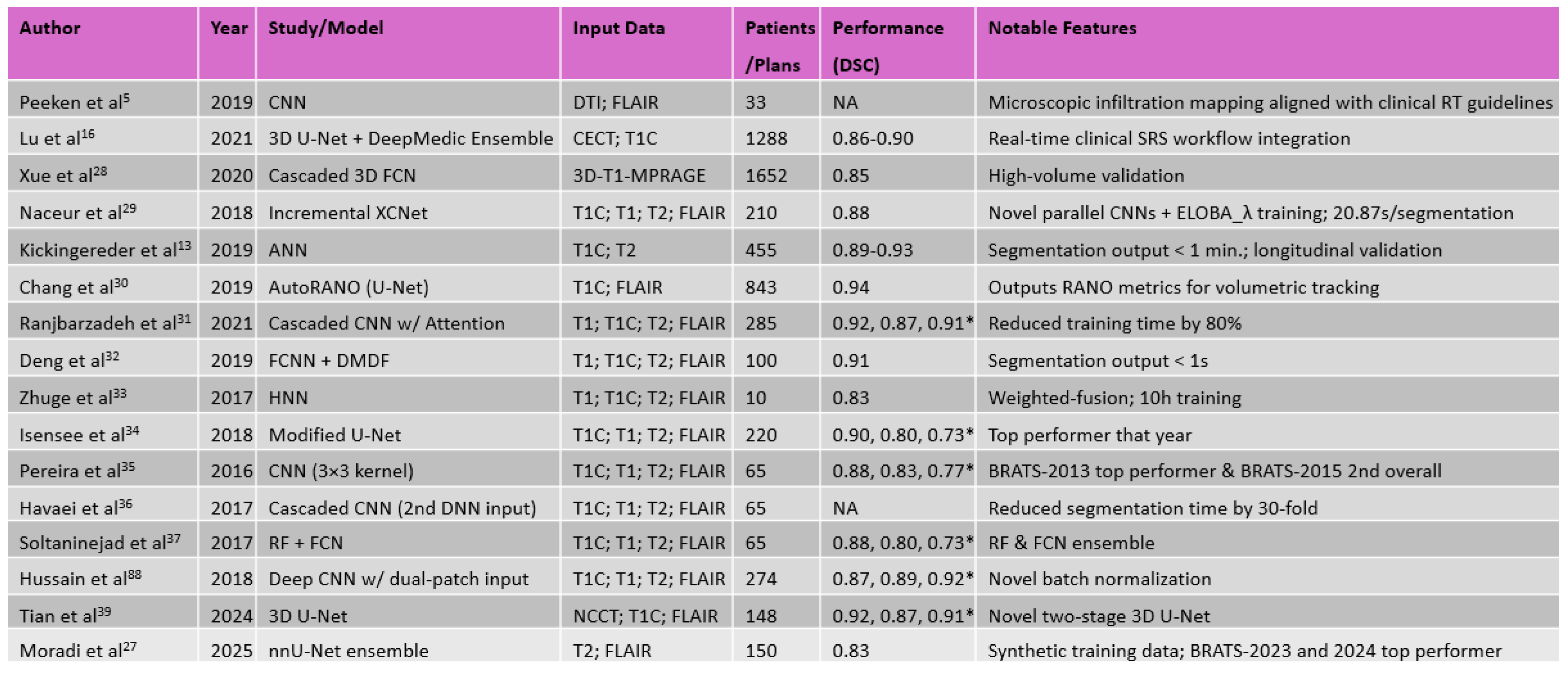 |
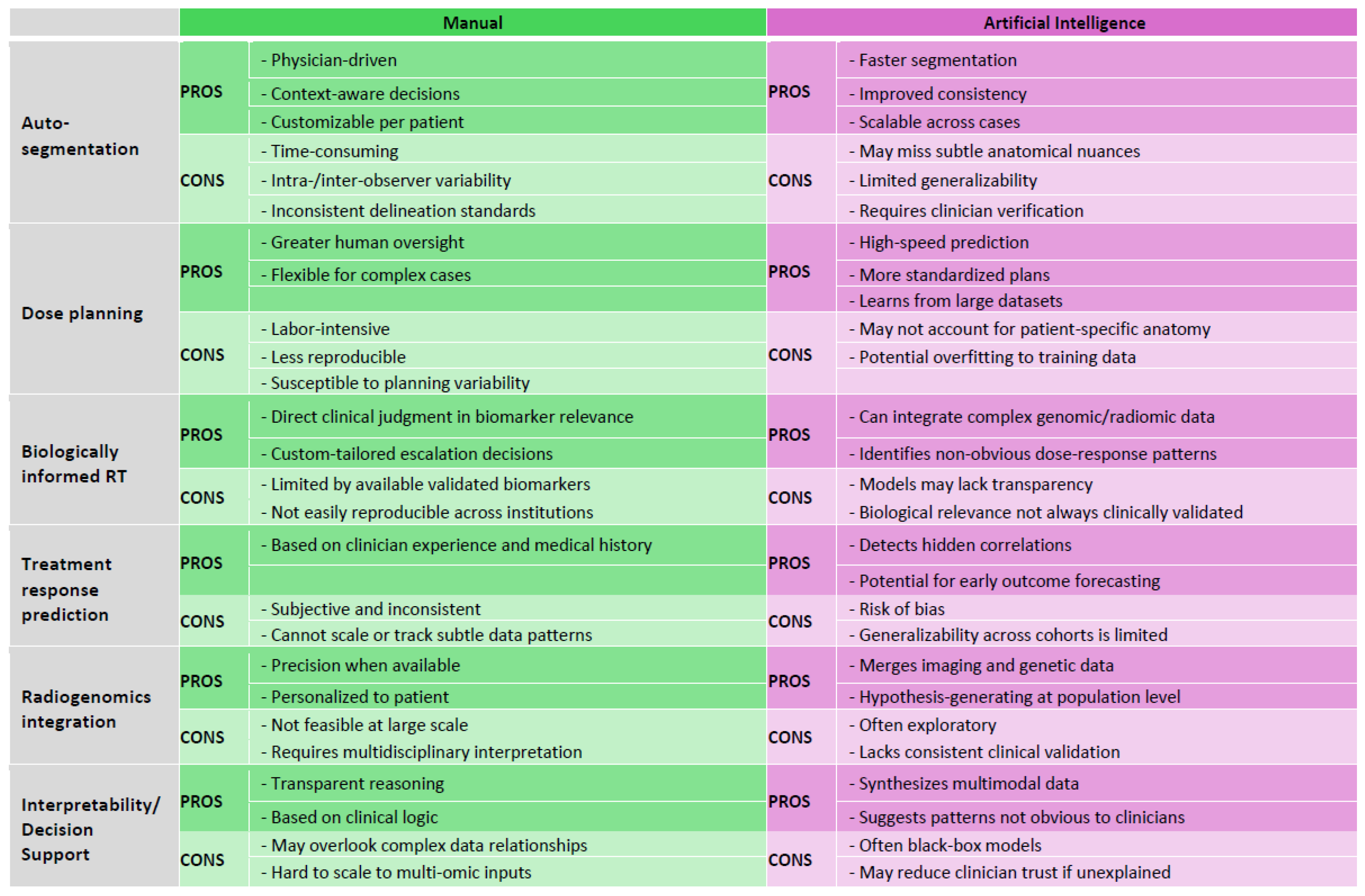 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





