1. Introduction
Liver cancer remains one of the most prevalent and deadly malignancies worldwide, ranking as the eighth most common cancer and the fourth leading cause of cancer-related deaths globally [
1]. Hepatocellular carcinoma (HCC) accounts for approximately 75–85% of primary liver cancer cases, with high morbidity and poor prognosis due to late diagnosis, high recurrence, and resistance to current therapies [
2,
3]. Despite advances in imaging and serological biomarkers such as alpha-fetoprotein (AFP), early detection and reliable prognostication remain elusive, limiting treatment success [
4].
MicroRNAs (miRNAs) have emerged as pivotal regulators of gene expression, orchestrating diverse cellular processes including proliferation, differentiation, apoptosis, and metabolism. These small (~19–25 nucleotides), non-coding RNAs regulate gene expression post-transcriptionally by binding to complementary sequences in target messenger RNAs (mRNAs), leading to mRNA degradation or translational repression [
5,
6]. Dysregulation of miRNAs contributes to oncogenesis and tumor progression across multiple cancers, including liver cancer [
7].
Exosomes, nanoscale extracellular vesicles secreted by most cell types, carry various molecular cargos, including proteins, lipids, DNAs, and RNAs. Of particular interest are exosomal microRNAs (exo-miRNAs), which serve as mediators of intercellular communication by transferring genetic information between cells [
8]. Exo-miRNAs in bodily fluids offer remarkable stability and reflect the physiological or pathological status of their cells of origin, positioning them as promising non-invasive biomarkers and potential therapeutic targets in liver cancer [
9,
10].
This review comprehensively examines the scientific evolution of exosomal microRNA research in liver cancer. We explore miRNA biogenesis, exosome biology, physiological and pathological roles of exosomal miRNAs, their implications in liver cancer diagnosis, progression, and therapy, and highlight current challenges and future directions in this burgeoning field.
2. Biogenesis and Functional Mechanisms of microRNAs
MicroRNAs originate from endogenous genes transcribed primarily by RNA polymerase II, yielding primary miRNAs (pri-miRNAs) that fold into hairpin structures [
11]. In the nucleus, the microprocessor complex comprising Drosha and DGCR8 cleaves pri-miRNAs into precursor miRNAs (pre-miRNAs), approximately 70 nucleotides long [
12]. Exportin-5 mediates their transport into the cytoplasm, where Dicer processes them into ~22 nucleotide double-stranded miRNA duplexes [
13]. One strand (the guide) incorporates into the RNA-induced silencing complex (RISC), while the passenger strand is typically degraded [
14].
Within RISC, miRNAs guide the complex to complementary sequences mainly in the 3’ untranslated regions (UTRs) of target mRNAs, inhibiting their translation or promoting degradation [
15]. Because individual miRNAs can target multiple mRNAs, and conversely, single mRNAs may be regulated by multiple miRNAs, these molecules act as critical regulators of gene expression networks [
16].
MiRNAs influence key biological processes, including development, metabolism, immune responses, and apoptosis. Their dysregulation is implicated in various diseases, particularly cancers, where they may function as oncogenes (oncomiRs) or tumor suppressors depending on their targets and context [
7,
17].
3. Exosomes and Exosomal microRNAs: Biogenesis and Selective Packaging
Exosomes are extracellular vesicles approximately 30–150 nm in diameter, formed within multivesicular bodies (MVBs) of the endosomal system [
18]. Upon fusion of MVBs with the plasma membrane, exosomes are secreted into the extracellular milieu, facilitating communication between cells [
19].
Exosome biogenesis involves the endosomal sorting complex required for transport (ESCRT) machinery, tetraspanins, and ceramide-dependent pathways, orchestrating cargo sorting into intraluminal vesicles [
20]. Selective packaging of miRNAs into exosomes is a tightly regulated process, influenced by sequence motifs, RNA-binding proteins such as hnRNPA2B1 and YBX1, and cellular context [
21,
22]. This selectivity enables exosomes to carry specific miRNA cargos reflective of the physiological or pathological status of the donor cells [
23].
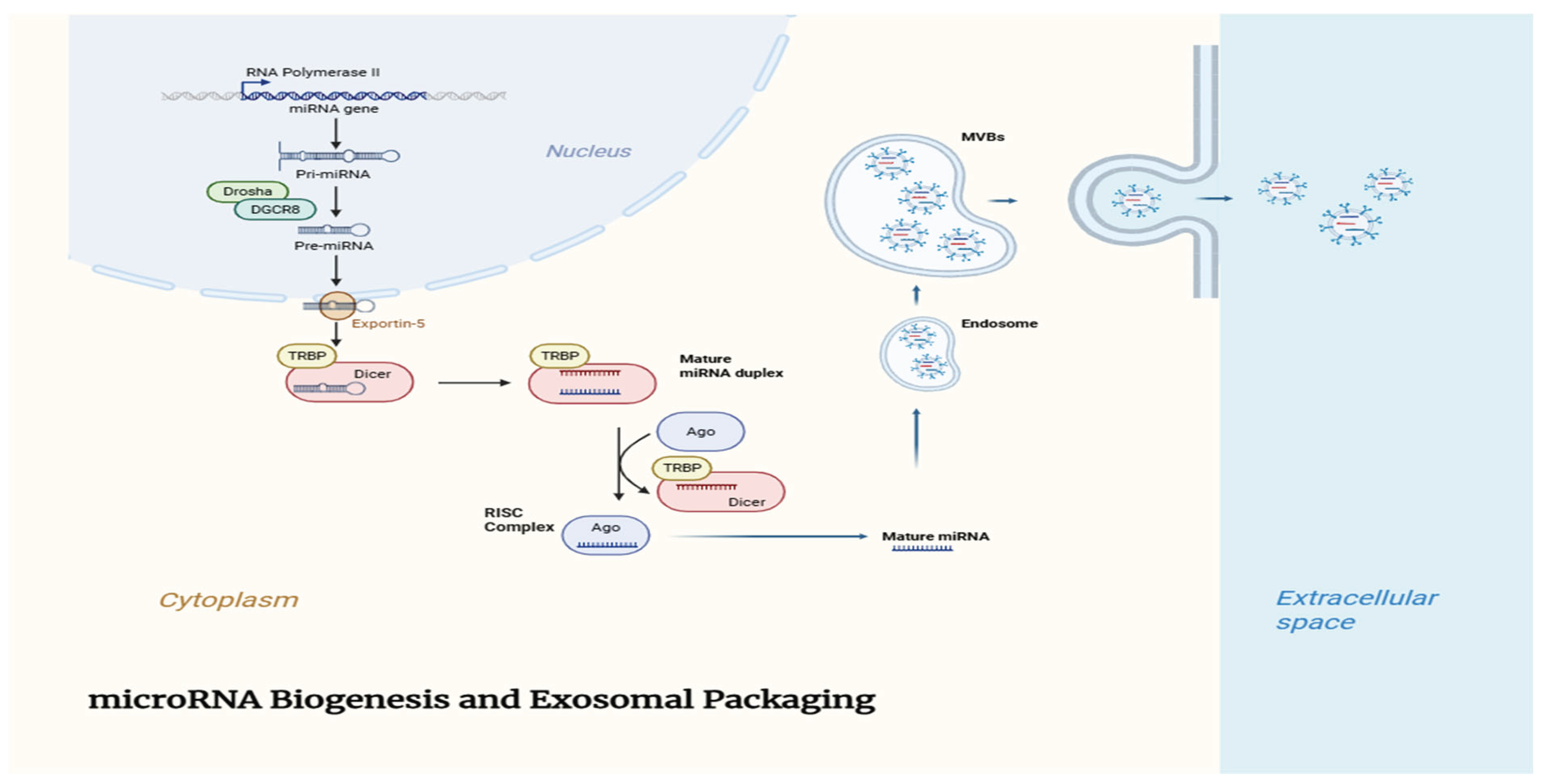
Figure 1.
Schematic Diagram of microRNA Biogenesis and Exosomal Packaging: Schematic representation of microRNA biogenesis and selective packaging into exosomes. MicroRNAs are transcribed and processed through nuclear and cytoplasmic steps, followed by selective incorporation into exosomes which mediate intercellular communication.
Figure 1.
Schematic Diagram of microRNA Biogenesis and Exosomal Packaging: Schematic representation of microRNA biogenesis and selective packaging into exosomes. MicroRNAs are transcribed and processed through nuclear and cytoplasmic steps, followed by selective incorporation into exosomes which mediate intercellular communication.
Exosomal miRNAs are remarkably stable in circulation, protected from RNase degradation by the lipid bilayer of exosomes [
24]. They can be taken up by recipient cells through endocytosis, phagocytosis, or membrane fusion, modulating gene expression and cellular phenotypes in target cells [
25]. This intercellular transfer plays crucial roles in maintaining homeostasis and in disease pathogenesis. The biogenesis of microRNAs involves a tightly regulated multistep process, including nuclear and cytoplasmic events that ultimately lead to the generation of mature miRNAs. The selective sorting of these miRNAs into exosomes further adds complexity to their regulatory roles.
Table 1 outlines the key molecules and steps involved in the biogenesis and exosomal packaging of microRNAs.
4. Physiological Roles of Exosomal miRNAs
Under physiological conditions, exosomal miRNAs participate in diverse biological processes. They contribute to tissue development, regeneration, and immune modulation by mediating crosstalk between different cell types [
26]. For example, mesenchymal stem cell-derived exosomes enriched with miRNAs support tissue repair and modulate inflammation [
27].
In the liver, exosomal miRNAs regulate hepatocyte function, liver regeneration, and responses to injury [
28]. They facilitate communication between hepatocytes, hepatic stellate cells (HSCs), Kupffer cells, and immune cells, orchestrating liver homeostasis and repair [
29]. Exosomal miRNAs also modulate lipid metabolism and insulin sensitivity, highlighting their role in metabolic regulation [
30]. Exosomal microRNAs have shown great promise as minimally invasive biomarkers for early diagnosis and prognosis in liver cancer.
Table 2 summarizes the most studied exosomal miRNAs with diagnostic and prognostic potential, along with their performance metrics in clinical samples.
5. Pathological Roles of Exosomal miRNAs in Diseases
Aberrant exosomal miRNA expression and signaling are implicated in various diseases, including inflammatory disorders, fibrosis, metabolic diseases, and cancer [
31]. In inflammatory and fibrotic diseases, exosomal miRNAs modulate immune responses and fibroblast activation, influencing disease progression [
32].
In cancer, tumor-derived exosomal miRNAs facilitate tumor growth and metastasis by remodeling the tumor microenvironment (TME), promoting angiogenesis, immune evasion, and metastasis [
33,
34]. For instance, exosomal miRNAs can suppress immune cell activity, induce epithelial-mesenchymal transition (EMT), and support metastatic niche formation [
35]. The transfer of miRNAs via exosomes can also contribute to drug resistance by altering drug metabolism or apoptotic pathways in recipient cells [
36].
Given their stability and accessibility in biofluids, exosomal miRNAs have emerged as promising biomarkers for disease diagnosis, prognosis, and therapeutic monitoring [
37].
6. Liver Cancer: Pathogenesis and Clinical Challenges
Hepatocellular carcinoma (HCC) arises primarily in the context of chronic liver disease, including viral hepatitis B and C, alcohol abuse, and non-alcoholic fatty liver disease (NAFLD) [
38]. The multistep pathogenesis involves chronic inflammation, fibrosis, cirrhosis, and genetic and epigenetic alterations that dysregulate key signaling pathways such as Wnt/β-catenin, PI3K/Akt, and TGF-β [
39].
Despite advances in treatment modalities including surgical resection, liver transplantation, ablation therapies, and systemic agents like sorafenib and lenvatinib, HCC prognosis remains poor due to late diagnosis, high recurrence rates, and therapeutic resistance [
40].
Current serum biomarkers, particularly AFP, lack sufficient sensitivity and specificity, especially for early-stage disease [
4]. This highlights an urgent need for novel biomarkers and therapeutic targets to improve HCC management.
7. Roles of Exosomal microRNAs in Liver Cancer
Recent studies have revealed critical roles for exosomal miRNAs in HCC pathogenesis and clinical management. Circulating exosomal miRNAs such as miR-21, miR-122, miR-148a, and miR-192 have been identified as potential diagnostic and prognostic biomarkers [
41,
42]. Panels of exosomal miRNAs often outperform single markers, improving sensitivity and specificity for early detection and monitoring [
43].
Mechanistically, tumor-derived exosomal miRNAs promote tumor progression by modulating the TME, inducing angiogenesis, facilitating EMT, and evading immune surveillance [
44,
45]. For example, exosomal miR-21 can activate hepatic stellate cells into cancer-associated fibroblasts, promoting fibrosis and tumor progression [
46]. Exosomal miR-93 and miR-199a modulate drug resistance pathways, influencing therapeutic response [
47,
48].
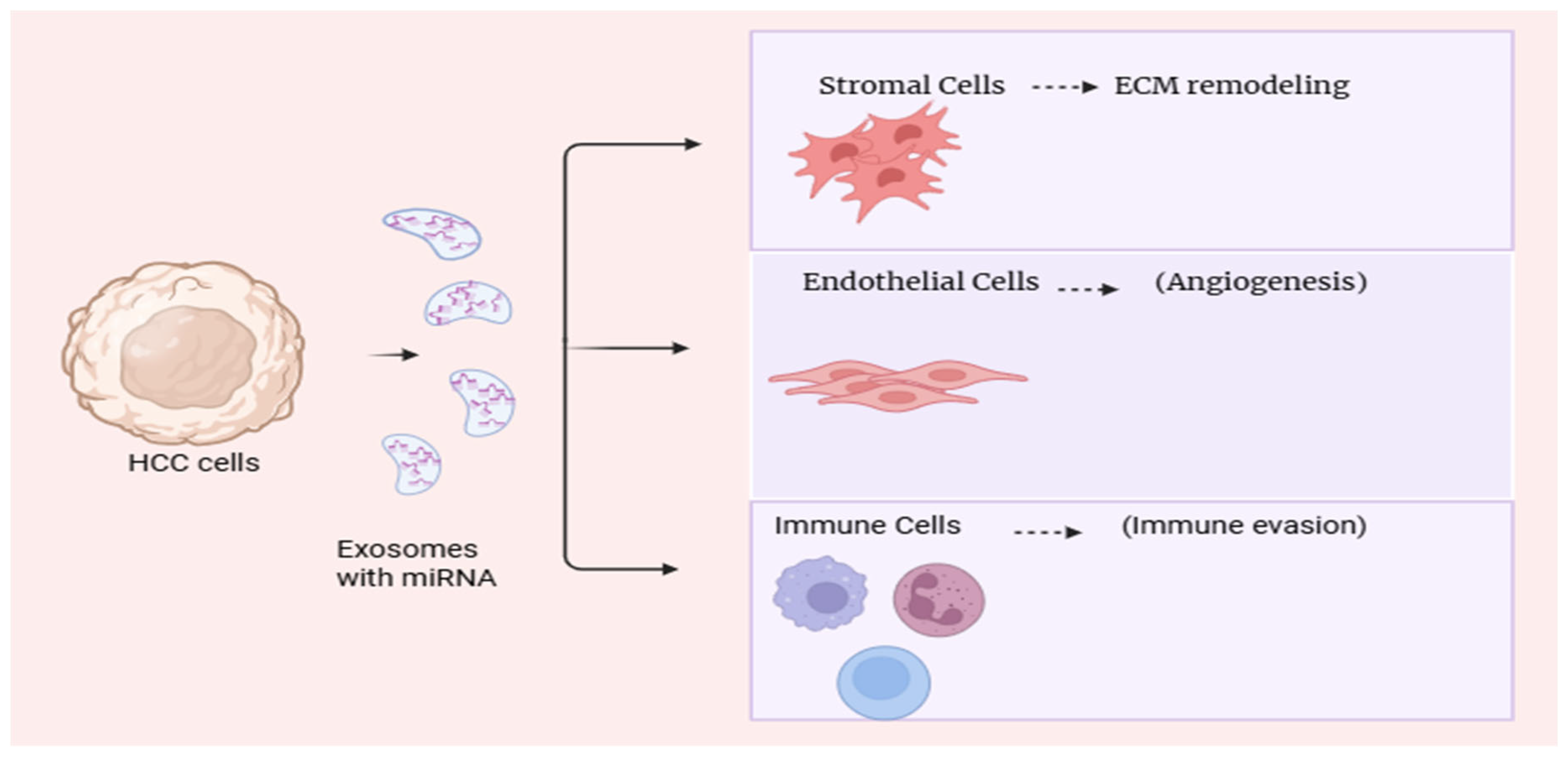
Figure 2.
Role of Exosomal miRNAs in Liver Cancer Progression: Role of exosomal microRNAs in modulating the liver cancer microenvironment. Tumor-derived exosomal miRNAs facilitate communication with stromal and immune cells to promote tumor progression and metastasis.
Figure 2.
Role of Exosomal miRNAs in Liver Cancer Progression: Role of exosomal microRNAs in modulating the liver cancer microenvironment. Tumor-derived exosomal miRNAs facilitate communication with stromal and immune cells to promote tumor progression and metastasis.
Furthermore, exosomal miRNAs contribute to liver fibrosis and cirrhosis, key precursors of HCC, by regulating HSC activation and inflammatory pathways [
49]. Several microRNAs have been identified as critical regulators of liver cancer progression, influencing tumor cell proliferation, invasion, and metastasis.
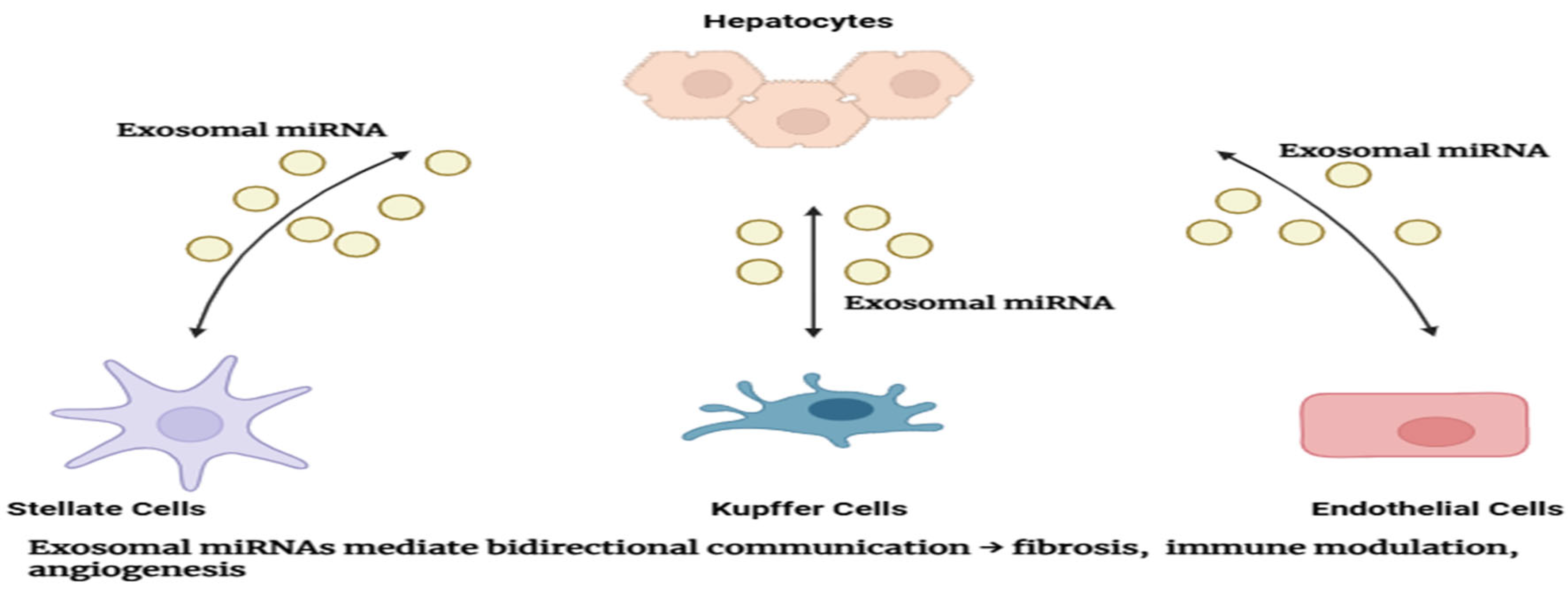
Figure 3.
Exosomal miRNA-mediated Communication in Liver Cancer Microenvironment: Intercellular communication within the liver cancer microenvironment mediated by exosomal microRNAs. Multiple cell types exchange exosomal miRNAs, influencing disease progression and therapeutic response.
Figure 3.
Exosomal miRNA-mediated Communication in Liver Cancer Microenvironment: Intercellular communication within the liver cancer microenvironment mediated by exosomal microRNAs. Multiple cell types exchange exosomal miRNAs, influencing disease progression and therapeutic response.
A summary of the key microRNAs implicated in hepatocellular carcinoma, along with their expression patterns and target pathways, is provided in
Table 3.
8. Therapeutic Implications and Potential
The therapeutic potential of exosomal miRNAs in HCC is multifaceted. Strategies include using exosomes as natural carriers for delivering tumor-suppressive miRNAs or miRNA inhibitors to modulate oncogenic pathways [
50]. Engineering exosomes derived from mesenchymal stem cells or other sources allows targeted delivery with minimal immunogenicity [
51].
Additionally, inhibiting exosome biogenesis or release may disrupt pathological intercellular communication in tumors, offering novel treatment avenues [
52]. However, clinical translation faces challenges including ensuring targeted delivery, understanding biodistribution and pharmacokinetics, avoiding off-target effects, and scalable manufacturing [
53].
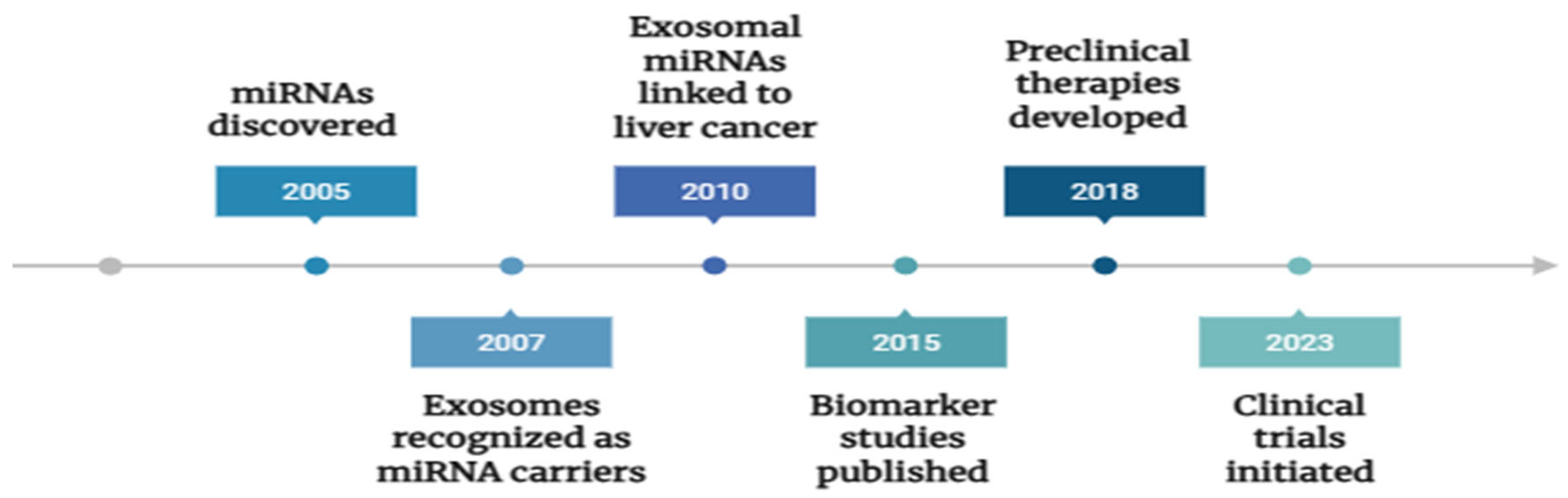
Figure 4.
Timeline of Key Discoveries and Therapeutic Developments in Exosomal miRNA Research for Liver Cancer: Timeline depicting significant advances in exosomal microRNA research related to liver cancer, highlighting major discoveries and therapeutic developments over the past two decades.
Figure 4.
Timeline of Key Discoveries and Therapeutic Developments in Exosomal miRNA Research for Liver Cancer: Timeline depicting significant advances in exosomal microRNA research related to liver cancer, highlighting major discoveries and therapeutic developments over the past two decades.
Ongoing clinical trials are exploring exosome-based diagnostics and therapeutics, although large-scale validation is required before routine clinical application [
54]. Targeting exosomal microRNAs offers a novel therapeutic strategy for liver cancer, with several approaches currently under investigation.
Table 4 provides an overview of existing therapeutic modalities focusing on miRNA mimics, inhibitors, and exosome-based delivery systems.
9. Emerging Technologies and Future Directions
Advances in high-throughput sequencing, single-vesicle analysis, and multi-omics integration are revolutionizing exosomal miRNA research, enabling comprehensive molecular profiling and biomarker discovery [
55]. Novel isolation methods and characterization technologies enhance the specificity and sensitivity of exosomal miRNA detection [
56].
Integration of artificial intelligence and bioinformatics facilitates pattern recognition and predictive modeling, accelerating translational research [
57]. Future work should focus on standardizing methodologies, validating biomarkers in large cohorts, elucidating mechanistic pathways, and optimizing therapeutic delivery systems. Regulatory frameworks and manufacturing standards must evolve to support clinical translation of exosome-based products [
58].
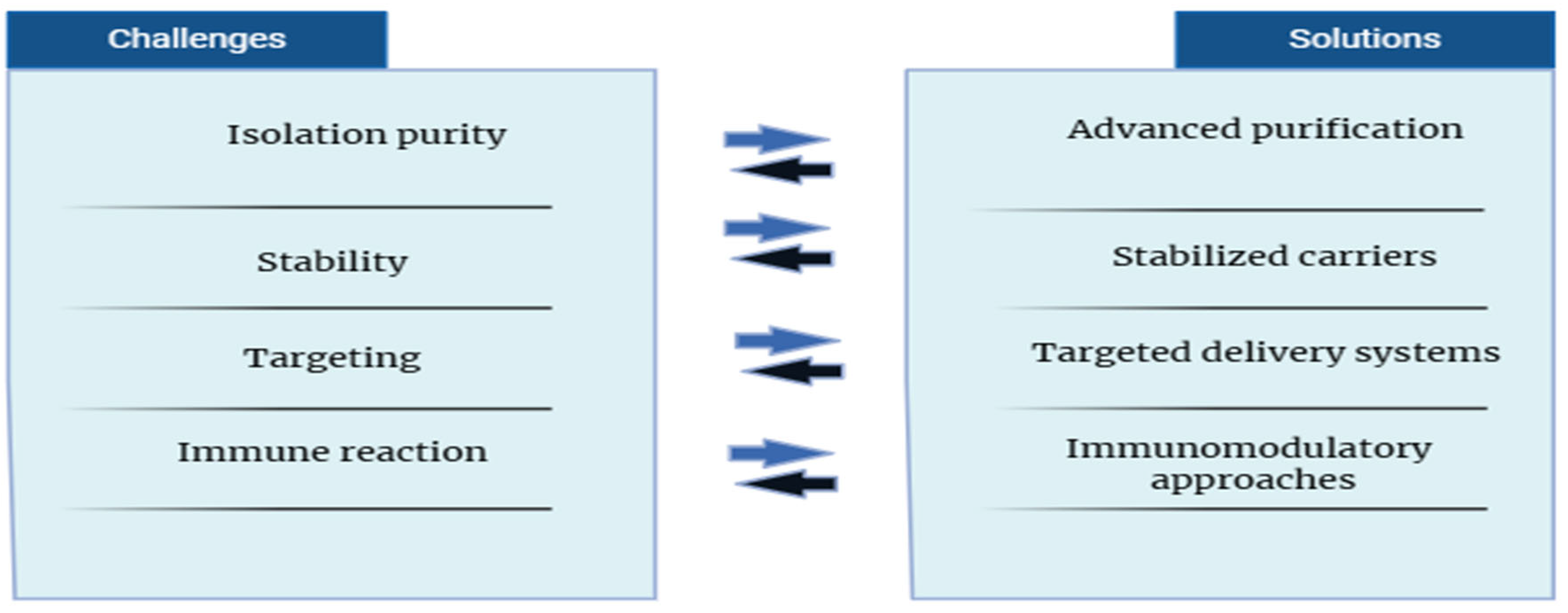
Figure 5.
Future Perspectives and Challenges in Exosomal miRNA-based Therapies: Overview of current challenges and future directions in the development of exosomal microRNA-based therapeutics for liver cancer.
Figure 5.
Future Perspectives and Challenges in Exosomal miRNA-based Therapies: Overview of current challenges and future directions in the development of exosomal microRNA-based therapeutics for liver cancer.
Conclusion
Exosomal microRNAs (miRNAs) represent a rapidly advancing domain with profound implications for elucidating the molecular pathogenesis of hepatocellular carcinoma (HCC), enhancing diagnostic accuracy, and informing the development of innovative therapeutic strategies. Owing to their stability, specificity, and role in mediating intercellular communication, exosomal miRNAs have emerged as promising non-invasive biomarkers and potential modulators of tumor behavior, positioning them as key components in the evolving landscape of precision oncology for HCC.
Realizing their full clinical potential, however, necessitates continued multidisciplinary efforts integrating molecular biology, clinical research, and bioengineering innovation. Overcoming current limitations related to isolation methods, functional characterization, and clinical validation will be critical to translating exosomal miRNA research into effective and clinically actionable tools for the management of liver cancer.::
Author Contributions
Conceptualisation & Supervision: Sameer Kumar V.B., Jordi Muntane, Manuscript Preparation: Ashutosh Kumar Maurya, Proofread & Edit: Ashish Kumar Maurya, Figure visualisation: Swetha E.
Acknowledgments
CMR, Ministry of Health, Govt. of India; KSCSTE, Govt. of Kerala.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [CrossRef]
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018.
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14.
- Trevisani F, D’Intino PE, Morselli-Labate AM, et al. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34(4):570–5. [CrossRef]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. [CrossRef]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. [CrossRef]
- Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle–Mediated Transfer of a Liver-Specific MicroRNA, miR-122, Regulates Hepatic Lipid Metabolism. Hepatology. 2011;54(4):1164–74.
- He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8(1):237–55. [CrossRef]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. [CrossRef]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. [CrossRef]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. [CrossRef]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell. 2003;115(2):199–208. [CrossRef]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–33. [CrossRef]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. [CrossRef]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69.
- Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–83. /: https. [CrossRef]
- Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–79. [CrossRef]
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89. [CrossRef]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [CrossRef]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276.
- Guduric-Fuchs J, O’Connor A, Camp B, O’Neill C, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. [CrossRef]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8.
- Montecalvo A, Larregina AT, Shufesky WJ, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119(3):756–66. [CrossRef]
- Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–35. [CrossRef]
- Bruno S, Grange C, Deregibus MC, et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20(5):1053–67. [CrossRef]
- Chen L, Zhang S, Wang J, et al. Exosomes derived from hepatocellular carcinoma cells induce activation of hepatic stellate cells through transferring miR-21. Cancer Sci. 2018;109(6):1965–76.
- Roderburg C, Luedde T. The role of the microRNA-29 family in liver fibrosis and hepatocellular carcinoma. J Hepatol. 2014;61(3):507–8.
- Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–5. [CrossRef]
- Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer — implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15(10):617–38. [CrossRef]
- Verma SK, Baliyan S, Patil V, et al. Role of extracellular vesicles in liver fibrosis: a concise review. Int J Mol Sci. 2020;21(21):7746.
- Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871(2):455–68. [CrossRef]
- Zeng Z, Li Y, Pan Y, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. [CrossRef]
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. [CrossRef]
- Chen WX, Liu XM, Lv MM, et al. Exosomal miR-21 regulates the sensitivity of breast cancer cells to doxorubicin by targeting PTEN. J Cell Mol Med. 2018;22(11):5385–97.
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–56. [CrossRef]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27.
- Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–62.
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90.
- Sugimachi K, Matsumura T, Hirata H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(2):532–8. [CrossRef]
- Lin XJ, Gao W, Wan J, et al. Serum exosomal miR-122 and miR-148a are promising biomarkers for early diagnosis of hepatocellular carcinoma. J Cancer. 2019;10(18):4582–9.
- Zhang X, Yang J, Li L, et al. A novel panel of serum exosomal microRNAs for early diagnosis of hepatocellular carcinoma. J Cell Biochem. 2019;120(10):17322–30.
- Lou G, Chen Z, Zheng M, Liu Y. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp Mol Med. 2017;49(6):e346. [CrossRef]
- Shi M, Zhang Z, Xu R, et al. Exosomal miR-103a-3p promotes hepatocellular carcinoma metastasis by targeting SFRP4 and activating Wnt/β-catenin signaling. Mol Ther Nucleic Acids. 2020;22:1–15.
- Wang B, Yao K, Huuskes BM, Shen HH, Zhuang J, Godson C. Exosomes from M2 macrophages promote angiogenesis in hepatocellular carcinoma by transferring miR-21. J Exp Clin Cancer Res. 2018;37(1):132.
- Xu H, Ma Q, Liu W, et al. Exosomal miR-199a-3p promotes sorafenib resistance in hepatocellular carcinoma. Mol Cancer. 2020;19(1):148.
- Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–68. [CrossRef]
- Kannan M, Kaur G, Haque SJ. Fibrosis and hepatocellular carcinoma: molecular connections and therapeutic targets. Front Pharmacol. 2019;10:994.
- Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. [CrossRef]
- Liang G, Zhu Y, Ali DJ, Tian T, Chen X. Engineered exosomes for targeted drug delivery. Theranostics. 2021;11(7):3183–95.
- Essandoh K, Li Y, Huo J, Fan GC. Exosomes as a nanocarrier for gene therapy: Progress and challenges. Nanomedicine. 2015;11(12):3219–32.
- Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38(6):754–63. [CrossRef]
- ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). Identifier NCT03608631, Exosomes in liver cancer (HCC) diagnosis and therapy; 2020 May 1 [cited 2025 Oct 3]. Available from: https://clinicaltrials.gov/ct2/show/NCT03608631.
- Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, et al. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide–polyethyleneimine coating. Lab Chip. 2016;16(16):3033–42.
- Nordin JZ, Lee Y, Vader P, Mäger I, Johansson HJ, Heusermann W, et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11(4):879–83. [CrossRef]
- Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–15. [CrossRef]
- Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. [CrossRef]
Table 1.
Biogenesis Pathway Components of microRNAs and Exosomal miRNAs.
Table 1.
Biogenesis Pathway Components of microRNAs and Exosomal miRNAs.
| Step |
Key Molecules/Proteins |
Description |
Relevance to Exosomal Packaging |
Reference(s) |
| Transcription |
RNA Polymerase II |
Primary miRNA (pri-miRNA) synthesis |
Initial step; source of all miRNAs |
[5,11] |
| Nuclear processing |
Drosha, DGCR8 |
Processing pri-miRNA to precursor miRNA (pre-miRNA) |
Generates pre-miRNA for export |
[12] |
| Nuclear export |
Exportin-5 |
Transports pre-miRNA to cytoplasm |
Enables cytoplasmic processing |
[13] |
| Cytoplasmic processing |
Dicer |
Converts pre-miRNA into mature miRNA duplex |
Produces mature miRNAs, ready for function |
[14] |
| RISC loading |
Argonaute proteins (Ago2) |
Assembly into RNA-induced silencing complex (RISC) |
Guides miRNA targeting; selective exosomal sorting |
[15,24] |
| Exosomal sorting |
hnRNPA2B1, YBX1 |
RNA-binding proteins mediate selective packaging |
Determines miRNA export via exosomes |
[21,22] |
Table 2.
Exosomal microRNAs as Diagnostic and Prognostic Biomarkers in Liver Cancer.
Table 2.
Exosomal microRNAs as Diagnostic and Prognostic Biomarkers in Liver Cancer.
| miRNA |
Sample Type |
Diagnostic/Prognostic Utility |
Sensitivity/Specificity (if available) |
Reference(s) |
| miR-21 |
Serum exosomes |
Early diagnosis, poor prognosis marker |
Sensitivity ~85%, Specificity ~80% |
[28,36] |
| miR-122 |
Plasma exosomes |
Early detection biomarker |
Sensitivity ~90%, Specificity ~85% |
[9,42] |
| miR-148a |
Serum exosomes |
Predicts recurrence after treatment |
Data limited |
[42,43] |
| miR-221 |
Serum exosomes |
Associated with aggressive tumor behavior |
Data limited |
[7] |
| miR-199a-3p |
Serum exosomes |
Predicts resistance to sorafenib therapy |
Data limited |
[47] |
Table 3.
Overview of Key microRNAs Implicated in Liver Cancer.
Table 3.
Overview of Key microRNAs Implicated in Liver Cancer.
| miRNA |
Expression Pattern |
Target Genes/Pathways |
Functional Role in HCC |
Reference(s) |
| miR-21 |
Upregulated |
PTEN, PDCD4 |
Promotes proliferation, invasion |
[28,46] |
| miR-122 |
Downregulated |
Cyclin G1, ADAM17 |
Tumor suppressor, regulates metabolism |
[9,42] |
| miR-199a |
Downregulated |
mTOR, c-Met |
Suppresses tumor growth |
[47] |
| miR-221 |
Upregulated |
CDKN1B, PTEN |
Enhances proliferation and survival |
[7] |
| miR-25-3p |
Upregulated |
Notch signaling pathway |
Promotes metastasis |
[34] |
| miR-148a |
Downregulated |
DNMT1 |
Tumor suppressor |
[42,43] |
| miR-103a |
Upregulated |
SFRP4, Wnt/β-catenin |
Promotes metastasis and EMT |
[45] |
Table 4.
Current Therapeutic Approaches Targeting Exosomal miRNAs in Liver Cancer.
Table 4.
Current Therapeutic Approaches Targeting Exosomal miRNAs in Liver Cancer.
| Therapeutic Strategy |
Target miRNA(s) |
Mode of Delivery |
Preclinical/Clinical Status |
Outcomes/Notes |
Reference(s) |
| miRNA mimics |
miR-122, miR-199a |
Lipid nanoparticles, exosomes |
Preclinical |
Suppression of tumor growth in vivo |
[44,45] |
| Anti-miRNA oligonucleotides |
miR-21, miR-221 |
Systemic administration |
Preclinical |
Reduced tumor proliferation and metastasis |
[28,46] |
| Exosome-based drug delivery |
Various miRNAs |
Engineered exosomes |
Early clinical/preclinical |
Improved targeting, reduced off-target effects |
[51,52,53] |
| Combination therapies |
miRNAs + Sorafenib |
Co-delivery via nanoparticles |
Preclinical |
Overcomes drug resistance |
[47,48] |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).