Submitted:
07 October 2025
Posted:
08 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Genomic DNA isolation
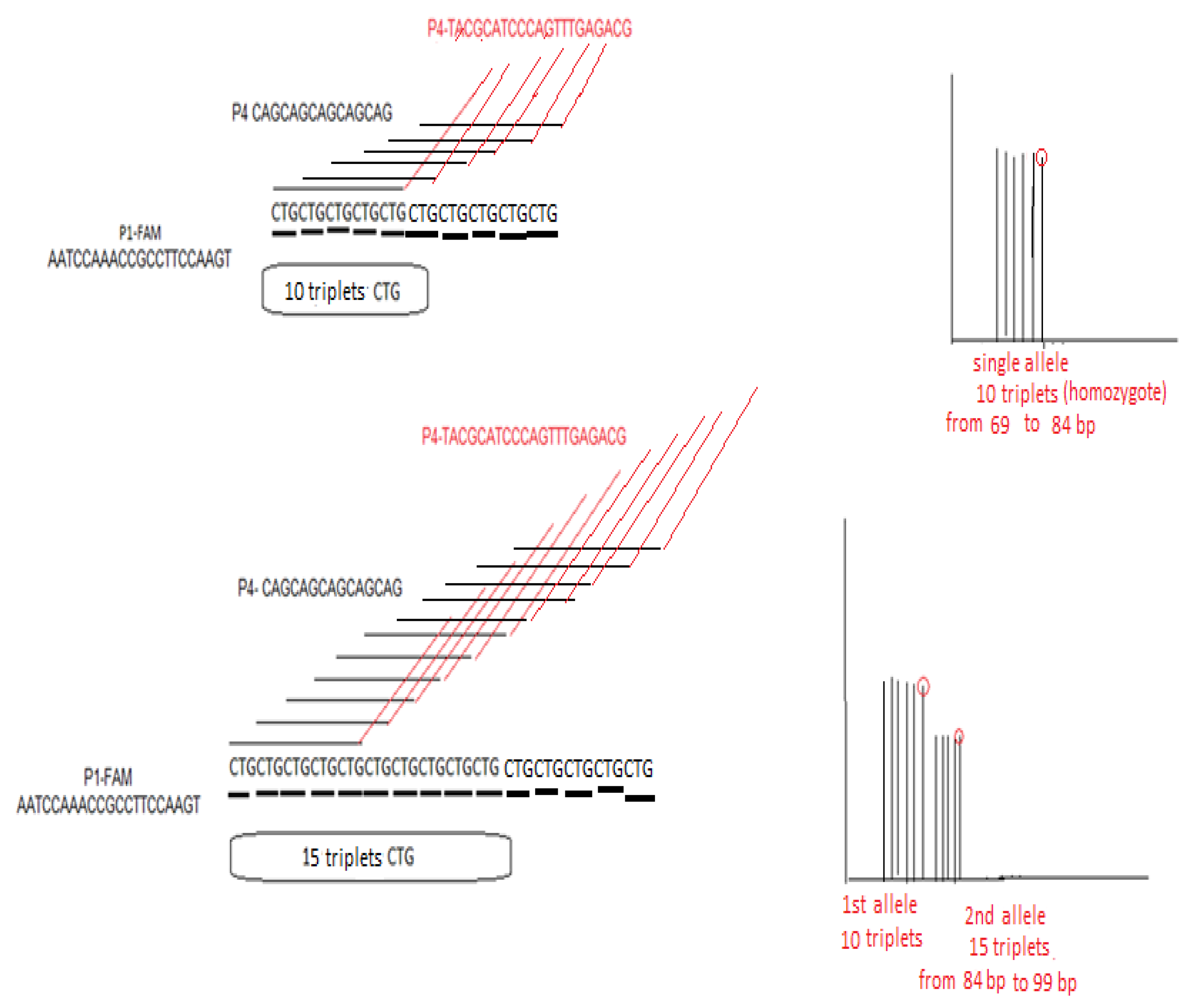
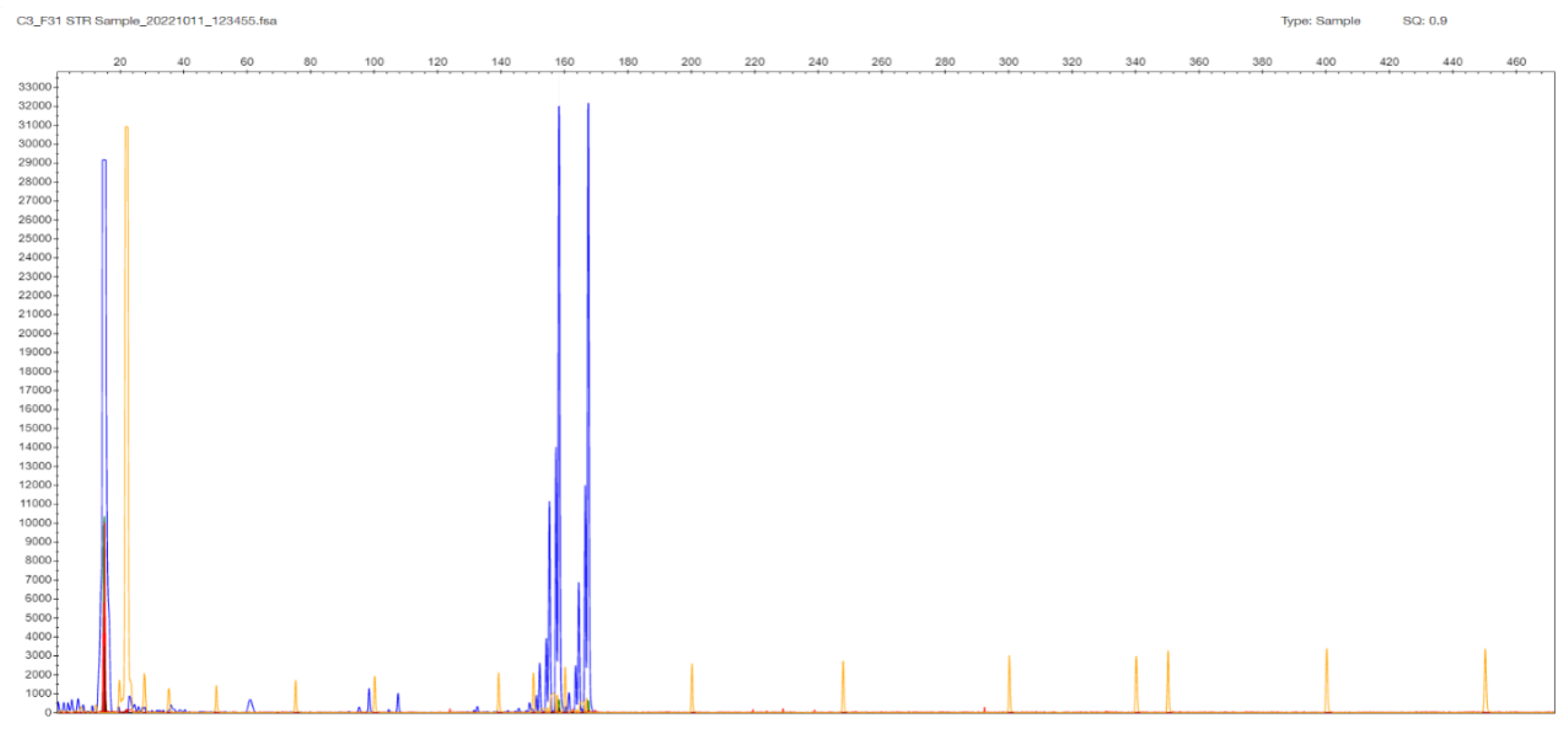
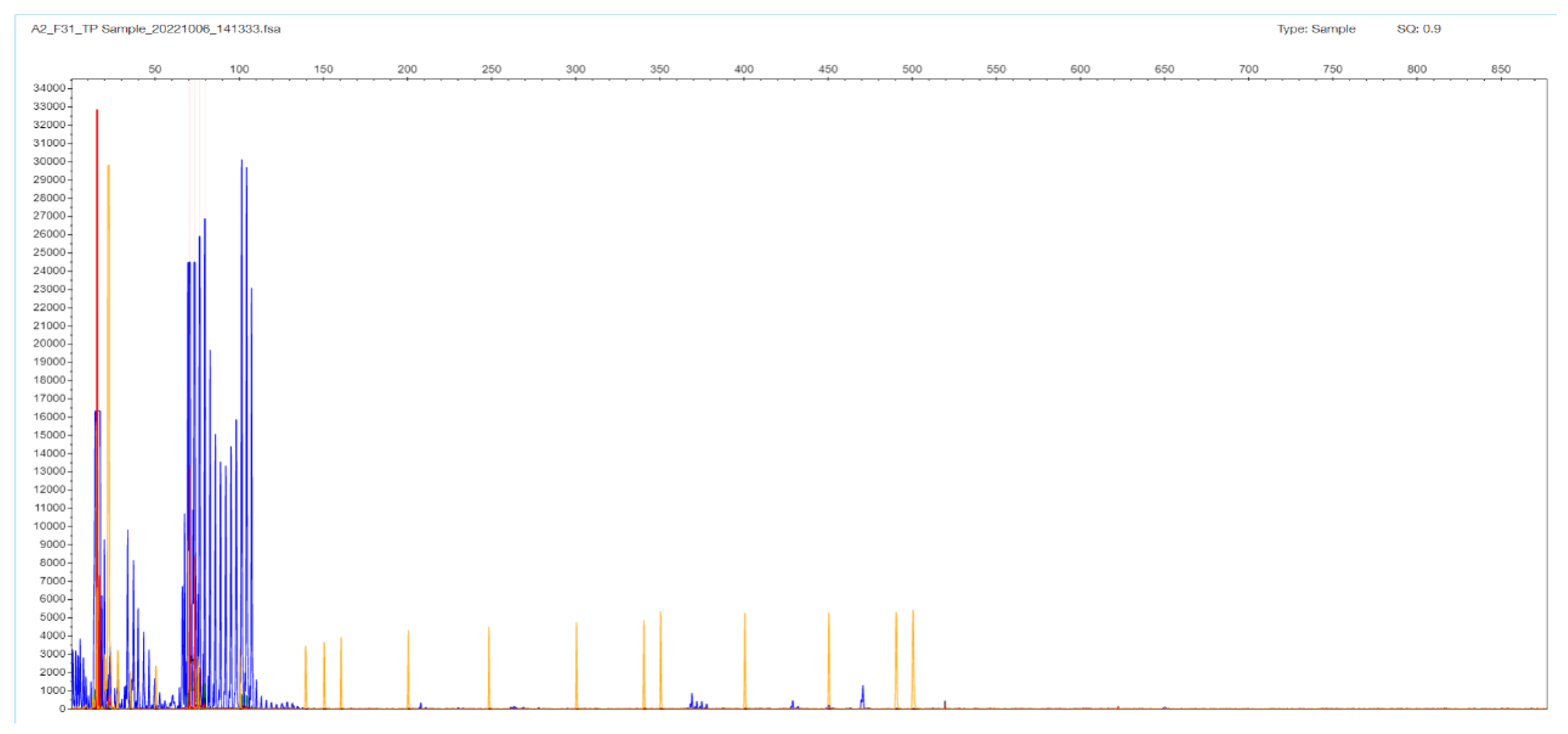
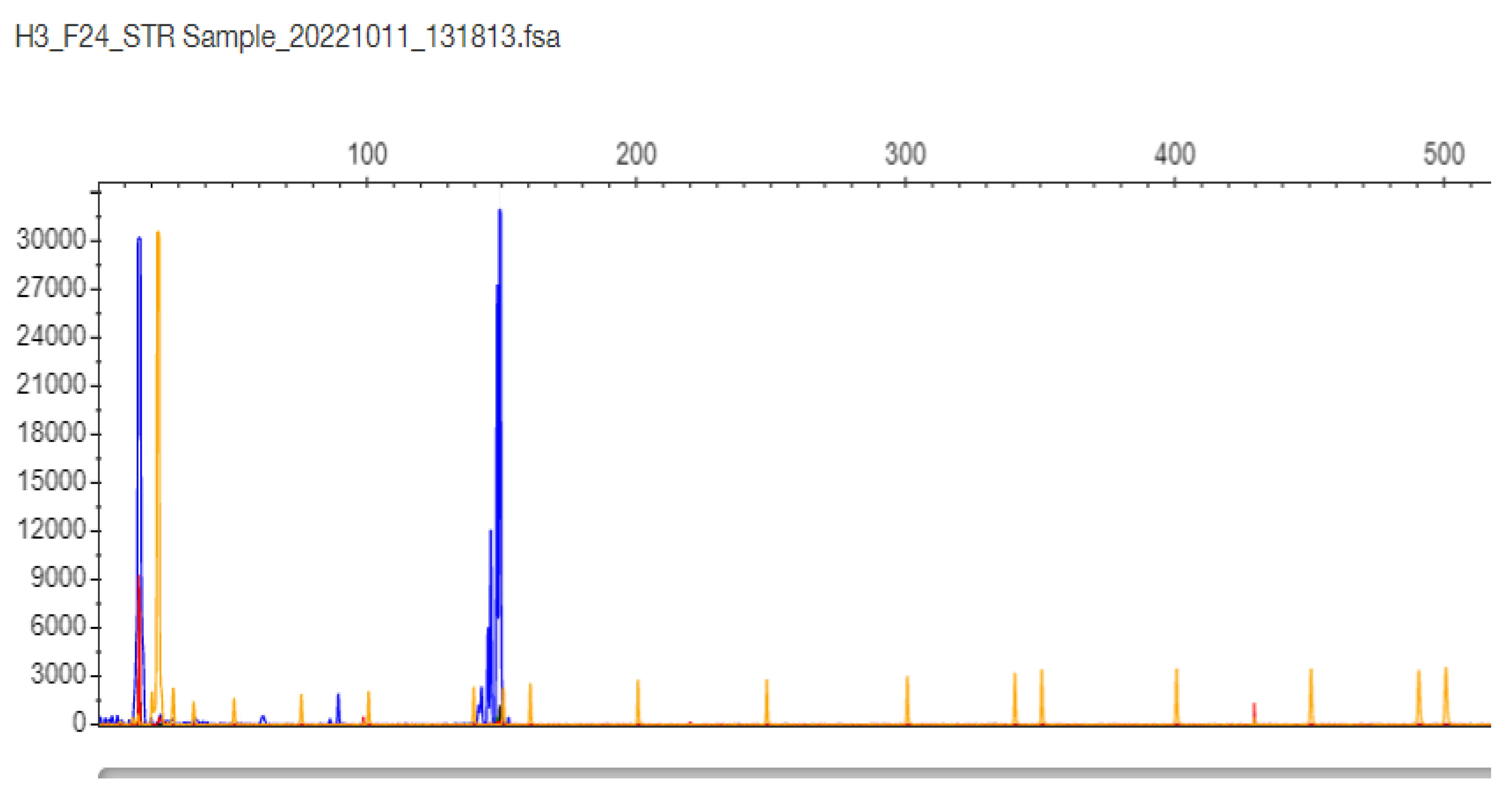
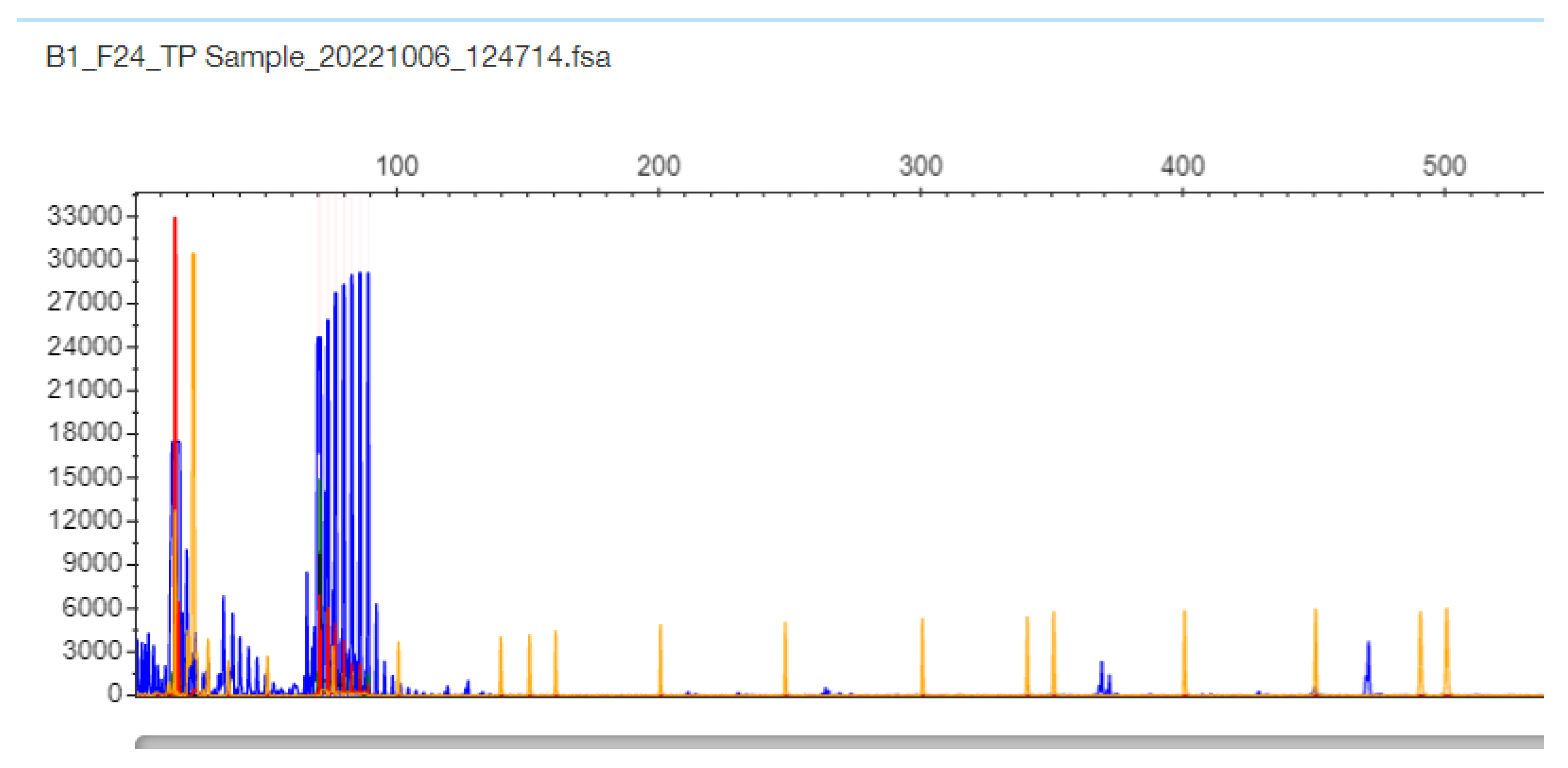
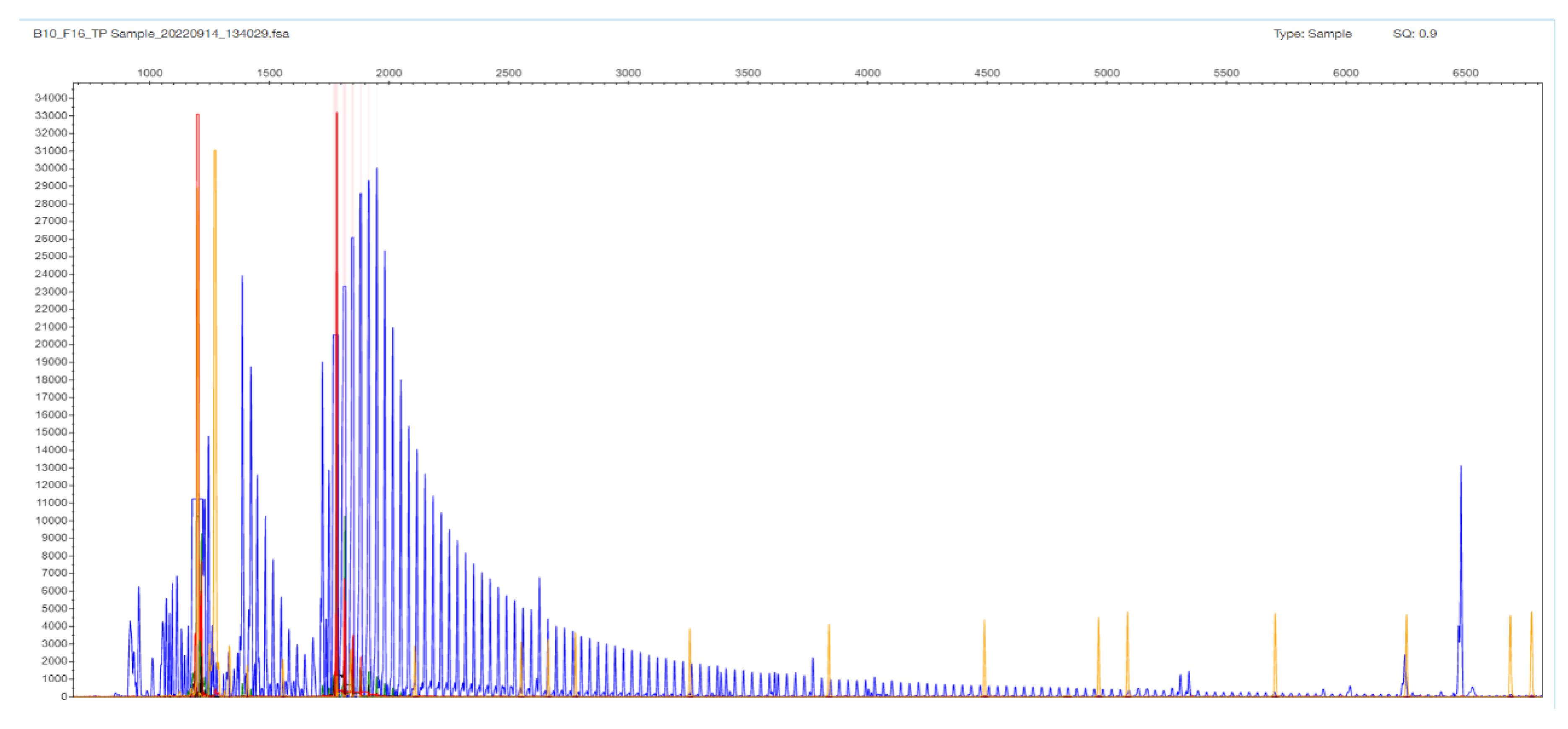
2.3. TCF4 CTG Expansion Evaluation by STR-PCR and TP-PCR Methods
2.4. DNA Sequencing
2.5. TCF4 rs 613872 real-time-qPCR assay
2.6. Statistics
3. Results
| Model | Genotype | STATUS=0 | STATUS=1 | OR (95% CI) | P-value | AIC |
|---|---|---|---|---|---|---|
| Codominant | N/N | 55 (94.8%) | 16 (44.4%) | 1.00 | <0.0001 | 103 |
| N/X | 3 (5.2%) | 17 (47.2%) | 19.41 (5.02-75.02) | |||
| X/X | 0 (0%) | 3 (8.3%) | NA (0.00-NA) | |||
| Dominant | N/N | 55 (94.8%) | 16 (44.4%) | 1.00 | <0.0001 | 102 |
| N/X-X/X | 3 (5.2%) | 20 (55.6%) | 22.83 (5.99-87.09) | |||
| Recessive | N/N-N/X | 58 (100%) | 33 (91.7%) | 1.00 | 0.015 | 127 |
| X/X | 0 (0%) | 3 (8.3%) | NA (0.00-NA) | |||
| Log-additive | --- | --- | --- | 19.85 (5.23-75.29) | <0.0001 | 101 |
| Model | Genotype | STATUS=0 | STATUS=1 | OR (95% CI) | P-value | AIC |
|---|---|---|---|---|---|---|
| Codominant | T/T | 38 (65.5%) | 14 (38.9%) | 1.00 | 2e-04 | 118 |
| G/T | 20 (34.5%) | 15 (41.7%) | 2.11 (0.81-5.45) | |||
| G/G | 0 (0%) | 7 (19.4%) | NA (0.00-NA) | |||
| Dominant | T/T | 38 (65.5%) | 14 (38.9%) | 1.00 | 0.0082 | 126 |
| G/T-G/G | 20 (34.5%) | 22 (61.1%) | 3.26 (1.33-8.02) | |||
| Recessive | T/T-G/T | 58 (100%) | 29 (80.6%) | 1.00 | 1e-04 | 118 |
| G/G | 0 (0%) | 7 (19.4%) | NA (0.00-NA) | |||
| Log-additive | --- | --- | --- | 3.59 (1.68-7.69) | 4e-04 | 120 |
4. Discussion
Supplementary Materials
References
- Sarnicola, 2019. 45, 1–10.
- Ong Tone, S.; Kocaba, V.; Böhm, M.; Wylegala, A.; White, T.L.; Jurkunas, U.V. Fuchs endothelial corneal dystrophy: The vicious cycle of Fuchs pathogenesis. Prog. Retin. Eye Res. 2021, 80, 100863. [CrossRef]
- Altamirano, F.; Ortiz-Morales, G.; Valdez-Garcia, J.E. Fuchs endothelial corneal dystrophy: An updated review. Int. Ophthalmol. 2024, 44, 61. [CrossRef]
- Kumar, V.; Deshpande, N.; Parekh, M.; Wong, R.; Ashraf, S.; Zahid, M.; et al. Estrogen genotoxicity causes preferential development of Fuchs endothelial corneal dystrophy in females. Redox Biol. 2024, 69, 102986. [CrossRef]
- Nanda, G.G.; Alone, D.P. Review: Current understanding of the pathogenesis of Fuchs’ endothelial corneal dystrophy. Mol. Vis. 2019, 25, 295–310.
- Thaung, C.; Davidson, A.E. Fuchs endothelial corneal dystrophy: Current perspectives on diagnostic pathology and genetics—Bowman Club Lecture. BMJ Open Ophthalmol. 2022, 7, e001103. [CrossRef]
- Dimtsas, G.S.; Moschos, M.M. Ultrathin-Descemet Stripping Automated Endothelial Keratoplasty versus Descemet Membrane Endothelial Keratoplasty: A systematic review and meta-analysis. In Vivo 2023, 37, 400–409. [CrossRef]
- Matthaei, M.; Hribek, A.; Clahsen, T.; Bachmann, B.; Cursiefen, C.; Jun, A.S. Fuchs endothelial corneal dystrophy: Clinical, genetic, pathophysiologic, and therapeutic aspects. Annu. Rev. Vis. Sci. 2019, 5, 151–175. [CrossRef]
- Liu, J.-X.; Chiang, T.-L.; Hung, K.-F.; Sun, Y.-C. Therapeutic future of Fuchs endothelial corneal dystrophy: An ongoing way to explore. Taiwan J. Ophthalmol. 2024, 14, 15–26. [CrossRef]
- Chan, M.F.; Pan, P.; Wolfreys, F.D. Novel mechanisms guide innovative molecular-based therapeutic strategies for Fuchs endothelial corneal dystrophy. Cornea 2023, 42, 929–933. [CrossRef]
- Liu, X.; Zheng, T.; Zhao, C.; Zhang, Y.; Liu, H.; Wang, L.; et al. Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy. Eye Vis. (Lond.) 2021, 8, 24. [CrossRef]
- Zhang, J.; McGhee, C.N.J.; Patel, D.V. The molecular basis of Fuchs’ endothelial corneal dystrophy. Mol. Diagn. Ther. 2019, 23, 97–112. [CrossRef]
- Iliff, B.W.; Riazuddin, S.A.; Gottsch, J.D. The genetics of Fuchs’ corneal dystrophy. Expert Rev. Ophthalmol. 2012, 7, 363–375. [CrossRef]
- Fuchs, B.; Zhang, K.; Bolander, M.E.; Sarkar, G. Differential mRNA fingerprinting by preferential amplification of coding sequences. Gene 2000, 258, 155–163. [CrossRef]
- Gottsch, J.D.; Bowers, A.L.; Margulies, E.H.; Seitzman, G.D.; Kim, S.W.; Saba, S.; et al. Serial analysis of gene expression in the corneal endothelium of Fuchs’ dystrophy. Invest. Ophthalmol. Vis. Sci. 2003, 44, 594–599. [CrossRef]
- Westin, I.M.; Viberg, A.; Golovleva, I.; Byström, B. CTG18.1 expansion in transcription factor 4 (TCF4) in corneal graft failure: Preliminary study. Cell Tissue Bank 2024, 25, (ahead of print). [CrossRef]
- Sahin Vural, G.; Bolat, H. Nanopore sequencing method for CTG18.1 expansion in TCF4 in late-onset Fuchs endothelial corneal dystrophy and a comparison of the structural features of cornea with first-degree relatives. Graefe’s Arch. Clin. Exp. Ophthalmol. 2024, 262, (ahead of print). [CrossRef]
- Vazirani, J.; Rambhia, M. Genetics in corneal dystrophy. In Genetics of Ocular Diseases; 2022; pp. 7–14.
- Qiu, J.; Gu, R.; Shi, Q.; Zhang, X. Long noncoding RNA ZFAS1: A novel anti-apoptotic target in Fuchs endothelial corneal dystrophy. Exp. Eye Res. 2024, 241, 109832. [CrossRef]
- Tsedilina, T.R.; Sharova, E.; Iakovets, V.; Skorodumova, L.O. Systematic review of SLC4A11, ZEB1, LOXHD1, and AGBL1 variants in the development of Fuchs’ endothelial corneal dystrophy. Front. Med. (Lausanne) 2023, 10, 1153122. [CrossRef]
- Baratz, K.H.; Tosakulwong, N.; Ryu, E.; Brown, W.L.; Branham, K.; Chen, W.; et al. E2-2 protein and Fuchs’s corneal dystrophy. N. Engl. J. Med. 2010, 363, 1016–1024. [CrossRef]
- Moschos, M.M.; Diamantopoulou, A.; Gouliopoulos, N.; Droutsas, K.; Kitsos, G.; et al. TCF4 and COL8A2 gene polymorphism screening in a Greek population of late-onset Fuchs endothelial corneal dystrophy. In Vivo 2019, 33, 963–971. [CrossRef]
- Li, Y.J.; Minear, M.A.; Rimmler, J.; Zhao, B.; Balajonda, E.; Hauser, M.A.; et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One 2011, 6, e18044. [CrossRef]
- Wieben, E.D.; Aleff, R.A.; Eckloff, B.W.; Atkinson, E.J.; Baheti, S.; Middha, S.; et al. Comprehensive assessment of genetic variants within TCF4 in Fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2014, 55, 6101–6107. [CrossRef]
- Wieben, E.D.; Aleff, R.A.; Tosakulwong, N.; Butz, M.L.; Highsmith, W.E. Jr; Edwards, A.O.; et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One 2012, 7, e49083. [CrossRef]
- Wieben, E.D.; Aleff, R.A.; Basu, S.; Sarangi, V.; Bowman, B.; McLaughlin, I.J.; et al. Amplification-free long-read sequencing of TCF4 expanded trinucleotide repeats in Fuchs endothelial corneal dystrophy. PLoS One 2019, 14, e0219446. [CrossRef]
- Fautsch, M.P.; Wieben, E.D.; Baratz, K.H.; Bhattacharyya, N.; Sadan, A.N.; Hafford-Tear, N.J.; et al. TCF4-mediated Fuchs endothelial corneal dystrophy: Insights into a common trinucleotide repeat-associated disease. Prog. Retin. Eye Res. 2021, 81, 100883. [CrossRef]
- Vasanth, S.; Eghrari, A.O.; Gapsis, B.C.; Wang, J.; Haller, N.F.; Stark, W.J.; et al. Expansion of CTG18.1 trinucleotide repeat in TCF4 is a potent driver of Fuchs’ corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2015, 56, 4531–4536. [CrossRef]
- Soh, Y.Q.; Lim, G.P.S.; Htoon, H.M.; Gong, X.; Mootha, V.V.; Vithana, E.N.; et al. Trinucleotide repeat expansion length as a predictor of the clinical progression of Fuchs’ endothelial corneal dystrophy. PLoS One 2019, 14, e0210996. [CrossRef]
- Eghrari, A.O.; Vasanth, S.; Wang, J.; Vahedi, F.; Riazuddin, S.A.; Gottsch, J.D. CTG18.1 expansion in TCF4 increases likelihood of transplantation in Fuchs corneal dystrophy. Cornea 2017, 36, 40–43. [CrossRef]
- Afshari, N.A.; Igo, R.P. Jr; Morris, N.J.; Stambolian, D.; Sharma, S.; Pulagam, V.L.; et al. Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Nat. Commun. 2017, 8, 14898.
- Mootha, V.V.; Gong, X.; Ku, H.C.; Xing, C. Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2014, 55, 33–42. [CrossRef]
- Warner, J.P.; Barron, L.H.; Goudie, D.; Kelly, K.; Dow, D.; Fitzpatrick, D.R.; et al. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J. Med. Genet. 1996, 33, 1022–1026. [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [CrossRef]
- Kuot, A.; Hewitt, A.W.; Snibson, G.R.; Souzeau, E.; Mills, R.; Craig, J.E.; et al. TGC repeat expansion in the TCF4 gene increases the risk of Fuchs’ endothelial corneal dystrophy in Australian cases. PLoS One 2017, 12, e0183719. [CrossRef]
- Kannabiran, C.; Chaurasia, S.; Ramappa, M.; Mootha, V.V. Update on the genetics of corneal endothelial dystrophies. Indian J. Ophthalmol. 2022, 70, 2126–2137.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





