1. Introduction
1.1. X-Ray Interpretation for Spinal Lesion
Conventional radiography or X-ray are used for assessment of spinal lesions [
4]. It has been the most widely-used tool used to identify and monitor various abnormalities of the spine. In clinical practice, radiologists usually interpret and evaluate the spine on X-ray scans stored in Digital Imaging and Communications in Medicine (DICOM) standard. Abnormal Findings indicate a difference in the appearance of lesions compared to normal areas. Sometimes these differences can be very small or negligible. This makes diagnosis of spinal lesions complex and time-consuming.
The swift progress in machine learning, particularly in deep neural networks, has shown significant promise in detecting diseases through medical imaging data [
3]. This study presents HealNNet-Lesions, a deep learning-based computer-aided diagnosis (CAD) framework. The system is designed to classify and localize abnormal findings from spine X-rays, aiming to enhance the efficiency and accuracy of clinical diagnosis and treatment. The development and validation of HealNNet-Lesions are conducted using VinDr-SpineXR dataset, where radiologist annotations serve as the ground truth [
1]. This research contributes to the integration of advanced computational systems into daily clinical routines.
1.2. Contributions
There are two major contributions of this work. First, we develop and evaluate HealNNet-Lesions – a deep learning framework that is able to classify and localize multiple spinal lesions. Second, we explore how AI and deep learning can help in replacing MRI or CT scans with X-Rays as the diagnosis of some specific spinal lesions and other medical diseases require an MRI or CT scans in some conditions [
4], which are far less accessible and more expensive than an X-Ray, especially in context of many low-middle income countries like India [
5,
6]. HealNNet-Lesions demonstrates that some spinal lesions, which might otherwise require an MRI or CT for diagnosis, can be successfully detected, from X-rays using a deep learning framework.
2. Proposed Method
2.1. The Framework
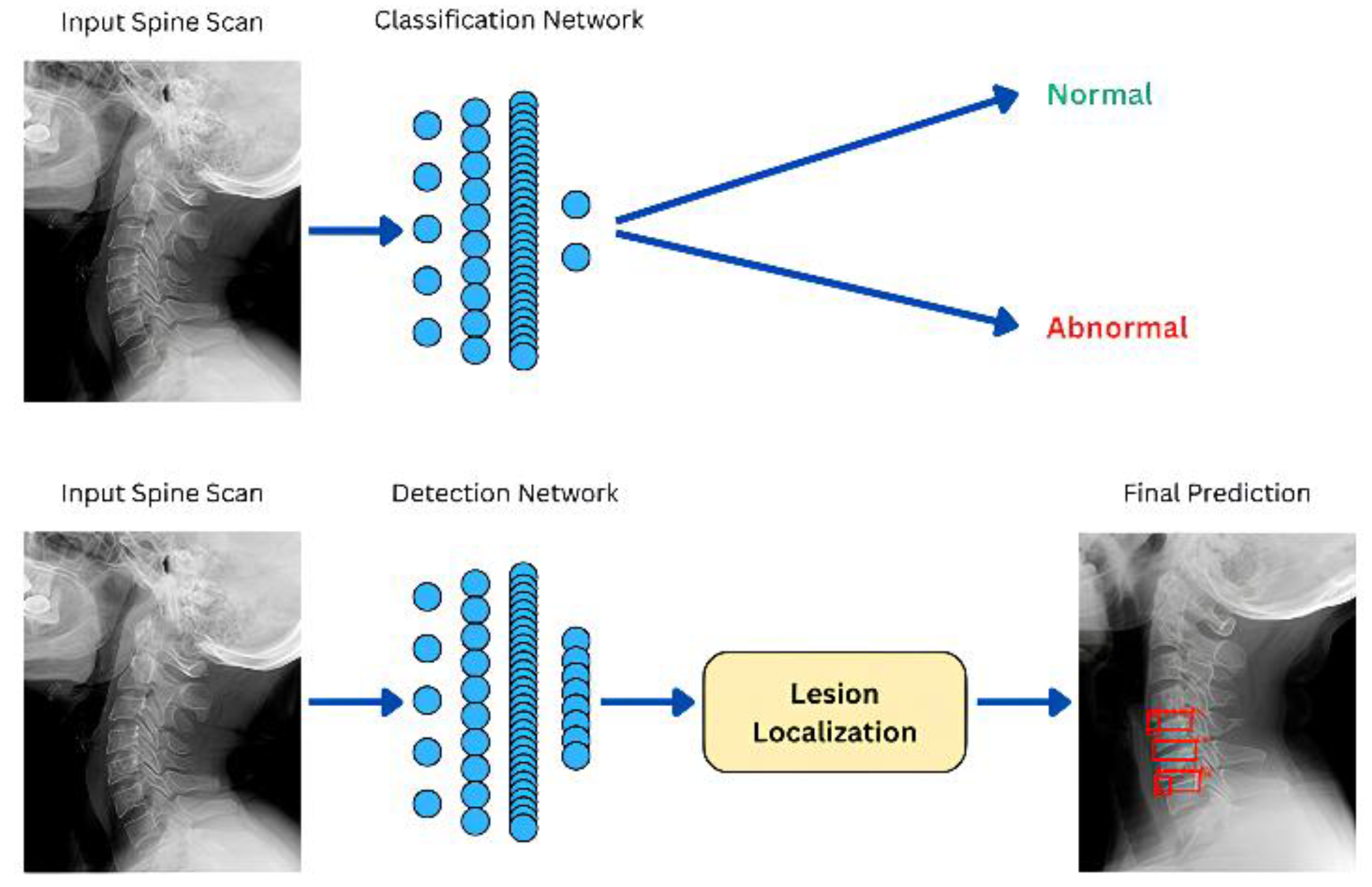
The proposed deep learning framework (see
Figure 1) shows (1) a classification network, which accepts a spine X-ray as input and classifies it as normal or abnormal. (2) a detection network that predicts the location of abnormal findings. If implemented in clinical practice, the classification network can speed up segregation of patients between those who possess an abnormal finding and those who do not. The detection network, HealNNet-Lesions, can further find abnormalities in scans of patients who were classified as abnormal. The DL framework proposed does not follow this approach and primarily focuses on separate and standalone detection and classification networks. The detection network identifies and draws bounding boxes around all lesion detected.
2.2. Dataset
Data Collection Spine X-ray with annotation of spinal lesions is required to develop a lesion detection and classification system. The data was collected from the VinDr-SpineXR Dataset [
1,
2]. The original dataset has more X-ray images than used in the development for this work. A total of 4502 X-ray images were used to train the detection and classification network and a total of 468 images for the testing dataset which was used to evaluate the deep learning network. To keep Protected Health Information (PHI) secure, all patient-identifiable information associated with the images was removed in the dataset. [
1].
Data Annotation In the VinDr-SpineXR dataset, all the images were already annotated. For the part of the dataset that was used develop this deep learning network, No. of annotations represent total annotations in each category, as there can be multiple annotation in a single image. The data characteristics including number of images used from each category in the training data is summarized in
Table 1 and for testing data, it is in
Table 2.
3. Model Development
The object detection framework was utilized for automated lesion detection in spinal X-Ray scans. The architecture is based on Faster R-CNN with a ResNet-50 backbone [
9] and Feature Pyramid Network (FPN), implemented using PyTorch [
15] and TorchVision [
16]. Faster R-CNN operates as a two-stage detector: the Region Proposal Network (RPN) first generates candidate object regions, followed by a region-of-interest (ROI) head that classifies these regions and refines their bounding box coordinates. The ResNet-50 backbone, pre-trained on ImageNet [
8], serves as a robust feature extractor, while the FPN enhances multi-scale feature representation, improving detection performance for lesions of varying sizes.
To adapt the model for the specific application, the final classification head was replaced to accommodate eight custom classes, corresponding to different lesion types and a "No finding" category. The model was initialized with pre-trained weights and subsequently fine-tuned on a custom dataset, which follows the COCO format [
10] and includes precise bounding box annotations for each lesion. Images were pre-processed by converting them to RGB and normalized to match the requirements of the pre-trained backbone. During training, stochastic gradient descent was used for optimization, and data augmentation techniques such as random horizontal flipping were applied to improve generalization and data imbalance handling.
4. Evaluation and Results
The model was evaluated on a test set of spinal X-ray images with COCO-format annotations. Each image was preprocessed and passed through the trained Faster R-CNN with ResNet-50 FPN. Predictions included bounding boxes, class labels, and confidence scores.
Quantitative performance for the localization model was measured using mean Average Precision (mAP) at an IoU threshold of 50%, computed with the torchmetrics library. The Classifier demonstrates an area under the receiver operating characteristic curve (AUROC) of 88.84% and an Accuracy of 71.37% for the image classification task. For HealNNet-Lesion, mean average precision (mAP@0.5) of 43.73% for the lesion localization task. These results serve as a PoC (Proof of Concept) for further work in this direction.
5. Accessibility and Affordability of MRI and CT in LMICs
The widespread reliance on Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) as the gold standards for diagnosing a multitude of spinal pathologies. Many findings including but not limited to, Compression fracture [
11], Osteoporosis [
12], Spinal stenosis [
13], Degenerative Disc Disease [
14] etc. may require an MRI or CT scan for diagnosis. The proposed model is designed to detect findings associated with the aforementioned conditions, such as Compression fracture, Osteoporosis, and others. The implementation of such deep learning models trained on a large enough dataset, thus, achieving a good performance may help in diagnosis or initial screening of such findings through X-Rays rather than an MRI or CT scan. Deep learning frameworks can help recognize patterns that are too small to be noticed by physicians. Although, it is X-Rays have their limitation including lack of soft tissue visualization, particularly in critical structures such as discs, ligaments, and the spinal cord [
17]. In Low-and Middle-Income Countries (LMICs) the high capital and operational costs of MRI and CT scans severely limit their availability. Research highlights a substantial deficit in number of MRI machines per capita in these regions, with specific data from India illustrating this disparity where the number of MRI Machines per million people in India was 3, far less than countries like USA and Japan with 55 and 40 machines per million people respectively [
6]. The cost of MRI and CTs are very high, a paper analysing the costs of MRI and CT scans in a hospital in India demonstrates how the patient charge of MRI and CT scans is around 2500 INR and 900 INR respectively [
7], posing a significant economic burden on for an average person in India. This is true for most of the LMICs, the costs for MRIs pose an economical challenge considering how expensive they are [
5].
6. Conclusions
This paper presents HealNNet-Lesions, a deep learning framework that successfully detects spinal lesions from X-ray images, achieving an AUROC of 88.84% and an Accuracy of 71.37% for the classification network and a mAP@0.5 of 43.73% for the detection network. By demonstrating the potential to diagnose complex spinal issues from more accessible X-rays, this work serves as a proof of concept for using Deep Learning to create affordable diagnostic tools. Although, X-Ray images do have their limitations compared to MRI and CTs, more performant deep learning networks for medical diagnosis trained on X-Ray images can allow for diagnosis or initial screening of such findings. The results set a benchmark demonstrating that in future, with a larger and more relevant dataset, more performant deep learning networks can be created for medical diagnosis. This work is particularly significant for improving healthcare accessibility and reducing the financial burden of expensive MRI and CT scans in LMICs.
References
- H. H. Pham, H. N. Trung, and H. Q. Nguyen, VinDr-SpineXR: A large annotated medical image dataset for spinal lesions detection and classification from radiographs, version 1.0.0. PhysioNet, 2021. [CrossRef]
- Goldberger et al., “PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals,” Circulation, vol. 101, no. 23, pp. e215–e220, 2000.
- Shen, D., Wu, G., Suk, H.I.: Deep learning in medical image analysis. Annual Review of Biomedical Engineering. [CrossRef]
- F. Priolo and A. Cerase, “The current role of radiography in the assessment of skeletal tumors and tumor-like lesions,” European Journal of Radiology, vol. 27, no. Suppl 1, pp. S77–S85, 1998. [CrossRef]
- B. S. Hilabi, S. A. Alghamdi, and M. Almanaa, "Impact of Magnetic Resonance Imaging on Healthcare in Low- and Middle-Income Countries," Cureus, vol. 15, no. 4, p. e37698, 2023. [CrossRef]
- Foundation for MSME Clusters (FMC), "Boosting the Indian Medical Devices Industry 2023," Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Government of India, 2023. Table 5.23, p. 102. Available: https://pharma-dept.gov.in/sites/default/files/Final%20Boosting%20of%20Medical%20Devices%20Industry%20-%20Report%20-%202023.pdf.
- K. Rehana, K. A. Malik, M. N. Jan, S. Yousuf, M. Ahmad, and S. A. Wani, "Unit cost of CT scan and MRI at a large tertiary care teaching hospital in North India," Health, vol. 5, no. 12, pp. 2059–2063, Dec. 2013. [CrossRef]
- Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., & Fei-Fei, L. (2009). ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (pp. 248-255).
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (pp. 770-778).
- T. -Y. Lin et al., "Microsoft COCO: Common Objects in Context," in European Conference on Computer Vision (ECCV), Zurich, Switzerland, 2014, pp. 740-755.
- "Compression fractures," Cleveland Clinic, Jun. 02, 2025. [Online]. Available: https://my.clevelandclinic.org/health/diseases/21950-compression-fractures. The article is medically reviewed.
- “Osteoporosis,” Cleveland Clinic, Jun. 02, 2025. Available: https://my.clevelandclinic.org/health/diseases/4443-osteoporosis. The article is medically reviewed.
- “Spinal stenosis,” Cleveland Clinic, Sep. 04, 2025. Available: https://my.clevelandclinic.org/health/diseases/17499-spinal-stenosis. The article is medically reviewed.
- “Degenerative disk Disease,” Cleveland Clinic, Sep. 04, 2025. Available: https://my.clevelandclinic.org/health/diseases/16912-degenerative-disk-disease. The article is medically reviewed.
- “PyTorch,” PyTorch. https://pytorch.org.
- TorchVision: PyTorch's Computer Vision library, Available: https://github.com/pytorch/vision, 2016.
- E. Levent et al., “Review article: Diagnostic Paradigm Shift in Spine Surgery,” Diagnostics, vol. 15, no. 5, p. 594, Feb. 2025. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).