Submitted:
01 October 2025
Posted:
01 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
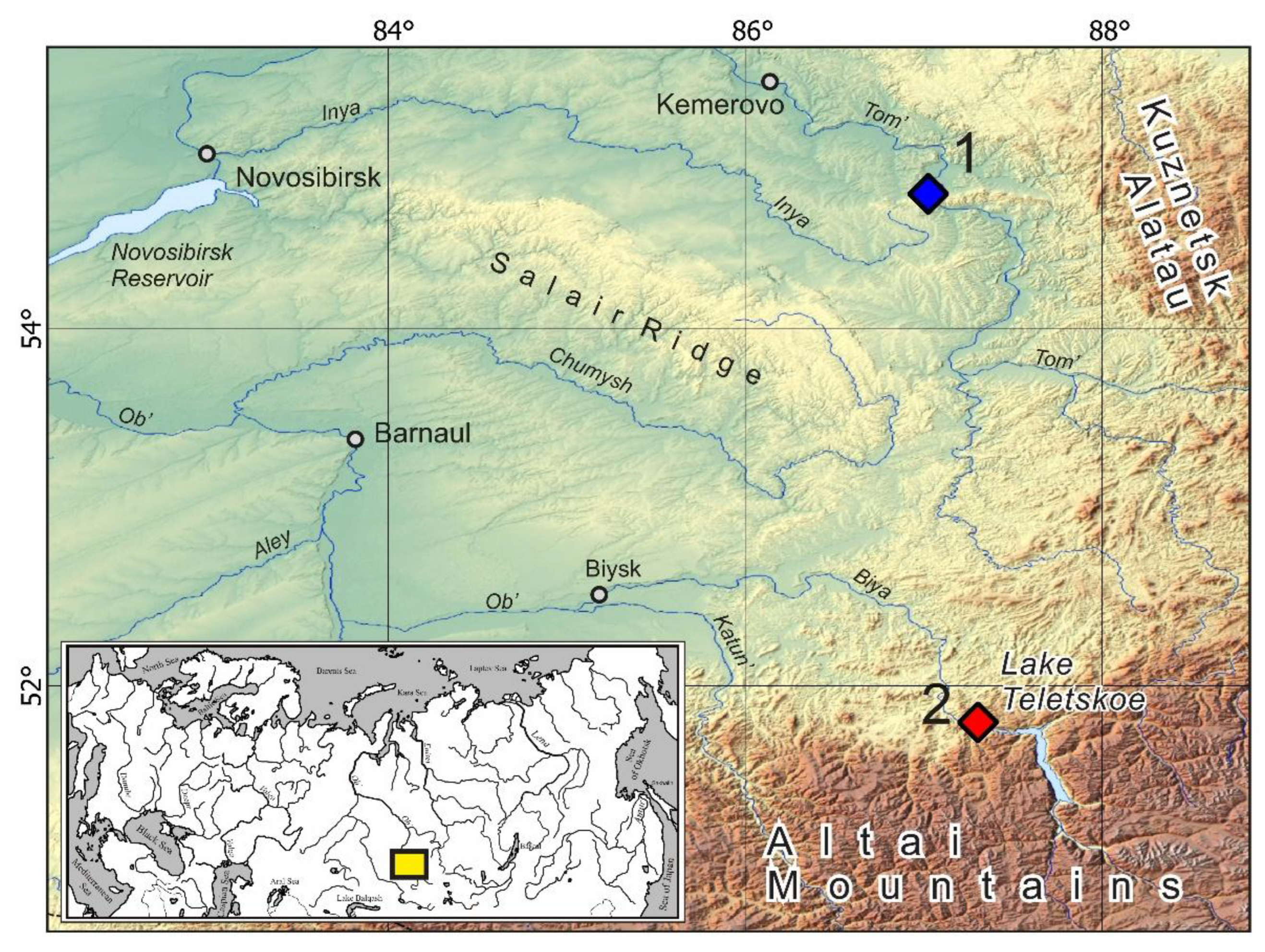
2.1. Trapping and Sample Collection
2.2. RNA Extraction and RT-PCR Analysis
2.3. Genetic and Phylogenetic Analysis
3. Results
3.1. Genetic Analysis
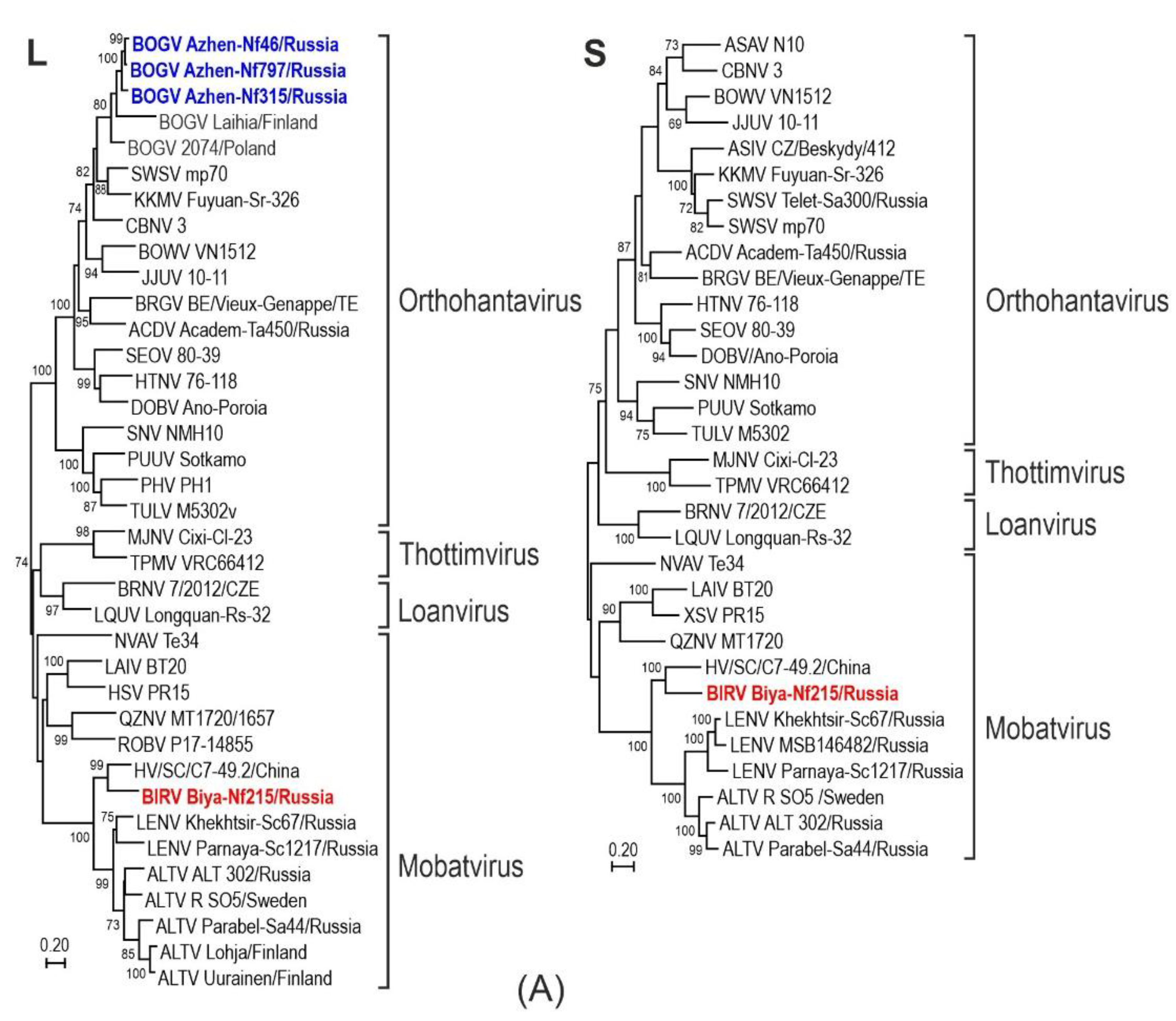
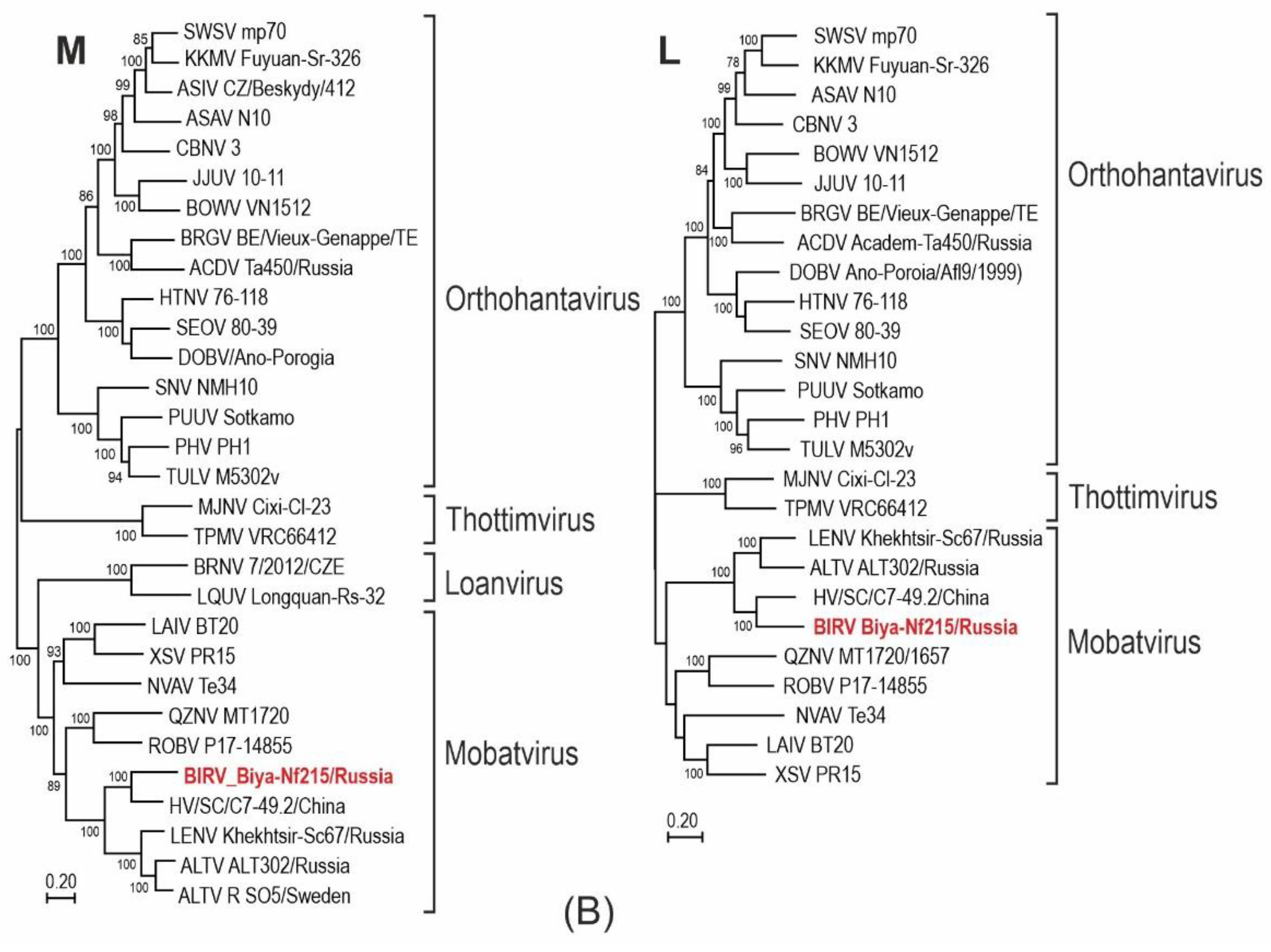
3.2. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Maes, P.; Adkins, S.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, E.; Blair, C.D.; Briese, T.; et al. Taxonomy of the order Bunyavirales: Second update 2018. Arch. Virol. 2019, 164, 927–941. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Figueiredo, L.T.; Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010, 23, 412–444. [Google Scholar] [CrossRef]
- Yanagihara, R.; Gu, S.H.; Arai, S.; Kang, H.J.; Song, J.-W. Hantaviruses: rediscovery and new beginnings. Virus Res. 2014, 187, 6–14. [Google Scholar] [CrossRef]
- Zhang, Y.Z. Discovery of hantaviruses in bats and insectivores and the evolution of the genus Hantavirus. Virus Res. 2014, 187, 15–21. [Google Scholar] [CrossRef]
- Ling, J.; Sironen, T.; Voutilainen, L.; Hepojoki, S.; Niemimaa, J.; Isoviita, V.M.; Vaheri, A.; Henttonen, H.; Vapalahti, O. Hantaviruses in Finnish soricomorphs: Evidence for two distinct hantaviruses carried by Sorex araneus suggesting ancient host-switch. Infect. Genet. Evol. 2014, 27, 51–61. [Google Scholar] [CrossRef]
- Kang, H.J.; Gu, S.H.; Yashina, L.N.; Cook, J.A.; Yanagihara, R. Highly divergent genetic variants of soricid-borne Altai virus (Hantaviridae) in Eurasia suggest ancient host-switching events. Viruses 2019, 11, 857. [Google Scholar] [CrossRef]
- Kang, H.J.; Bennett, S.N.; Dizney, L.; Sumibcay, L.; Arai, S.; Ruedas, L.A.; Song, J.W.; Yanagihara, R. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 2009, 388, 8–14. [Google Scholar] [CrossRef]
- Klempa, B. Reassortment events in the evolution of hantaviruses. Virus Genes 2018, 54, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Liphardt, S.W.; Kang, H.J.; Dizney, L.J.; Ruedas, L.A.; Cook, J.A.; Yanagihara, R. Complex history of codiversification and host switching of a newfound soricid-borne orthohantavirus in North America. Viruses 2019, 11, 637. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, P.; Tia, M.; Alabi, A.; Anon, J.C.; Auste, B.; Essbauer, S.; Gnionsahe, A.; Kigninlman, H.; Klempa, B.; Kraef, C.; et al. Human infections by non-rodent-associated hantaviruses in Africa. J. Infect. Dis. 2016, 214, 1507–1511. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Shimizu, K.; Nishigami, K.; Tsuda, Y.; Sarathukumara, Y.; Muthusinghe, D.S.; Gamage, C.D.; Granathne, L.; Lokupathirage, S.M.W.; Nanayakkara, N.; et al. Serological methods for detection of infection with shrew-borne hantaviruses: Thottapalayam, Seewis, Altai, and Asama viruses. Arch. Virol. 2021, 166, 275–280. [Google Scholar] [CrossRef]
- Song, J.-W.; Gu, S.H.; Bennett, S.N.; Arai, S.; Puorger, M.; Hilbe, M.; Yanagihara, R. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol. J. 2007, 4, 114. [Google Scholar] [CrossRef] [PubMed]
- Yashina, L.; Abramov, S.; Gutorov, V.; Dupal, T.; Krivopalov, A.; Panov, V.; Danchinova, G.; Vinogradov, V.; Luchnikova, E.; Hay, J.; Kang, H.J.; Yanagihara, R. Seewis virus: Phylogeography of a shrew-borne hantavirus in Siberia, Russia. Vector-Borne Zoonotic Dis. 2010, 10, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, M.; Radosa, L.; Rosenfeld, U.M.; Schmidt, S.; Triebenbacher, C.; Löhr, P.W.; Fuchs, D.; Heroldová, M.; Jánová, E.; Stanko, M.; Mošanský, L.; Fričová, J.; Pejčoch, M.; Suchomel, J.; Purchart, L.; Groschup, M.H.; Krüger, D.H.; Klempa, B.; Ulrich, R.G. Broad geographical distribution and high genetic diversity of shrew-borne Seewis hantavirus in Central Europe. Virus Genes 2012, 45, 48–55. [Google Scholar] [CrossRef]
- Yashina, L.N.; Abramov, S.A.; Zhigalin, A.V.; Smetannikova, N.A.; Dupal, T.A.; Krivopalov, A.V.; Kikuchi, F.; Senoo, K.; Arai, S.; Mizutani, T.; et al. Geographic distribution and phylogeny of soricine shrew-borne Seewis virus and Altai virus in Russia. Viruses 2021, 13, 1286. [Google Scholar] [CrossRef]
- Kang, H.J.; Bennett, S.N.; Sumibcay, L.; Arai, S.; Hope, A.G.; Mocz, G.; Song, J.-W.; Cook, J.A.; Yanagihara, R. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS ONE 2009, 4, e6149. [Google Scholar] [CrossRef]
- Laenen, L.; Vergote, V.; Kafetzopoulou, L.E.; Wawina, T.B.; Vassou, D.; Cook, J.A.; Hugot. J.P.; Deboutte, W.; Kang, H.J.; Witkowski, P.T.; Köppen-Rung, P.; et al. A novel hantavirus of the European mole, Bruges virus, is involved in frequent Nova virus coinfections. Genome Biol. Evol. 2018, 10, 45–55. [Google Scholar] [CrossRef]
- Gu, S.H.; Miñarro, M.; Feliu, C.; Hugot, J.-P.; Forrester, N.L.; Weaver, S.C.; Yanagihara, R. Multiple lineages of hantaviruses harbored by the Iberian mole (Talpa occidentalis) in Spain. Viruses 2023, 15, 1313. [Google Scholar] [CrossRef]
- Yashina, L.N.; Kartashov, M.Y.; Wang, W.; Li, K.; Zdanovskaya, N.I.; Ivanov, L.I.; Zhang, Y.Z. Co-circulation of distinct shrewborne hantaviruses in the far east of Russia. Virus Res. 2019, 272, 197717. [Google Scholar] [CrossRef]
- Kang, H.J.; Arai, S.; Hope, A.G.; Cook, J.A.; Yanagihara, R. Novel hantavirus in the flat-skulled shrew (Sorex roboratus). Vector Borne Zoonotic Dis. 2010, 10, 593–597. [Google Scholar] [CrossRef]
- Arai, S.; Kang, H.J.; Gu, S.H.; Ohdachi, S.D.; Cook, J.A.; Yashina, L.N.; Tanaka-Taya, K.; Abramov, S.A.; Morikawa, S.; Okabe, N.; et al. Genetic diversity of Artybash virus in the Laxmann’s shrew (Sorex caecutiens). Vector Borne Zoonotic Dis. 2016, 16, 468–475. [Google Scholar] [CrossRef]
- Yashina, L.N.; Ivanov, L.I.; Kompanets, G.G.; Zdanovskaya, N.I.; Kartashov, M.Yu. Shrew-borne hantaviruses (Hantaviridae: Orthohantavirus) in the Far East of Russia. Problems of Virology (Voprosy Virusologii). 2023, 68, 79–85. [Google Scholar] [CrossRef]
- Yashina, L.N.; Zinich, L.S.; Smetannikova, N.A.; Kartashov, M.; Kovalenko, I.S.; Yusupova, Z.S.; Konashenko, E.V.; Tikhonov, S.N. Hantaviruses (Hantaviridae) in Republic of Crimea. Mol. Genet. Microbiol. Virol. (In Russ.). 2024, 42, 37–42. [Google Scholar] [CrossRef]
- Gu, S.H.; Markowski, J.; Kang, H.J.; Hejduk, J.; Sikorska, B.; Liberski, P.P.; Yanagihara, R. Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol. J. 2013, 10, 160. [Google Scholar] [CrossRef]
- Tkachenko, E.A.; Ivanov, A.P.; Donets, M.A.; Miasnikov, Y.A.; Ryltseva, E.V.; Gaponova, L.K.; Bashkirtsev,V. N.; Okulova, N.M.; Drozdov, S.G.; Slonova, R.A.; et al. Potential reservoir and vectors of haemorrhagic fever with renal syndrome (HFRS) in the U.S.S.R. Ann. Soc. Belg. Med. Trop. 1983, 63, 267–269. [Google Scholar] [PubMed]
- Klempa, B.; Fichet-Calvet, E.; Lecompte, E.; Auste, B.; Aniskin, V.; Meisel, H.; Barrier, P.; Koivogue, L.; Meulen, J.; Krüger, D.H. Novel hantavirus sequences in shrew, Guinea. Emerg. Infect. Dis. 2007, 13, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013, 16, 1303. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Cui, X.; Fan, K.; Liang, X.; Gong, W.; Chen, W.; He, B.; Chen, X.; Wang, H.; Wang, X.; Zhang, P.; et al. Virus diversity, wildlife-domestic animal circulation and potential zoonotic viruses of small mammals, pangolins and zoo animals. Nat. Commun. 2023, 14, 2488. [Google Scholar] [CrossRef]
- Yashina, L.N.; Panov, V.V.; Abramov, S.A.; Smetannikova, N.A.; Luchnikova, E.M.; Dupal, T.A.; Krivopalov, A.V.; Arai, S.; Yanagihara, R. Academ virus, a novel hantavirus in the Siberian mole (Talpa altaica) from Russia. Viruses 2022, 14, 309. [Google Scholar] [CrossRef]
- Pavelková Řičánková, V.; Robovský, J.; Riegert, J. Ecological structure of recent and last glacial mammalian faunas in northern Eurasia: The case of Altai-Sayan Refugium. PLoS ONE 2014, 9, e85056. [Google Scholar] [CrossRef] [PubMed]
- Provan, J.; Bennett, K.D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2006, 23, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Marková, S.; Horníková, M.; Lanier, H.C.; Henttonen, H.; Searle, J.B.; Weider, L.J.; Kotlík, P. High genomic diversity in the bank vole at the northern apex of a range expansion: The role of multiple colonizations and end-glacial refugia. Mol. Ecol. 2020, 29, 1730–1744. [Google Scholar] [CrossRef] [PubMed]
- Shchipanov, N.A. , Litvinov, Y.N.; Sheftel, B.I. Rapid method for estimating local biodiversity of a community of small mammals. Contemp. Probl. Ecol. 2008, 1, 596–602. [Google Scholar] [CrossRef]
- Ilyashenko, V.B.; Luchnikova, E.M.; Skalon, N.S.; Grebentschikov, I.S.; Kovalevsky, A.V. Long-term dynamics of small-mammal communities in anthropogenically disturbed territories in the south-east of West Siberia. IOP Conf. Ser.: Earth Environ. Sci. 2019, 224, 012055. [Google Scholar] [CrossRef]
- Bannikova, A.A.; Lebedev, V.S. Order Eulipotyphla. In The Mammals of Russia: A taxonomic and geographic reference; Pavlinov, I.Ya., Lissovsky, A.A., Eds.; KMK Sci. Press: Moscow, Russia, 2012; pp. 25–73. [Google Scholar]
| Positive/tested | GenBank no. | |||||
| Capture site | Year | shrews | Virus strain | S | M | L |
| Altai Republic, Teletskoye | 2019 | 1/2 | BIRV_Biya-Nf215 | PQ355537 | PQ355538 | PQ355539 |
| Kemerovo Oblast, Azhendarovo | 2021 | 1/13 | BOGV_Azhen-Nf315 | - | - | PQ355534 |
| 2022 | 2/17 | BOGV_Azhen-Nf46 | - | - | PQ355535 | |
| BOGV_Azhen-Nf797 | - | - | PQ355536 | |||
| 2023 | 0/1 | - | - | - | - | |
| 2024 | 0/6 | - | - | - | - | |
| 2025 | 0/25 | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





