1. Problem Statement
The skin cancer occurrence, melanoma and non-melanoma, has increased over the last decades. Currently, the World Health Organization (WHO) [
1] estimates that 2-3 million non-melanoma cancers and 132,000 melanomas occur every year in the world. According to the Brazilian Cancer National Institute (INCA) [
2], one in every three cancer diagnosis is a skin cancer. So, approaches to automatically detect/recognize skin cancer are very desired by means of Computer Aided Diagnosis (CAD). In the late 2017, we started a partnership with the Dermatological Assistance Program (PAD), hosted at the hospital of the Federal University of Espirito Santo (UFES). From a smartphone application (App), was possible to obtaian skin lesion images using the device’s camera and patient lesion information. The PAD-UFES-20 dataset [
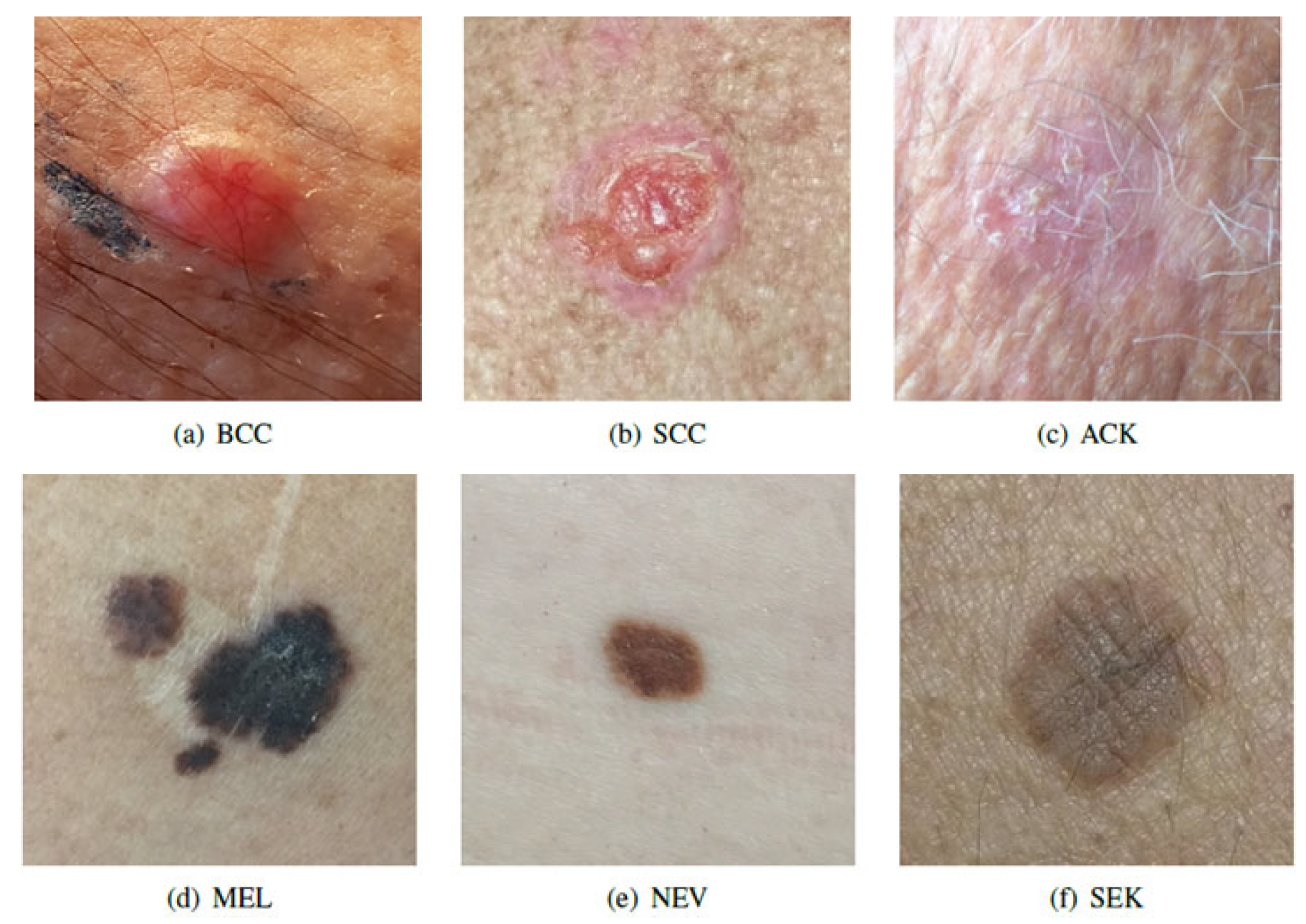
3], is one of the first dataset made up of clinical images and patient information available in the world. It is composed of the six most common skin diseases: 1)
cancer: Melanoma (MEL), Basal Cell Carcinoma (BCC), and Squamous Cell Carcinoma (SCC); and 2)
non-cancer: Nevus (NEV), Actinic Keratosis (ACK) and Seborrheic Keratosis (SEK) as shown in
Figure 1.
With those information, it has been developed a methodology based on deep learning [
4] to fuse such information for skin cancer of the main skin lesions with circa 7% improvement [
5,
7], and a smartphone-based application to automatically recognize melanoma [
6]. Nonetheless, it was not possible to correctly discriminate all kinds of skin lesions yet, specially between MEL and NEV and between BCC and SCC. Our findings indicate that is not possible to solve this problem with only these two kinds of information. So, looking for an alternative way to solve this problem there are 2 possibilities: 1)
a data centric approach, and 2)
an algorithmic approach. Based on our previous experience and lessons learned, it is not possible to solve this problem only looking at the state of the art in deep learning algorithms. We focus on a data-centric approach inspired by multi and cross-modal learning [
8], i.e., we propose to use the information from the histopathological image to the automated diagnostic. Some kinds of skin lesions like NEV and MEL or BCC and SCC, which are the most difficult ones, are not possible to differentiate with only clinical image and patient lesion information. In this direction, we propose a non-invasive flexible approach to allow a fast and most reliable diagnosis of the main skin cancer using the information provided by the histopathological image.
2. Proposed Methodology
Dermatologists do not trust only on visual aspect of the lesion (image) and anamnese; in case of doubts they may ask the patient to a biopsy examination in order to certify their diagnostic. So, it is necessary to look for additional source of information beyond the visual image in order to improve the classification metrics (accuracy, precision, and recall) in CAD systems. A valuable information is the gold-standard, i.e. the histopathological result of the biopsy. The main issue is how to use this information in CAD systems to operate online?. The skin lesions image of the PAD-UFES-20 dataset are labeled by experts during the clinical diagnosis and confirmed by biopsy (gold-standard).
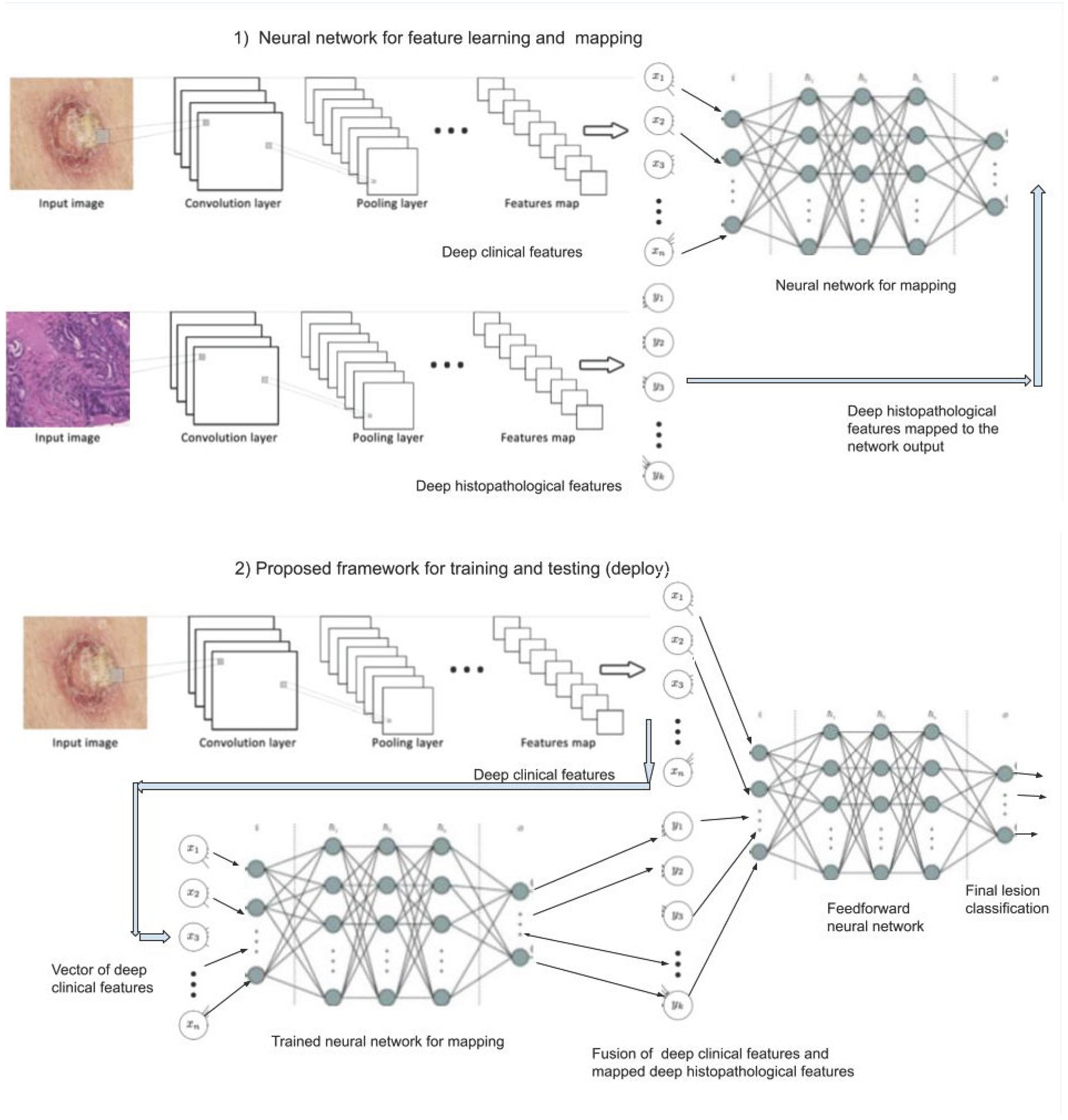
In order to develop our novel approach to automatically recognize the main skin lesions, we have been exploring convolutional neural networks (CNN) to extract features of images [
4]. We propose a novel framework as illustrated in
Figure 2, composed of two stages: 1) in the first one, we train a neural network for feature learning and mapping and 2) in the second stage we train the entire framework. Firstly, the features of the clinical and histopathological images are extracted by two CNNs, respectively. Both feature maps are flattened and then used to train a neural network for mapping features from the clinical to histopathological domain. Next, with only the clinical images available, the clinical features are extracted from the clinical image by the same CNN, and the histopathological features are obtained by the previous trained neural network for feature mapping. Both features are combined via a fusion algorithm for instance based on concatenation [
5] or based on attention mechanism [
7]. Next, the fused features are inputted to a feedforward neural network providing the final automated classification diagnosis. All training of the neural networks is end-to-end using the backpropagation algorithm. To the best of our knowledge, there is no such an approach yet in the literature using this kind of mapping (virtual biopsy) to improve CAD.
3. Concluding Remarks
PAD-UFES-20 dataset is a reliable patient data and clinical image database of skin cancer with skin lesions image obtained via smartphone and assessed by experts providing the gold-standard of biopsy, i.e., this dataset is certified by pathologists, making possible to use the clinical image and the corresponding histopathological image. While works in the literature are using only clinical images for classification of skin lesions using deep learning, this approach offer an alternative approach to get a more reliable diagnosis of skin cancer. The main limitation is the public availability of reliable open datasets in this topic involving clinical and histopathological images. This approach is obviously applicable to other kind of cancer, where the multimodal images are available, i.e. the mapping from one domain to another.
Acknowledgment
The author thanks the Brazilian research agency Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil – grant no. 304688/2021-5.
References
- WHO - World Health Organization. (2019) How common is the skin cancer? [Online]. Available: https://www.who.int/uv/faq/skincancer/en/index1.html.
- INCA - Instituo Nacional do Cancer José de Alencar. (2019) The cancer incidence in Brazil. [Online]. Available: http://www1.inca.gov.br/estimativa/2018/estimativa-2018.pdf.
- Pacheco, A.G.C. et al. (2020). PAD-UFES-20: a skin lesion benchmark composed of patient data and clinical images collected from smartphones, June 2020, https://data.mendeley.com/datasets/zr7vgbcyr2/1, available at https://arxiv.org/pdf/2007.00478.pdf.
- LeCun, Y., Bengio, Y. and Hinton, G. (2015). Deep learning. Nature 521, p. 436–444. [CrossRef]
- Pacheco, A.G.C., Krohling, R.A. (2020). The impact of patient clinical information on automated skin cancer detection. Computers in Biology and Medicine. Vol. 116, 103545,2020. [CrossRef]
- Castro, P.B.C., Krohling, B.A., Pacheco, A.G.C., Krohling, R.A. An App to detect melanoma using deep learning: An approach to handle imbalanced data Based on evolutionary algorithms. In: International Joint Conference on Neural Networks, IJCNN 2020 (virtual conference), Glasgow, July, 2020.
- Pacheco, A.G.C., Krohling, R.A. (2021) An attention-based mechanism to combine images and metadata in deep learning models applied to skin cancer classification, IEEE Journal of Biomedical and Health Informatics, 1-10, 2021. [CrossRef]
- Beltrán Beltrán, L. V. et al. Deep multimodal learning for cross-modal retrieval: One model for all tasks, Pattern Recognition Letters, vol. 146, pp. 38-45, June 2021. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).