Submitted:
12 September 2025
Posted:
17 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
3. Discussion and Conclusions
4. Materials and Methods
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Holtedahl, K.; Borgquist, L.; Donker, G.A.; Buntinx, F.; et al. Symptoms and signs of urogenital cancer in primary care. BMC Primary Care, 2023, 24, 107. [Google Scholar] [CrossRef]
- Bray, F. ; Ferlay, J; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin, 2018; 68, 394–424. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10(2), 63–89. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.; Poole, E.M. Ovarian Cancer Prevention, Screening, and Early Detection: Report From the 11th Biennial Ovarian Cancer Research Symposium. Int. J. Gynecol. Cancer 2017, 27 (Suppl.5), S20–S22. [Google Scholar] [CrossRef]
- Sung, H. , et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians, 2021; 71, 209–249. [Google Scholar] [CrossRef]
- Maek, M. Penis-preserving surgery in patients with primary penile urethral cancer (Ger). Urologe A 2014, 53, 1800–1804. [Google Scholar] [CrossRef]
- Funston, G.; O’Flynn, H.; Ryan, N.A.J.; Hamilton, W. , Crosbie, E.J. Recognizing Gynecological Cancer in Primary Care: Risk Factors, Red Flags, and Referrals. Adv. Ther. 2018; 35, 577–589. [Google Scholar] [CrossRef]
- Rissin, D. , et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. , et al. Translational Bioinformatics for Diagnostic and Prognostic Prediction of Prostate Cancer in the Next-Generation Sequencing Era. BioMed Res. Int. 2013; 2013, 901578. [Google Scholar] [CrossRef]
- Lumbreras, B. , et al. Variables Associated with False-Positive PSA Results: A Cohort Study with Real-World Data. Cancers 2023, 15, 261. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef]
- Zhu, C.-Z.; Ting, H.-N.; Ng, K.-H.; Ong, T.-A. A Review on the Accuracy of Bladder Cancer Detection Methods. J. Cancer 2019, 10(17), 4038–4044. [Google Scholar] [CrossRef]
- Yoo, J. W.; Koo, K. C.; Chung, B. H.; Baek, S. Y.; Lee, S. J.; Park, K. H.; Lee, K. S. Deep Learning Diagnostics for Bladder Tumor Identification and Grade Prediction Using RGB Method. Sci. Rep. 2022, 12(1), 17699. [Google Scholar] [CrossRef]
- Chang, L.; Ni, J.; Zhu, Y.; Pang, B.; Graham, P.; Zhang, H.; Li, Y. Liquid Biopsy in Ovarian Cancer: Recent Advances in Circulating Extracellular Vesicle Detection for Early Diagnosis and Monitoring Progression. Theranostics 2019, 9(14), 4130–4140. [Google Scholar] [CrossRef]
- Hafeez, S.; Huddart, R. Advances in Bladder Cancer Imaging. BMC Med. 2013, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Trau, M.; Wuethrich, A. Digital platforms enabling single-molecule analysis for cancer detection. Trends Anal. Chem. 2024, 171, 117502. [Google Scholar] [CrossRef]
- Waseem, M.; Ahmad, M.K.; Serajuddin, M.; Bhaskar, V.; Sankhwar, S.N.; Mahdi, A.A. MicroRNA-183-5p: A New Potential Marker for Prostate Cancer. Ind. J. Clin. Biochem. 2019, 34, 207–212. [Google Scholar] [CrossRef]
- Xu, Y.-Z.; Xi, Q.-H.; Ge, W.-L.; Zhang, X.-Q. Identification of Serum MicroRNA-21 as a Biomarker for Early Detection and Prognosis in Human Epithelial Ovarian Cancer. Asian Pac. J. Cancer Prev. 2013, 14, 1057–1060. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Zhang, Y.; Zheng, S.F.; Feng, T.; Tian, X.; Abudurexiti, M.; Wang, Z.D.; Zhu, W.K.; Su, J.Q.; Zhang, H.L.; Shi, G.H.; Wang, Z.L.; Cao, D.L. , Ye, D.W. The function and mechanisms of action of circular RNAs in Urologic Cancer. Mol. Cancer 2023, 22, 61. [Google Scholar] [CrossRef]
- Wang, H. , Feng, Y., Zheng, X., Xu, X. The Diagnostic and Therapeutic Role of snoRNA and lincRNA in Bladder Cancer. Cancers (Basel) 2023, 15(4), 1007. [Google Scholar] [CrossRef]
- Crea, F.; Quagliata, L.; Michael, A.; Liu, H. H.; Frumento, P.; Azad, A. A.; Xue, H.; Pikor, L.; Watahiki, A.; Morant, R.; Eppenberger-Castori, S.; Wang, Y.; Parolia, A.; Lennox, K. A.; Lam, W. L.; Gleave, M.; Chi, K. N.; Pandha, H.; Wang, Y.; Helgason, C. D. Integrated Analysis of the Prostate Cancer Small-Nucleolar Transcriptome Reveals SNORA55 as a Driver of Prostate Cancer Progression. Mol. Oncol. 2016, 10(5), 693–703. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D. H.; Park, P. K.; Diao, L.; Putkey, J. A.; Yang, L.; Guo, A.-Y.; Lin, C.; Han, L. A Pan-Cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21(7), 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, P. , Ladame, S., O’Hare, D. Molecular Methods in Electrochemical MicroRNA Detection. Analyst 2019, 144(1), 114–129. [Google Scholar] [CrossRef]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research Advances in the Detection of MiRNA. J. Pharm. Analysis 2019, 9(4), 217–226. [Google Scholar] [CrossRef] [PubMed]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10(11), 186. [Google Scholar] [CrossRef]
- Yang, T. , Zhang M., Zhang N. Modified Northern blot protocol for easy detection of mRNAs in total RNA using radiolabeled probes. BMC Genomics 2022, 23(1), 66. [Google Scholar] [CrossRef]
- Crampton, N. , Bonass, W.A., Kirkham, J., Thomson, N.H. Formation of aminosilane-functionalized mica for atomic force microscopy imaging of DNA. Langmuir, 7884. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Smelov, M.V.; Ivanova, N.D.; Ziborov, V.S. AFM imaging of protein aggregation in studying the impact of knotted electromagnetic field on a peroxidase. Sci. Rep. 2020, 10, 9022. [Google Scholar] [CrossRef] [PubMed]
- Stern, E.; Klemic, J.; Routenberg, D.; Wyrembak, P.; Turner-Evans, D.; Hamilton, A.; LaVan, D.; Fahmy, T.; Reed, M.A. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature 2007, 445, 519–522. [Google Scholar] [CrossRef]
- Stern, E.; Vlacic, A.; Rajan, N,K. ; Criscione, J.M.; Park, J.; Ilic, B.R.; Mooney, D.J.; Reed, M.A.; Fahmy, T.M. Label-free biomarker detection from whole blood. Nature Nanotech. 2010, 5, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Elfström, N.; Juhasz, R.; Sychugov, I.; Engfeldt, T.; Eriksson Karlström, A.; Linnros, J. Surface Charge Sensitivity of Silicon Nanowires: Size Dependence. Nano Lett. 2007, 7, 2608–2612. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Pleshakova, T.O.; Kozlov, A.F.; Malsagova, K.A.; Krohin, N.V.; Shumyantseva, V.V.; Shumov, I.D.; Popov, V.P.; Naumova, O.V.; Fomin, B.I.; et al. SOI nanowire for the high-sensitive detection of HBsAg and α-fetoprotein. Lab Chip. 2012, 12, 5104–5111. [Google Scholar] [CrossRef]
- Ivanov, Y.; et al. Detection of Marker miRNAs, Associated with Prostate Cancer, in Plasma Using SOI-NW Biosensor in Direct and Inversion Modes. Sensors 2019, 19, 5248. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Malsagova, K.A.; Goldaeva, K.V.; Kapustina, S.I.; Pleshakova, T.O.; Popov, V.P.; Kozlov, A.F.; Galiullin, R.A.; Shumov, I.D.; Enikeev, D.V.; et al. Nanoribbon Biosensor-Based Detection of microRNA Markers of Prostate Cancer. Sensors 2023, 23, 7527. [Google Scholar] [CrossRef]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical detection of single viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 14017–14022. [Google Scholar] [CrossRef]
- Rajan, N.K.; Duan, X.; Reed, M.A. Performance limitations for nanowire/nanoribbon biosensors. WIREs Nanomed Nanobiotechnol 2013. [Google Scholar] [CrossRef] [PubMed]
- Miao, L. , Liu, H. Y., Zhou, C., He, X. LINC00612 Enhances the Proliferation and Invasion Ability of Bladder Cancer Cells as CeRNA by Sponging MiR-590 to Elevate Expression of PHF14. J. Exp. Clin. Cancer Res, 2019; 38, 143. [Google Scholar] [CrossRef]
- Jin, L.; et al. Identification of MiR-195-3p as an Oncogene in RCC. Mol. Medicine Rep. 2017, 15(4), 1916–1924. [Google Scholar] [CrossRef] [PubMed]
- Larne, O.; et al. miR-183 in Prostate Cancer Cells Positively Regulates Synthesis and Serum Levels of Prostate-specific Antigen. Eur. Urol. 2015, 68(4), 581–588. [Google Scholar] [CrossRef]
- Chen, S.-N.; et al. MicroRNA in Ovarian Cancer: Biology, Pathogenesis, and Therapeutic Opportunities. Int. J. Environ. Res. Public Health 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Salomo, K.; Huebner, D.; Boehme, M.U.; Herr, A.; Brabetz, W.; Heberling, U.; Hakenberg, O.W.; Jahn, D.; Grimm, M.-O.; Steinbach, D.; Horstmann, M.; Froehner, M.; Wirth, M. P.; Fuessel, S. Urinary Transcript Quantitation of CK20 and IGF2 for the Non-Invasive Bladder Cancer Detection. J. Cancer Res. Clin. Oncol. 2017, 143(9), 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, Y.; Jia, L.; Liu, C.; Xu, F. Circular RNA ABCB10 Promotes Tumor Progression and Correlates with Pejorative Prognosis in Clear Cell Renal Cell Carcinoma. Int. J. Biol. Markers 2019, 34(2), 176–183. [Google Scholar] [CrossRef]
- Xia, Q.; et al. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific circRNAs in Prostate Cancer. Cell. Physiol. Biochem. 2018, 50(5), 1903–1915. [Google Scholar] [CrossRef]
- Gan, X. , et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer, 2020; 19, 1–13. [Google Scholar] [CrossRef]
- Chow, R. D.; Chen, S. Sno-Derived RNAs Are Prevalent Molecular Markers of Cancer Immunity. Oncogene 2018, 37(50), 6442–6462. [Google Scholar] [CrossRef]
- Hirasawa, Y.; Pagano, I.; Chen, R.; Sun, Y.; Dai, Y.; Gupta, A.; Tikhonenkov, S.; Goodison, S.; Rosser, C.J.; Furuya, H. Diagnostic performance of Oncuria™, a urinalysis test for bladder cancer. J. Transl. Med. 2021, 19, 141. [Google Scholar] [CrossRef]
- Fischerova, D.; Burgetova, A. Imaging techniques for the evaluation of ovarian cancer. Best Practice & Research Clinical Obstetrics & Gynaecology, 2014; 28, 697–720. [Google Scholar] [CrossRef]
- Dewey, M.; Schink, T.; Dewey, C. F. Claustrophobia during Magnetic Resonance Imaging: Cohort Study in over 55,000 Patients. J. Magn. Reson. Imaging 2007, 26(5), 1322–1327. [Google Scholar] [CrossRef]
- Galvão-Lima, L.J.; et al. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. BioMed Eng. OnLine 2021, 20, 21. [Google Scholar] [CrossRef]
- Kosaka N., Yoshioka Y., Fujita Y., Ochiya T. Versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 126(4), 1163-1172. [CrossRef]
- Ma, S. , Kong, S., Wang, F., Ju, S. CircRNAs: biogenesis, functions, and role in drug-resistant Tumours. Mol. Cancer 2020, 19, 119. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, L.; Wu, P.; Wu, Y.; Zhang, T.; Zhang, D.; Tian, J. The Potential Role of Small Nucleolar RNAs in Cancers – An Evidence Map. Int. J. General Medicine, 2022; 2022, 3851–3864. [Google Scholar] [CrossRef]
- Wajahat, M. , Bracken, C. P., Orang, A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yoshii, S.; Kumagai, S.; Fujiwara, I.; Nishio, K.; Okuda, M.; Matsukawa, N.; Yamashita, I. High-Density and Highly Surface Selective Adsorption of Protein–Nanoparticle Complexes by Controlling Electrostatic Interaction. Jpn. J. Appl. Phys. 2006, 45, 4259–4264. [Google Scholar] [CrossRef]
- Stern, E. E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [PubMed]
- Laborde, C.; Pittino, F.; Verhoeven, H.A.; Lemay, S.G.; Selmi, L.; Jongsma, M.A.; Widdershoven, F.P. Real-Time Imaging of Microparticles and Living Cells with CMOS Nanocapacitor Arrays. Nature Nanotech. 2015, 10, 791–795. [Google Scholar] [CrossRef]
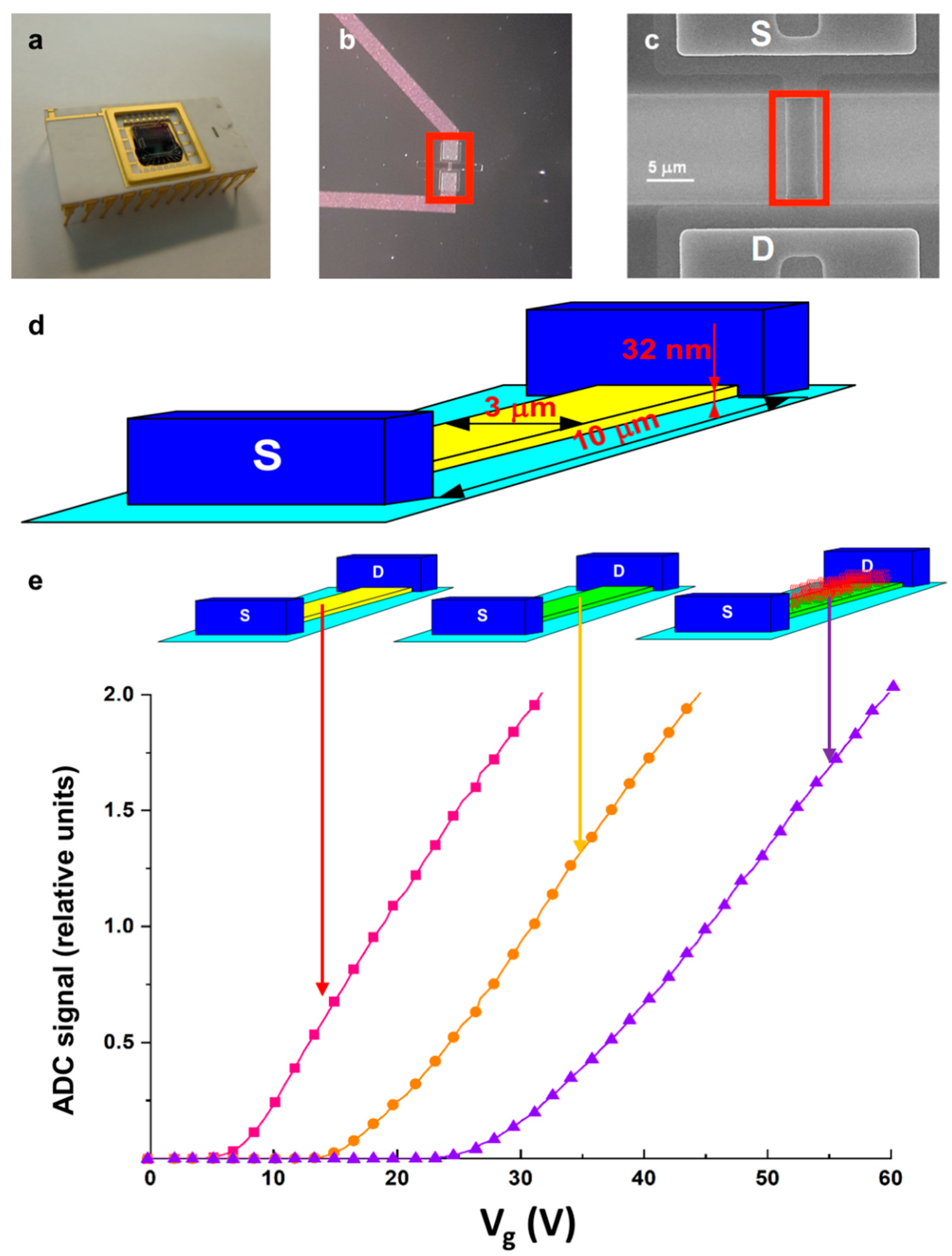
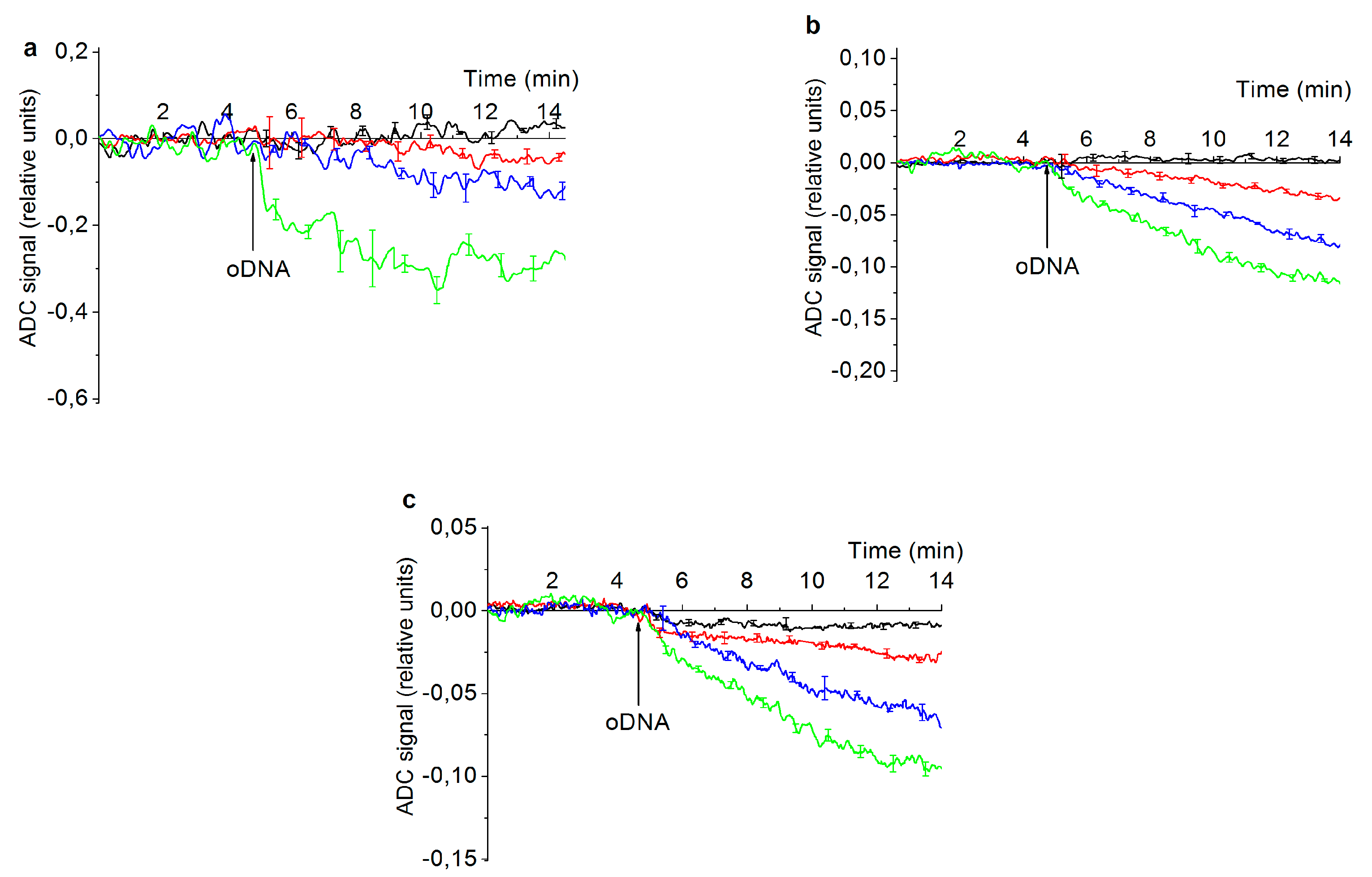
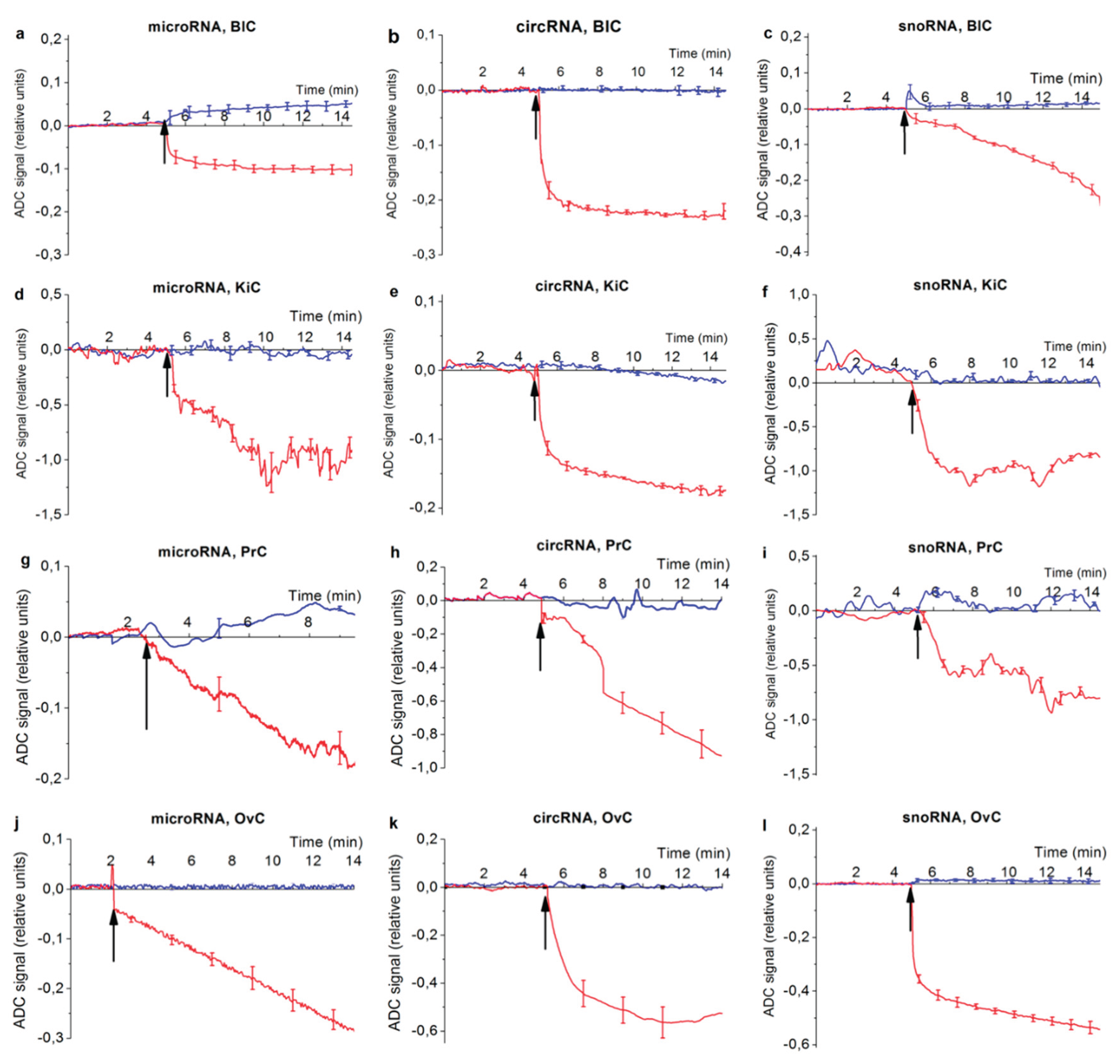
| Pathology | ncRNA type | ncRNA name | Ref. |
| BlC | microRNA | miRNA-590-3p | [37] |
| circRNA | circRNA_0020792 | [41] | |
| snoRNA | snoRNA _SNORD58С | [22] | |
| KiC | microRNA | miRNA-195-3p | [38] |
| circRNA | circRNA_0016825 | [42] | |
| snoRNA | snoRNА_SNORА77 | [45] | |
| PrC | microRNA | hsa-mir-198 | [39] |
| circRNA | circRNA 57558 | [43] | |
| snoRNA | SNORA55 | [21] | |
| OvC | microRNA | hsa-mir-21 | [40] |
| circRNA | hsa_circ_0049116 | [44] | |
| snoRNA | snoRNA _SNORD58С | [22] |
| ncRNA name | oDNA analogue sequence | Nanoribbon-immobilised oDNA probe sequence |
| miRNA-590-3p | TAATTTTATGTATAAGCTAGT | (NH2)-TTTTTTTTTT ACTAGCTTATACATAAAATTA |
| miRNA-195-3p | CCAATATTGGCTGTGCTGCTCC | 5’- (NH2)-(T)10-GGAGCAGCACAGCCAATATTGG |
| hsa-mir-198 | CCGCAGAGTGTGACTCCTGTTCT GTGTATGG CACTGGTAGAATTCACTGTGAAC AGTCTCAG TCAGTGAATTACCGAAGGGCCAT AAACAGAGCAGAGACAGATCCACGA | (NH2)-(T)10-TCGTGGATCTGTCTCTGCTCTGTTTA TGGCCCTTCGGTAATTCACTGACTGAGA CTGTTCACAGTGATTTCTACCAGTGCCA TACACAGAACAGGAGTCACACTCTGCGG |
| hsa-mir-21 | TGTCGGGTAGCTTATCAGACTG ATGTTGACTGTTGAATCTCATGGCAA CACCAGTCGATGGGCTGTCTGACA | 5’- (NH2)-(T)10-TGTCAGACAGCCCATCGACTGGT GTTGCCATGAGATTCAACAGT CAACATCAGTCTGATAAGCTACCCG |
| circRNA_0020792 | AGCCTCCTGGGGGGCACT GGCCACTGAGCCCCCTTGGA GAAGTCAGAGGG | (NH2)-(T)10-CCCTCTGACTTCTCCAA GGGGGCTCAGTGGCCAGTGCCCCCCA GGAGGCT |
| circ_0016825 | TATTTATAGTCTCAAAA TTCCTAAAGCAATGCTACAACCA TTGAATTTGC | (NH2)-(T)10-GCAAATTCAATGGTTG TAGCATTGCTTTAGGAATTTTGAGA CTATAAATA |
| circRNA 57558 | TCACTGCAGGCATGT | (NH2)-(T)10-ACATGCCTGCAGTGA |
| hsa_circ_0049116 | TGGAGTGGATGCCAT | (NH2)-(T)10-ATGGCATCCACTCCA |
| SNORA55 | GCAGAGGAAATCCAG | (NH2)-(T)10-CTGGATTTCCTCTGC |
| snoRNA _SNORD58С | TTAGGACACCTTTGG | (NH2)-(T)10-CCAAAGGTGTCCTAA |
| snorRNА_SNORА77 | TCCAGGGTGCTGTGG | (NH2)-(T)10-CCACAGCACCCTGGA |
| snoRNA _SNORD58С | TTAGGACACCTTTGG | (NH2)-T10-CCAAAGGTGTCCTAA |
| Plasma Sample No. | Diagnosis |
| #1 | Bladder cancer (T3N0M0) |
| #2 | Right kidney stone (T0N0M0, control sample) |
| #33 | Bladder cancer (T3N0M0) |
| #3c | Left kidney cyst (T0N0M0, control sample) |
| #4 | Renal cell carcinoma (T1aN0M0) |
| #3 | Urolithiasis (T0N0M0, control sample) |
| #8pr | Prostate cancer (T2N0M0) |
| #38 | Left kidney cyst (T0N0M0, control sample) |
| #6ov | Ovarian cancer (T2N0M0) |
| #10 | Urolithiasis (T0N0M0, control sample) |
| #44 | Prostate cancer (T1cN0M0) |
| #8 | Ovarian cancer (T2N0M0) |
| #7 | Urethritis (T0N0M0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





