Submitted:
11 September 2025
Posted:
12 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Downloading Reference Genomes
2.2. Cas Proteins Detection
2.3. Analysis of the Biological Function of Cas Proteins
2.4. Classification of Cas Proteins into Functional Stages
2.5. Conservation Patterns of Direct Repeats in the Leptospira Genus
2.6. Bioinformatic Identification of the Immunological Memory (Spacer)
2.7. Identification of Unique Effector Proteins
2.8. Bioinformatic Detection of Intact Bacteriophages in Genomes
3. Results
3.1. Downloading Reference Genomes
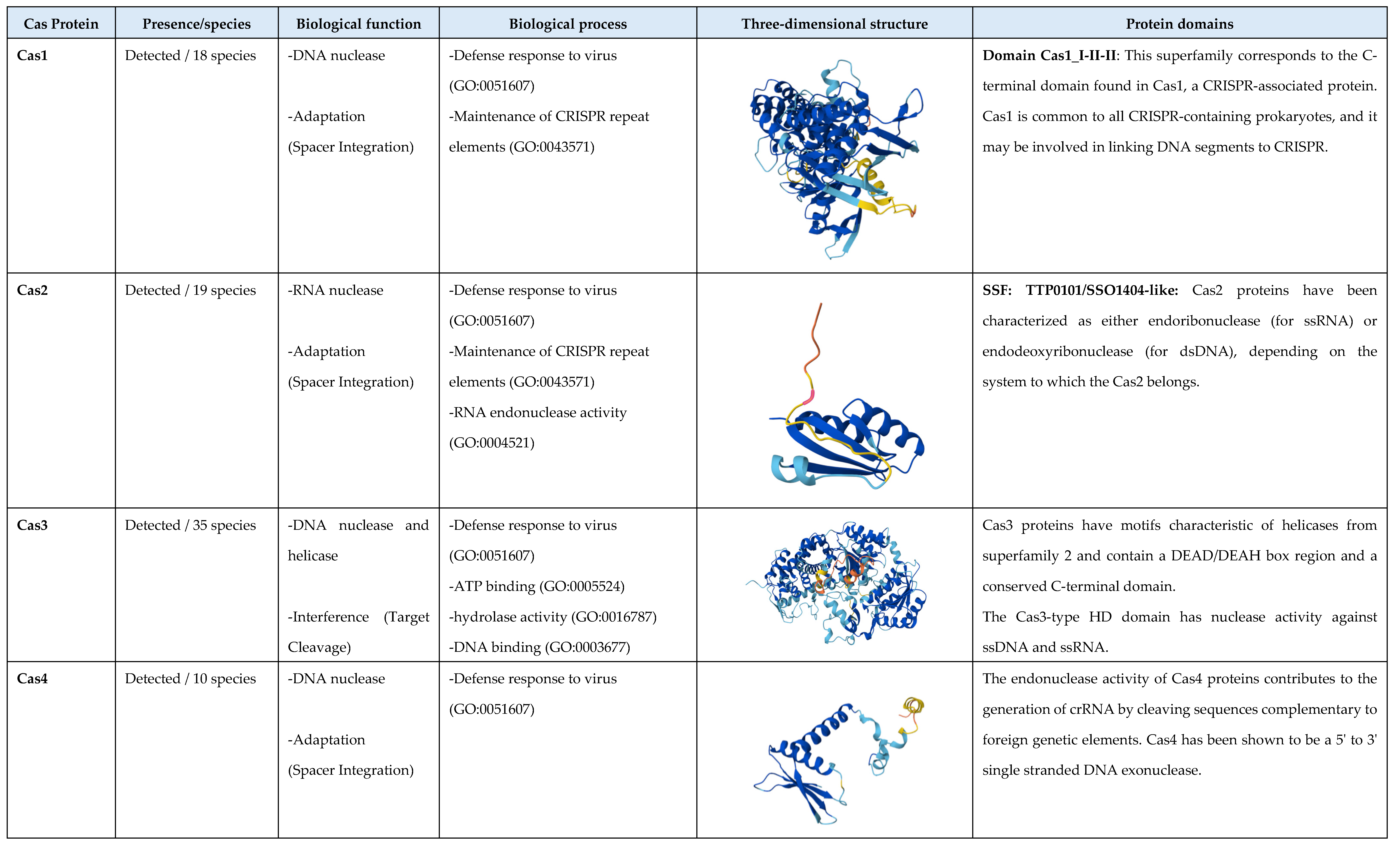
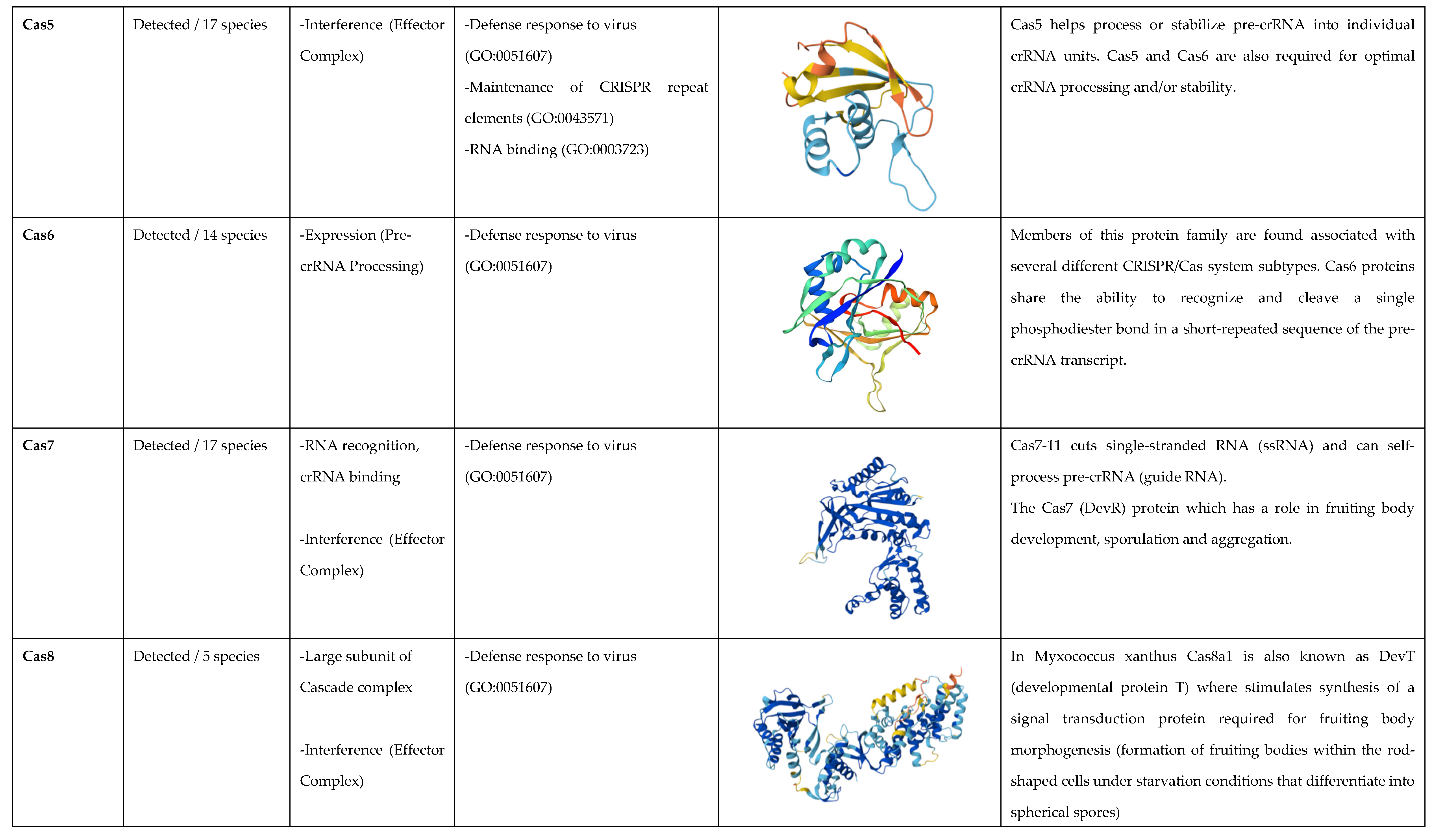
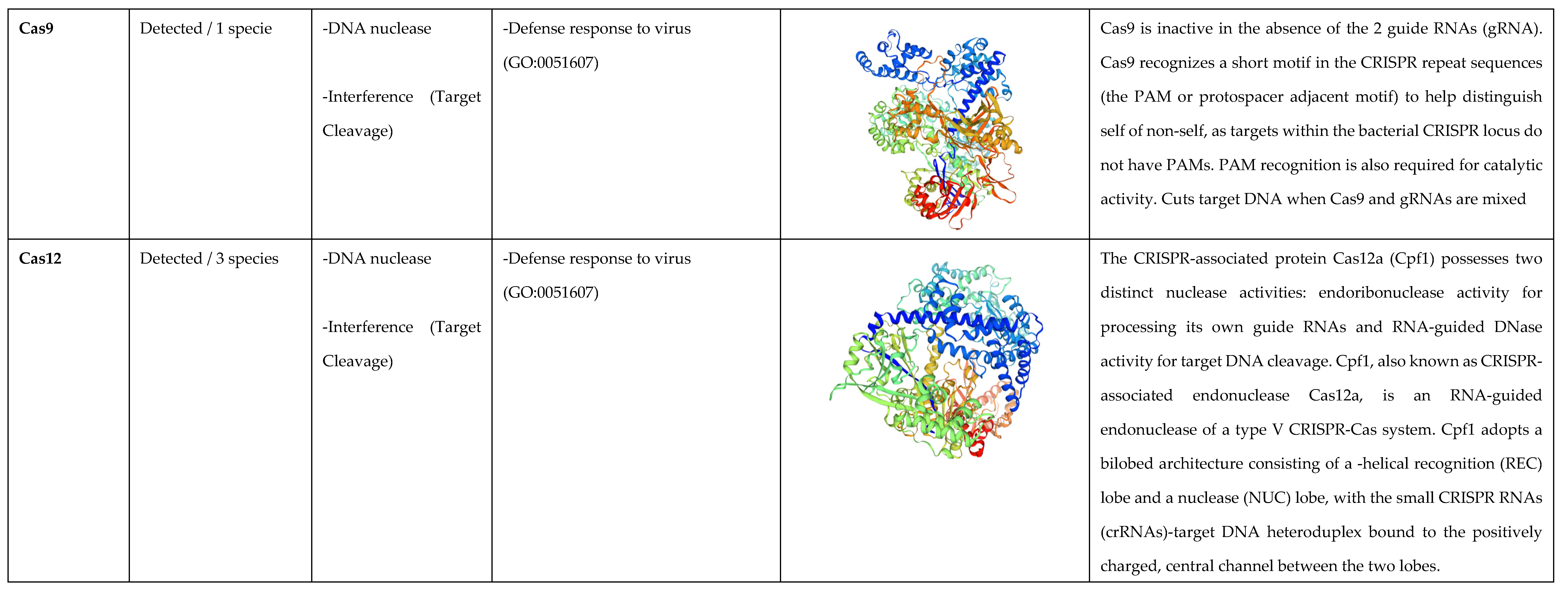
3.2. Cas Proteins Detection
3.3. Analysis of the Biological Function of Cas Proteins
3.4. Classification of Cas Proteins into Functional Stages
3.5. Conservation Patterns of the Direct Repeats in the Leptospira Genus
3.6. Bioinformatic Identification of the Immunological Memory (Spacer Sequences)
3.7. Identification of Unique Effector Proteins
3.8. Bioinformatic Detection of Intact Bacteriophages in Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adler B, De La Peña Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010 Jan;140(3–4):287–96. [CrossRef]
- Haake DA. Spirochaetal lipoproteins and pathogenesis. Microbiol Read Engl. 2000 July;146(Pt 7):1491–504. [CrossRef]
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001 Apr;14(2):296–326.
- Vijayachari P, Sugunan AP, Shriram AN. Leptospirosis: an emerging global public health problem. J Biosci. 2008 Nov;33(4):557–69. [CrossRef]
- McBride AJA, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005 Oct;18(5):376–86.
- Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis. 2015 Sept 17;9(9):e0003898.
- Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019 May;13(5):e0007270. [CrossRef]
- Cerqueira GM, Picardeau M. A century of Leptospira strain typing. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2009 Sept;9(5):760–8. [CrossRef]
- Fernandes LGV, Stone NE, Roe CC, Goris MGA, van der Linden H, Sahl JW, et al. Leptospira sanjuanensis sp. nov., a pathogenic species of the genus Leptospira isolated from soil in Puerto Rico. Int J Syst Evol Microbiol. 2022 Oct;72(10). [CrossRef]
- Korba AA, Lounici H, Kainiu M, Vincent AT, Mariet JF, Veyrier FJ, et al. Leptospira ainlahdjerensis sp. nov., Leptospira ainazelensis sp. nov., Leptospira abararensis sp. nov. and Leptospira chreensis sp. nov., four new species isolated from water sources in Algeria. Int J Syst Evol Microbiol. 2021 Dec;71(12). [CrossRef]
- Hamond C, Tibbs-Cortes B, Fernandes LGV, LeCount K, Putz EJ, Anderson T, et al. Leptospira gorisiae sp. nov, L. cinconiae sp. nov, L. mgodei sp. nov, L. milleri sp. nov and L. iowaensis sp. nov: five new species isolated from water sources in the Midwestern United States. Int J Syst Evol Microbiol. 2025;75(1):006595. [CrossRef]
- Dos Santos Ribeiro P, Carvalho NB, Aburjaile F, Sousa T, Veríssimo G, Gomes T, et al. Environmental Biofilms from an Urban Community in Salvador, Brazil, Shelter Previously Uncharacterized Saprophytic Leptospira. Microb Ecol. 2023 Nov;86(4):2488–501. [CrossRef]
- Fernandes LGV, Hornsby RL, Nascimento ALTO, Nally JE. Genetic manipulation of pathogenic Leptospira: CRISPR interference (CRISPRi)-mediated gene silencing and rapid mutant recovery at 37 °C. Sci Rep. 2021 Jan 19;11(1):1768. [CrossRef]
- Pappas CJ, Xu H, Motaleb MA. Creating a Library of Random Transposon Mutants in Leptospira. In: Koizumi N, Picardeau M, editors. Leptospira spp: Methods and Protocols [Internet]. New York, NY: Springer US; 2020 [cited 2025 Aug 12]. p. 77–96. Available from: . [CrossRef]
- Bourhy P, Louvel H, Saint Girons I, Picardeau M. Random Insertional Mutagenesis of Leptospira interrogans , the Agent of Leptospirosis, Using a mariner Transposon. J Bacteriol. 2005 May;187(9):3255–8. [CrossRef]
- Murray GL, Morel V, Cerqueira GM, Croda J, Srikram A, Henry R, et al. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect Immun. 2009 Feb;77(2):810–6. [CrossRef]
- Lourdault K, Matsunaga J, Evangelista KV, Haake DA. High-throughput Parallel Sequencing to Measure Fitness of Leptospira interrogans Transposon Insertion Mutants During Golden Syrian Hamster Infection. J Vis Exp JoVE. 2017 Dec 18;(130):56442.
- Liao S, Sun A, Ojcius DM, Wu S, Zhao J, Yan J. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogansstrain Lai. BMC Microbiol. 2009 Dec 9;9(1):253. [CrossRef]
- Croda J, Figueira CP, Wunder EA, Santos CS, Reis MG, Ko AI, et al. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 2008 Dec;76(12):5826–33. [CrossRef]
- Pappas CJ, Benaroudj N, Picardeau M. A Replicative Plasmid Vector Allows Efficient Complementation of Pathogenic Leptospira Strains. Appl Environ Microbiol. 2015 May;81(9):3176–81. [CrossRef]
- Pacesa M, Pelea O, Jinek M. Past, present, and future of CRISPR genome editing technologies. Cell. 2024 Feb 29;187(5):1076–100. [CrossRef]
- Nidhi S, Anand U, Oleksak P, Tripathi P, Lal JA, Thomas G, et al. Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int J Mol Sci. 2021 Mar 24;22(7):3327. [CrossRef]
- Chehelgerdi M, Chehelgerdi M, Khorramian-Ghahfarokhi M, Shafieizadeh M, Mahmoudi E, Eskandari F, et al. Correction: Comprehensive review of CRISPR-based gene editing: mechanisms, challenges, and applications in cancer therapy. Mol Cancer. 2024 Feb 27;23(1):43.
- Hillary VE, Ceasar SA. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol Biotechnol. 2023 Mar;65(3):311–25. [CrossRef]
- Sternberg SH, Richter H, Charpentier E, Qimron U. Adaptation in CRISPR-Cas Systems. Mol Cell. 2016 Mar;61(6):797–808.
- Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc B Biol Sci. 2016 Nov 5;371(1707):20150496. [CrossRef]
- Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015 Feb;30:100–11. [CrossRef]
- Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc B Biol Sci. 2019 May 13;374(1772):20180087. [CrossRef]
- Alkhnbashi OS, Meier T, Mitrofanov A, Backofen R, Voß B. CRISPR-Cas bioinformatics. Methods. 2020 Feb;172:3–11. [CrossRef]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017 June;37:67–78. [CrossRef]
- Li C, Chu W, Gill RA, Sang S, Shi Y, Hu X, et al. Computational Tools and Resources for CRISPR/Cas Genome Editing. Genomics Proteomics Bioinformatics. 2023 Feb 1;21(1):108–26. [CrossRef]
- Mojica FJM, Montoliu L. On the Origin of CRISPR-Cas Technology: From Prokaryotes to Mammals. Trends Microbiol. 2016 Oct;24(10):811–20. [CrossRef]
- Strich JR, Chertow DS. CRISPR-Cas Biology and Its Application to Infectious Diseases. Kraft CS, editor. J Clin Microbiol. 2019 Apr;57(4):e01307-18.
- Bhatia S, Pooja, Yadav SK. CRISPR-Cas for genome editing: Classification, mechanism, designing and applications. Int J Biol Macromol. 2023 May;238:124054. [CrossRef]
- Mougiakos I, Bosma EF, De Vos WM, Van Kranenburg R, Van Der Oost J. Next Generation Prokaryotic Engineering: The CRISPR-Cas Toolkit. Trends Biotechnol. 2016 July;34(7):575–87.
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020 Feb;18(2):67–83. [CrossRef]
- Dixit B, Ghosh KK, Fernandes G, Kumar P, Gogoi P, Kumar M. Dual nuclease activity of a Cas2 protein in CRISPR –Cas subtype I-B of Leptospira interrogans. FEBS Lett. 2016 Apr;590(7):1002–16.
- Anand V, Prabhakaran HS, Gogoi P, Kanaujia SP, Kumar M. Structural and functional characterization of Cas2 of CRISPR-Cas subtype I-C lacking the CRISPR component. Front Mol Biosci. 2022 Sept 12;9:988569. [CrossRef]
- Anand V, Prabhakaran HS, Prakash A, Hussain MS, Kumar M. Differential processing of CRISPR RNA by LinCas5c and LinCas6 of Leptospira. Biochim Biophys Acta BBA - Gen Subj. 2023 Dec;1867(12):130469. [CrossRef]
- Prakash A, Kumar M. Transcriptional analysis of CRISPR I-B arrays of Leptospira interrogans serovar Lai and its processing by Cas6. Front Microbiol. 2022 July 29;13:960559. [CrossRef]
- Hussain MS, Anand V, Kumar M. Functional PAM sequence for DNA interference by CRISPR-Cas I-B system of Leptospira interrogans and the role of LinCas11b encoded within lincas8b. Int J Biol Macromol. 2023 May;237:124086. [CrossRef]
- Prakash A, Kumar M. Characterizing the transcripts of Leptospira CRISPR I-B array and its processing with endoribonuclease LinCas6. Int J Biol Macromol. 2021 July;182:785–95. [CrossRef]
- Dixit B, Prakash A, Kumar P, Gogoi P, Kumar M. The core Cas1 protein of CRISPR-Cas I-B in Leptospira shows metal-tunable nuclease activity. Curr Res Microb Sci. 2021 Dec;2:100059. [CrossRef]
- Dixit B, Anand V, Hussain MdS, Kumar M. The CRISPR-associated Cas4 protein from Leptospira interrogans demonstrate versatile nuclease activity. Curr Res Microb Sci. 2021 Dec;2:100040. [CrossRef]
- Fernandes LGV, Nascimento ALTO. A Novel Breakthrough in Leptospira spp. Mutagenesis: Knockout by Combination of CRISPR/Cas9 and Non-homologous End-Joining Systems. Front Microbiol. 2022 May 26;13:915382. [CrossRef]
- Fernandes LGV, Hamond C, Tibbs-Cortes BW, Putz EJ, Olsen SC, Palmer MV, et al. CRISPR-prime editing, a versatile genetic tool to create specific mutations with a single nucleotide resolution in Leptospira. Norris SJ, editor. mBio. 2024 Sept 11;15(9):e01516-24. [CrossRef]
- Xiao G, Yi Y, Che R, Zhang Q, Imran M, Khan A, et al. Characterization of CRISPR-Cas systems in Leptospira reveals potential application of CRISPR in genotyping of Leptospira interrogans. APMIS. 2019 Apr;127(4):202–16. [CrossRef]
- Natarajan S, Joseph J, Vinayagamurthy B, Estrela P. A Lateral Flow Assay for the Detection of Leptospira lipL32 Gene Using CRISPR Technology. Sensors. 2023 July 20;23(14):6544. [CrossRef]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014 July 15;30(14):2068–9. [CrossRef]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008 Dec;9(1):75.
- Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015 Feb 10;5(1):8365. [CrossRef]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014 Jan;42(D1):D206–14. [CrossRef]
- Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018 July 2;46(W1):W246–51. [CrossRef]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct;215(3):403–10. [CrossRef]
- Zdobnov EM, Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001 Sept 1;17(9):847–8. [CrossRef]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 July 2;46(W1):W296–303. [CrossRef]
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024 June 13;630(8016):493–500. [CrossRef]
- Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics. 2012 Dec;13(1):173.
- DeLano WL, Scientific D, Carlos S. PyMOL: An Open-Source Molecular Graphics Tool.
- Kumar S, Stecher G, Suleski M, Sanderford M, Sharma S, Tamura K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Battistuzzi FU, editor. Mol Biol Evol. 2024 Dec 6;41(12):msae263. [CrossRef]
- Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008 May 19;36(Web Server):W35–41. [CrossRef]
- Russell DA, Hatfull GF. PhagesDB: the actinobacteriophage database. Wren J, editor. Bioinformatics. 2017 Mar 1;33(5):784–6. [CrossRef]
- Wishart DS, Han S, Saha S, Oler E, Peters H, Grant JR, et al. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023 July 5;51(W1):W443–50. [CrossRef]
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016 July 8;44(W1):W16–21.
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011 July 1;39(suppl):W347–52. [CrossRef]
- Samaha Th S. In-Silico Characterisation of a Hypothetical Protein (LA_1016) of Leptospira Interrogans Serovar Lai Strain 56601. Austin J Proteomics Bioinform & Genomics. 2017;4(2).
- Ijaq J, Chandrasekharan M, Poddar R, Bethi N, Sundararajan VS. Annotation and curation of uncharacterized proteins- challenges. Front Genet [Internet]. 2015 Mar 31 [cited 2025 Aug 12];6. Available from: http://www.frontiersin.org/Bioinformatics_and_Computational_Biology/10.3389/fgene.2015.00119/abstract. [CrossRef]
- Takeshita D, Sato M, Inanaga H, Numata T. Crystal Structures of Csm2 and Csm3 in the Type III-A CRISPR–Cas Effector Complex. J Mol Biol. 2019 Feb;431(4):748–63. [CrossRef]
- Campbell A. The future of bacteriophage biology. Nat Rev Genet. 2003 June 1;4(6):471–7. [CrossRef]
- Hussain W, Yang X, Ullah M, Wang H, Aziz A, Xu F, et al. Genetic engineering of bacteriophages: Key concepts, strategies, and applications. Biotechnol Adv. 2023 May;64:108116. [CrossRef]
- Jia HJ, Jia PP, Yin S, Bu LK, Yang G, Pei DS. Engineering bacteriophages for enhanced host range and efficacy: insights from bacteriophage-bacteria interactions. Front Microbiol. 2023 May 31;14:1172635. [CrossRef]
- Chen Y, Batra H, Dong J, Chen C, Rao VB, Tao P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front Microbiol. 2019 May 3;10:954. [CrossRef]
- Segundo-Arizmendi N, Arellano-Maciel D, Rivera-Ramírez A, Piña-González AM, López-Leal G, Hernández-Baltazar E. Bacteriophages: A Challenge for Antimicrobial Therapy. Microorganisms. 2025 Jan 7;13(1):100. [CrossRef]
- Girma A. Bacteriophages as an alternative strategy for the treatment of drug resistant bacterial infections: Current approaches and future perspectives. Cell Surf. 2025 Dec;14:100149. [CrossRef]
- Subramanian A. Emerging roles of bacteriophage-based therapeutics in combating antibiotic resistance. Front Microbiol. 2024 July 5;15:1384164. [CrossRef]
- Summers WC. Bacteriophage Therapy. Annu Rev Microbiol. 2001 Oct;55(1):437–51.
- Girons IS, Margarita D, Amouriaux P, Baranton G. First isolation of bacteriophages for a spirochaete: Potential genetic tools for Leptospira. Res Microbiol. 1990 Nov;141(9):1131–8. [CrossRef]
- Bourhy P, Frangeul L, Couvé E, Glaser P, Saint Girons I, Picardeau M. Complete Nucleotide Sequence of the LE1 Prophage from the Spirochete Leptospira biflexa and Characterization of Its Replication and Partition Functions. J Bacteriol. 2005 June 15;187(12):3931–40. [CrossRef]
- Schiettekatte O, Vincent AT, Malosse C, Lechat P, Chamot-Rooke J, Veyrier FJ, et al. Characterization of LE3 and LE4, the only lytic phages known to infect the spirochete Leptospira. Sci Rep. 2018 Aug 6;8(1):11781. [CrossRef]

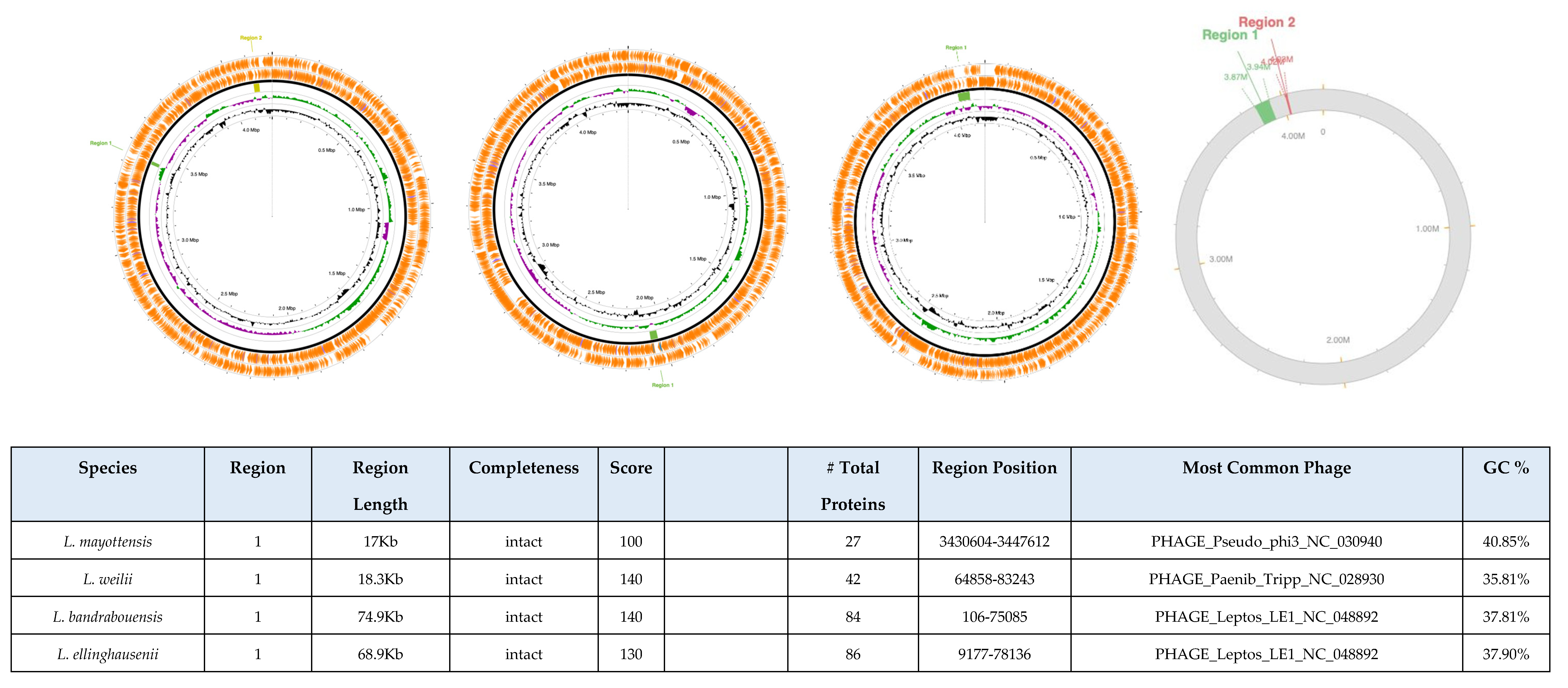

| Species | subgroup | Access Number | Sequencing level | Genome Size (Mb) | GC (%) | Genes | Proteins | Non-Coding |
|---|---|---|---|---|---|---|---|---|
| L. adleri | p1 | GCA_002811985.1 | Scaffold | 4.8 | 43.5 | 4789 | 4626 | 163 |
| L. ainazelensis | p1 | GCA_016918785.1 | Contig | 4.9 | 42.5 | 4417 | 4310 | 107 |
| L. ainlahdjerensis | p1 | GCA_016919175.1 | Contig | 4.8 | 42.5 | 4409 | 4310 | 99 |
| L. alexanderi | p1 | GCA_000243815.3 | Contig | 4.2 | 40 | 4582 | 4541 | 41 |
| L. alstonii | p1 | GCA_000347175.1 | Contig | 4.4 | 42.5 | 4423 | 4380 | 43 |
| L. barantonii | p1 | GCA_002811925.1 | Contig | 4.4 | 44 | 4135 | 4033 | 102 |
| L. borgpetersenii | p1 | GCA_003516145.1 | Chromosome | 4 | 40 | 3792 | 3463 | 329 |
| L. ellisii | p1 | GCA_002811955.2 | Contig | 4.3 | 48 | 3998 | 3896 | 102 |
| L. gomenensis | p1 | GCA_004770155.1 | Contig | 4.3 | 46 | 3931 | 3802 | 129 |
| L. kirschneri | p1 | GCA_000243695.3 | Contig | 4.4 | 36 | 4029 | 3986 | 43 |
| L. interrogans | p1 | GCA_002073495.2 | Chromosome | 4.6 | 35 | 4049 | 3772 | 277 |
| L. kmetyi | p1 | GCA_003722295.1 | Chromosome | 4.4 | 45 | 4160 | 4035 | 125 |
| L. mayottensis | p1 | GCA_000306675.3 | Chromosome | 4.2 | 39.5 | 3944 | 3592 | 352 |
| L. noguchii | p1 | GCA_000306255.2 | Contig | 4.7 | 35.5 | 4565 | 4520 | 45 |
| L. sanjuanensis | p1 | GCA_022267325.1 | Contig | 4.5 | 45 | 4152 | 4052 | 100 |
| L. santarosai | p1 | GCA_000313175.2 | Chromosome | 4 | 42 | 4191 | 4080 | 111 |
| L. stimsonii | p1 | GCA_003545875.1 | Contig | 4.7 | 42.5 | 4747 | 4599 | 148 |
| L. tipperaryensis | p1 | GCA_001729245.1 | Chromosome | 4.6 | 42.5 | 4342 | 4255 | 87 |
| L. weilii | p1 | GCA_006874765.1 | Chromosome | 4.4 | 41 | 4312 | 3965 | 347 |
| L. yasudae | p1 | GCA_003545925.1 | Contig | 4.4 | 45.5 | 4286 | 4162 | 124 |
| L. gorisiae | p1 | GCA_040833975.1 | Chromosome | 4.5 | 41.5 | 4104 | 3939 | 165 |
| L. andrefontaineae | p2 | GCA_004770105.1 | Contig | 4.3 | 40 | 3969 | 3894 | 75 |
| L. broomii | p2 | GCA_000243715.3 | Contig | 4.4 | 43 | 4249 | 4205 | 44 |
| L. dzoumogneensis | p2 | GCA_004770895.1 | Contig | 4.1 | 41 | 3814 | 3724 | 90 |
| L. fainei | p2 | GCA_000306235.2 | Contig | 4.3 | 43.5 | 4157 | 4113 | 44 |
| L. fletcheri | p2 | GCA_004769195.1 | Contig | 3.7 | 47.5 | 3409 | 3326 | 83 |
| L. fluminis | p2 | GCA_004771275.1 | Contig | 3.7 | 47.5 | 3427 | 3342 | 85 |
| L. haakeii | p2 | GCA_002812225.1 | Scaffold | 4.2 | 40 | 3920 | 3814 | 106 |
| L. hartskeerlii | p2 | GCA_002811475.1 | Scaffold | 4.1 | 40.5 | 3787 | 3708 | 79 |
| L. inadai | p2 | GCA_000243675.3 | Contig | 4.5 | 44.5 | 4314 | 4264 | 50 |
| L. johnsonii | p2 | GCA_003112675.1 | Contig | 4.1 | 41.5 | 3754 | 3713 | 41 |
| L. koniambonensis | p2 | GCA_004769555.1 | Contig | 4.3 | 39 | 4000 | 3929 | 71 |
| L. langatensis | p2 | GCA_004770615.1 | Contig | 4.1 | 45 | 3751 | 3671 | 80 |
| L. licerasiae | p2 | GCA_000526875.1 | Contig | 4.2 | 41 | 3899 | 3834 | 65 |
| L. neocaledonica | p2 | GCA_002812205.1 | Scaffold | 4.2 | 40 | 3978 | 3889 | 89 |
| L. perolatii | p2 | GCA_002811875.1 | Contig | 4 | 42.5 | 3712 | 3631 | 81 |
| L. saintgironsiae | p2 | GCA_002811765.1 | Contig | 4.1 | 39 | 3830 | 3736 | 94 |
| L. sarikeiensis | p2 | GCA_004769615.1 | Contig | 4.4 | 40.5 | 4026 | 3928 | 98 |
| L. selangorensis | p2 | GCA_004769405.1 | Contig | 4.2 | 40 | 3894 | 3814 | 80 |
| L. semungkisensis | p2 | GCA_004770055.1 | Contig | 3.9 | 43 | 3626 | 3562 | 64 |
| L. venezuelensis | p2 | GCA_002150035.1 | Contig | 4.3 | 39 | 4030 | 3973 | 57 |
| L. wolffii | p2 | GCA_004770635.1 | Contig | 4.2 | 46 | 3851 | 3771 | 80 |
| L. cinconiae | p2 | GCA_040833995.1 | Chromosome | 4.1 | 42 | 3801 | 3741 | 60 |
| L. abararensis | s1 | GCA_016918735.1 | Contig | 4.2 | 39 | 3968 | 3900 | 68 |
| L. bandrabouensis | s1 | GCA_004770555.1 | Contig | 4.2 | 37.5 | 3928 | 3857 | 71 |
| L. biflexa | s1 | GCA_000017685.1 | Chromosome | 4 | 39 | 3775 | 3726 | 49 |
| L. bourretii | s1 | GCA_004770145.1 | Contig | 4.2 | 38 | 3923 | 3840 | 83 |
| L. bouyouniensis | s1 | GCA_004770625.1 | Contig | 4.1 | 37 | 3833 | 3746 | 87 |
| L. brenneri | s1 | GCA_004769295.1 | Contig | 3.9 | 38.5 | 3662 | 3593 | 69 |
| L. chreensis | s1 | GCA_016919165.1 | Contig | 4.5 | 40 | 4176 | 4086 | 90 |
| L. congkakensis | s1 | GCA_004770265.1 | Contig | 4 | 38 | 3712 | 3647 | 65 |
| L. ellinghausenii | s1 | GCA_003114815.1 | Contig | 4.2 | 37.5 | 3960 | 3921 | 39 |
| L. harrisiae | s1 | GCA_002811945.1 | Scaffold | 3.9 | 38 | 3726 | 3651 | 75 |
| L. jelokensis | s1 | GCA_004769775.1 | Contig | 4.1 | 39 | 3833 | 3755 | 78 |
| L. kanakyensis | s1 | GCA_004769235.1 | Contig | 4.1 | 38.5 | 3884 | 3810 | 74 |
| L. kemamanensis | s1 | GCA_004769665.1 | Contig | 3.8 | 39 | 3533 | 3436 | 97 |
| L. levettii | s1 | GCA_002812085.1 | Scaffold | 3.9 | 37.5 | 3655 | 3579 | 76 |
| L. meyeri | s1 | GCA_004368965.1 | Contig | 4.2 | 38 | 4028 | 3930 | 98 |
| L. montravelensis | s1 | GCA_004770045.1 | Contig | 4 | 37.5 | 3760 | 3686 | 74 |
| L. mtsangambouensis | s1 | GCA_004770475.1 | Contig | 4.1 | 38 | 3836 | 3764 | 72 |
| L. noumeaensis | s1 | GCA_004770765.1 | Contig | 4.1 | 38.5 | 3832 | 3758 | 74 |
| L. perdikensis | s1 | GCA_004769575.1 | Contig | 4 | 38.5 | 3740 | 3680 | 60 |
| L. terpstrae | s1 | GCA_000332495.2 | Contig | 4.1 | 38 | 3932 | 3889 | 43 |
| L. vanthielii | s1 | GCA_004770365.1 | Contig | 4.1 | 39 | 3839 | 3753 | 86 |
| L. wolbachii | s1 | GCA_000332515.2 | Contig | 4.1 | 39 | 3956 | 3912 | 44 |
| L. yanagawae | s1 | GCA_004769275.1 | Contig | 4 | 38.5 | 3704 | 3631 | 73 |
| L. mgodei | s1 | GCA_040833985.1 | Chromosome | 4 | 39 | 3807 | 3752 | 55 |
| L. milleri | s1 | GCA_040833955.1 | Chromosome | 3.9 | 38.5 | 3628 | 3555 | 73 |
| L. iowaensis | s1 | GCA_040833965.1 | Chromosome | 4.1 | 37 | 3867 | 3812 | 55 |
| L. paudalimensis | s1 | GCA_026151345.1 | Contig | 4.1 | 37.5 | 3769 | 3711 | 58 |
| L. soteropolitanensis | s1 | GCA_026151335.1 | Contig | 4.1 | 37.5 | 3934 | 3863 | 71 |
| L. limi | s1 | GCA_026151395.1 | Contig | 3.9 | 37.5 | 3679 | 3619 | 60 |
| L. idonii | s2 | GCA_004770995.1 | Contig | 4.1 | 41 | 3797 | 3724 | 73 |
| L. ilyithenensis | s2 | GCA_004771005.1 | Contig | 4.2 | 40.5 | 3950 | 3849 | 101 |
| L. kobayashii | s2 | GCA_003114835.3 | Chromosome | 4.3 | 40.5 | 3945 | 3902 | 43 |
| L. ognonensis | s2 | GCA_004770745.1 | Scaffold | 4 | 39.5 | 3733 | 3660 | 73 |
| L. ryugenii | s2 | GCA_003114855.1 | Scaffold | 4 | 40 | 3698 | 3659 | 39 |
| Species | Clade | Proteins | Class | Type | Subtype | Variant | Native target |
|---|---|---|---|---|---|---|---|
| L. adleri | p1 | cas1_TypeIA, cas4_TypeI-II, cas3_TypeI, cas3a_TypeI, cas2_TypeI-II-III, cas5a2_TypeIA, cas6_TypeIA, cas7b_TypeIB | Class 1 | I, II, III | IA, IB | cas3a, cas5a2, cas7b | DNA |
| L. ainazelensis | p1 | Not detected | - | - | - | - | - |
| L. ainlahdjerensis | p1 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. alexanderi | p1 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas4_TypeI-II , cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I, II | IE | - | DNA |
| L. alstonii | p1 | cas1_TypeIA , cas2_TypeI-II-III, cas3_TypeI, cas5a2_TypeIA, cas6_TypeIE, cas7_TypeI, cas4_TypeI-II, cas5_TypeIE | Class 1 | I, II, III | IA, IE | Cas5a | DNA |
| L. barantonii | p1 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. borgpetersenii | p1 | cse2_TypeIE, cse1_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeI, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. ellisii | p1 | cas3a_TypeI | Class 1 | I | - | - | DNA |
| L. gomenensis | p1 | Not detected | - | - | - | - | - |
| L. kirschneri | p1 | cas1_TypeIC, cas2_TypeI-II-III, cas3_TypeI, cas4_TypeI-II, cas5_TypeIA, cas5c_TypeIC, cas7c_TypeIC, cas8c_TypeIC | Class 1 | I, II, III | IC, IA | Cas5c, cas7c, cas8c | DNA |
| L. interrogans | p1 | cas1_TypeIC, cas2_TypeI-II-III, cas3_TypeI, cas3a_TypeI, cas4_TypeI-II, cas5c_TypeIC, cas7c_TypeIC, cas8c_TypeIC | Class 1 | I, II, III | IC | Cas3a | DNA |
| L. kmetyi | p2 | cas1_TypeIA, cas2_TypeI-II-III, cas3_TypeI, cas5a2_TypeIA, cas6_TypeIA, cas7_TypeI, cas8a1a3_TypeIA | Class 1 | I, II, III | IA | Cas5a2, cas8a1a3 | DNA |
| L. mayottensis | p1 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. noguchii | p1 | cas2_TypeI-II-III, cas3_TypeI, cas3a_TypeI, cas4_TypeI-II, cas5c_TypeIC, cas7c_TypeIC, cas8c_TypeIC | Class 1 | I, II, III | IC | Cas3a, cas5c, cas7c,cas8c | DNA |
| L. sanjuanensis | p1 | Not detected | - | - | - | - | - |
| L. santarosai | p1 | cse1_TypeIE, cas1_TypeIE, cas1_TypeIA, cas2_TypeI-II-III, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas5a2_TypeIA, cas6_TypeIE, cas6_TypeIA, cas7_TypeIE, cas7_TypeI, cas8a1a3_TypeIA | Class 1 | I, II, III | IE, IA | Cas5a2, cas8a1a3 | DNA |
| L. stimsonii | p1 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. tipperaryensis | p1 | Not detected | - | - | - | - | - |
| L. weilii | p1 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. yasudae | p1 | Not detected | - | - | - | - | - |
| L. gorisiae | p1 | cse1_TypeIE, cpf1_TypeU (Cas12a), cas1_TypeIE, cas1_TypeU, cas2_TypeIE, cas2_Type I-II-III, cas3_TypeI, cas4_TypeU, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE |
Class 1 Class 2 |
II, V | IE | - | DNA |
| L. andrefontaineae | p2 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. broomii | p2 | Not detected | - | - | - | - | - |
| L. dzoumogneensis | p2 | cas3_TypeI, cas3a_TypeI | Class 1 | I | - | Cas3a | DNA |
| L. fainei | p2 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. fletcheri | p2 | cas1_TypeI-II-III, cas2_TypeI-II-III, cas4_TypeI-II, cas9_TypeIIB | Class 2 | II | IIB | - | DNA |
| L. fluminis | p2 | Not detected | - | - | - | - | - |
| L. haakeii | p2 | csm2_TypeIIIA, cas3_TypeI | Class 1 | I, III | IIIA | - | DNA and RNA |
| L. hartskeerlii | p2 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. inadai | p2 | cas1_TypeIB, cas1_TypeU, cpf1_TypeU (Cas12a), cas1_TypeI-II-III, cas2_TypeI-II-III, cas3_TypeI, cas4_TypeI-II, cas4_TypeU, cas4_TypeI-II, cas5b_TypeIB, cas6_TypeI-III, cas7b_TypeIB |
Class 1 Class 2 |
I, II, II, V | IB | Cas5b, cas7b | DNA |
| L. johnsonii | p2 | Not detected | - | - | - | - | - |
| L. koniambonensis | p2 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| L. langatensis | p2 | Not detected | - | - | - | - | - |
| L. licerasiae | p2 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. neocaledonica | p2 | Not detected | - | - | - | - | - |
| L. perolatii | p2 | Not detected | - | - | - | - | - |
| L. saintgironsiae | p2 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. sarikeiensis | p2 | Not detected | - | - | - | - | - |
| L. selangorensis | p2 | Not detected | - | - | - | - | - |
| L. semungkisensis | p2 | Not detected | - | - | - | - | - |
| L. venezuelensis | p2 | Not detected | - | - | - | - | - |
| L. wolffii | p2 | Not detected | - | - | - | - | - |
| L. cinconiae | p2 | Not detected | - | - | - | - | - |
| L. abararensis | s1 | cas3_TypeI, cas3a_TypeI | Class 1 | I | - | Cas3a | DNA |
| L. bandrabouensis | s1 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. biflexa | s1 | Not detected | - | - | - | - | - |
| L. bourretii | s1 | Not detected | - | - | - | - | - |
| L. bouyouniensis | s1 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. brenneri | s1 | Not detected | - | - | - | - | - |
| L. chreensis | s1 | cas3a_TypeI | Class 1 | I | - | - | DNA |
| L. congkakensis | s1 | cas3a_TypeI | Class 1 | I | - | - | DNA |
| L. ellinghausenii | s1 | Not detected | - | - | - | - | - |
| L. harrisiae | s1 | Not detected | - | - | - | - | - |
| L. jelokensis | s1 | Not detected | - | - | - | - | - |
| L. kanakyensis | s1 | Not detected | - | - | - | - | - |
| L. kemamanensis | s1 | cas3_TypeI | Class 1 | I | - | - | DNA |
| L. levettii | s1 | Not detected | - | - | - | - | - |
| L. meyeri | s1 | Not detected | - | - | - | - | - |
| L. montravelensis | s1 | Not detected | - | - | - | - | - |
| L. mtsangambouensis | s1 | Not detected | - | - | - | - | - |
| L. noumeaensis | s1 | Not detected | - | - | - | - | - |
| L. perdikensis | s1 | Not detected | - | - | - | - | - |
| L. terpstrae | s1 | Not detected | - | - | - | - | - |
| L. vanthielii | s1 | cas3a_TypeI | Class 1 | I | - | - | DNA |
| L. wolbachii | s1 | Not detected | - | - | - | - | - |
| L. yanagawae | s1 | Not detected | - | - | - | - | - |
| L. mgodei | s1 | Not detected | - | - | - | - | - |
| L. milleri | s1 | Not detected | - | - | - | - | - |
| L. iowaensis | s1 | Not detected | - | - | - | - | - |
| L. paudalimensis | s1 | Not detected | - | - | - | - | - |
| L. soteropolitanensis | s1 | Not detected | - | - | - | - | - |
| L. limi | s1 | Not detected | - | - | - | - | - |
| L. idonii | s2 | cas3_TypeI, cas3a_TypeI | Class 1 | I | - | cas3a | DNA |
| L. ilyithenensis | s2 | cas1_TypeU, cas2_TypeI-II-III, cas3_TypeI, cas4_TypeU, cpf1_TypeU(Cas12a) | Class 2 | I, II, III, V | - | - | DNA |
| L. kobayashii | s2 | Not detected | - | - | - | - | - |
| L. ognonensis | s2 | Not detected | - | - | - | - | - |
| L. ryugenii | s2 | cse1_TypeIE, cse2_TypeIE, cas1_TypeIE, cas2_TypeIE, cas3_TypeI, cas5_TypeIE, cas6_TypeIE, cas7_TypeIE | Class 1 | I | IE | - | DNA |
| Species | Adaptation Spacer Integration |
Expression Pre-crRNA Processing |
Interference | Taxonomic classification |
|
|---|---|---|---|---|---|
| Effector Complex | Target Cleavage | ||||
| L. adleri | Cas1_TypeIA, Cas2_TypeI-II-III, Cas4_TypeI-II | Cas6_TypeIA | Cas5a2_TypeIA, Cas7b_TypeIB | Cas3_TypeI,Cas3a_TypeI | Class 1 type IF2 |
| L. alexanderi | Cas1_TypeIE, Cas2_TypeIE, Cas4_TypeI-II | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. alstonii | Cas1_TypeIA , Cas2_TypeI-II-III, Cas4_TypeI-II | Cas6_TypeIE | Cas5a2_TypeIA, Cas5_TypeIE, Cas7_TypeI | Cas3_TypeI | Class 1 type IF2 |
| L. borgpetersenii | Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeI, Cas7_TypeIE, Cse1_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. kirschneri | Cas1_TypeIC, Cas2_TypeI-II-III, Cas4_TypeI-II | Cas5_TypeIA, Cas5c_TypeIC, Cas7c_TypeIC, Cas8c_TypeIC | Cas3_TypeI | Class 1 type IC | |
| L. interrogans | Cas1_TypeIC, Cas2_TypeI-II-III, Cas4_TypeI-II | Cas5c_TypeIC, Cas7c_TypeIC, Cas8c_TypeIC | Cas3_TypeI, Cas3a_TypeI | Class 1 type IC | |
| L. mayottensis | Cas1_TypeIE, Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. noguchii | Cas2_TypeI-II-III, Cas4_TypeI-II | Cas5c_TypeIC, Cas7c_TypeIC, Cas8c_TypeIC | Cas3_TypeI, Cas3a_TypeI | Class 1 type IC | |
| L. santarosai | Cas1_TypeIA,Cas1_TypeIE,Cas2_TypeI-II-III, Cas2_TypeIE | Cas6_TypeIA, Cas6_TypeIE | Cas5_TypeIE,Cas5a2_TypeIA,Cas7_TypeIE,Cas7_TypeI, Cas8a1a3_TypeIA, Cse1_TypeIE | Cas3_TypeI | Class 1 type B |
| L. stimsonii | Cas1_TypeIE, Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. weilii | Cas1_TypeIE, Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. gorisiae | Cas1_TypeU, Cas1_TypeIE, Cas2_Type_I-II-III, Cas2_TypeIE, Cas4_TypeU | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE | Cas12a, Cas3_TypeI | Class 1, type IE Class 2 type V |
| L. kmetyi | Cas1_TypeIA, Cas2_TypeI-II-III | Cas6_TypeIA | Cas5a2_TypeIA, Cas7_TypeI, Cas8a1a3_TypeIA | Cas3_TypeI | Class 1 type B |
| L. fainei | Cas1_TypeIE, cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. fletcheri | Cas1_TypeI-II-III, Cas2_TypeI-II-III, Cas4_TypeI-II | RNAse III | Cas9_TypeIIB | Class 2 type IIB | |
| L. inadai | Cas1_TypeIB, Cas1_TypeU, Cas1_TypeI-II-III, Cas2_TypeI-II-III, Cas4_TypeI-II,Cas4_TypeU, Cas4_TypeI-II | Cas6_TypeI-III | Cas5b_TypeIB, Cas7b_TypeIB | Cas3_TypeI, Cas12a | Class 1 type IF2 Class 2 type V |
| L. koniambonensis | Cas1_TypeIE, Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
| L. ilyithenensis | Cas1_TypeU, Cas2_TypeI-II-III, Cas4_TypeU | Cas3_TypeI,Cas12a | Class 2 type V | ||
| L. ryugenii | Cas1_TypeIE, Cas2_TypeIE | Cas6_TypeIE | Cas5_TypeIE, Cas7_TypeIE, Cse1_TypeIE, Cse2_TypeIE | Cas3_TypeI | Class 1, type IE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





