Submitted:
11 September 2025
Posted:
12 September 2025
You are already at the latest version
Abstract
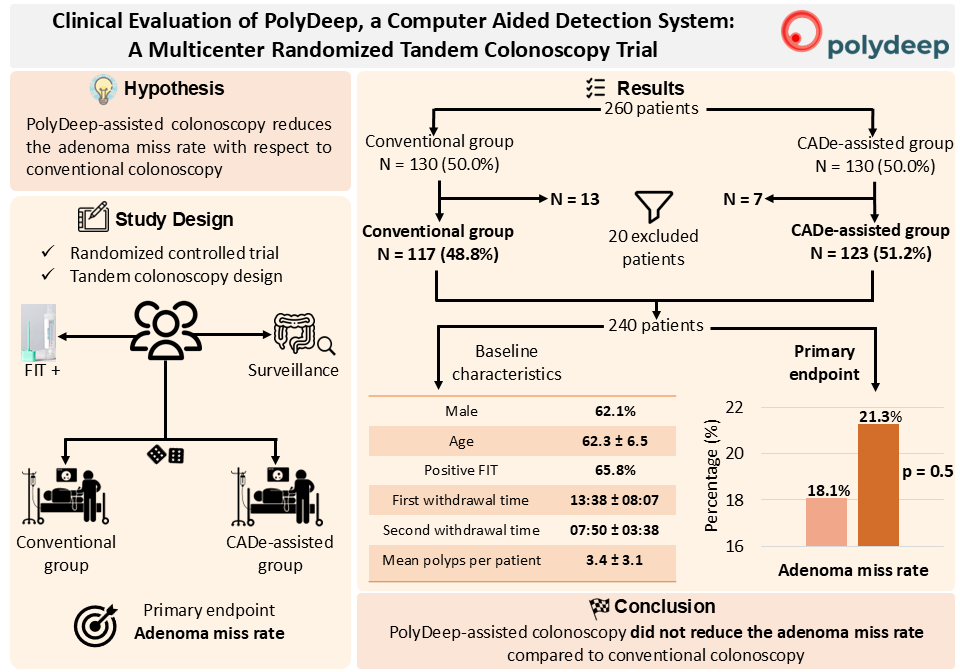
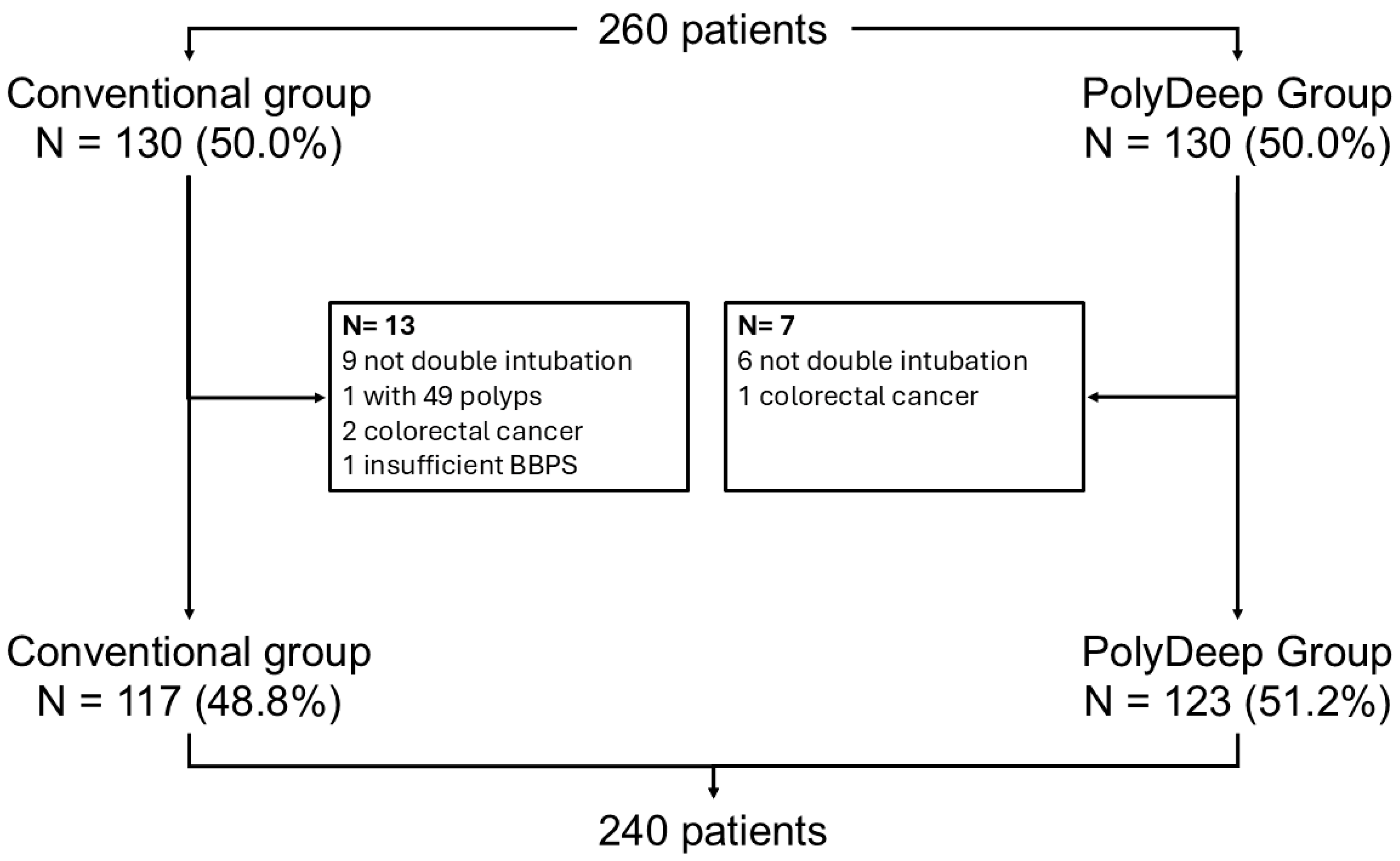
Background/Objectives: Computer-aided detection (CADe) systems are increasingly used in endoscopy to enhance lesion recognition. PolyDeep is a CADe/x tool previously assessed in an observational study. The aim of our study is to determine if PolyDeep-assisted colonoscopy reduces the adenoma miss rate (AMR) compared with conventional colonoscopy. Methods: We carried out a multicenter randomized controlled trial with a tandem colonoscopy design in participants from a colorectal cancer screening program (positive fecal immunochemical test-FIT or surveillance). Expert endoscopists performed all colonoscopies, and patients were allocated to groups by a computer-generated sequence. The primary endpoint was AMR; secondary endpoints included polyp miss rate (PMR), serrated lesion miss rate (SLMR) and advanced polyp miss rate (APMR). Results: From May to November 2023, we recruited 260 patients and excluded 20, leaving 240 for analysis. Baseline characteristics were balanced between groups (62.1% male; mean age 62.3 ± 6.5 years; 65.8% FIT-positive; mean first withdrawal time 13:38 ± 08:07 minutes; mean second withdrawal time 07:50 ± 03:38 minutes; lesion detection rate 76.6%; mean polyps per patient 3.4 ± 3.1). We did not find statistically significant differences between PolyDeep-assisted and conventional colonoscopy groups in AMR (21.3% vs 18.1%, p = 0.5), PMR (21.8% vs 20.3%, p = 0.7), SLMR (23.4% vs 25.6%, p = 0.9) or APMR (7.3% vs 11.3%, p = 0.5). In the subgroup analysis according to indication, we did not find any statistically significant differences. Conclusions: In the context of a CRC screening program, PolyDeep-assisted colonoscopy did not reduce AMR.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Participants of the Study
2.3. Randomization Process
2.4. Clinical Setting
2.5. Endpoints
2.6. Sample Size
2.7. Statistical Analysis
3. Results
3.1. Population Description
3.2. Diagnostic Performance: Adenoma Miss Rate, Polyp Miss Rate, Serrated Lesion Miss Rate
3.3. Sub-Analysis by Size, Location and Advanced Lesions
3.4. Sub-Analysis by Colonoscopy Indication
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Xia, R.; Si, W.; Zhang, W.; Zhang, Y.; Zhuang, G. Cost Effectiveness of Colorectal Cancer Screening Strategies in Middle- and High-Income Countries: A Systematic Review. Journal of Gastroenterology and Hepatology (Australia) 2025.
- Lee, J.Y.; Cha, J.M.; Yoon, J.Y.; Kwak, M.S.; Lee, H.H. Association between Colonoscopy and Colorectal Cancer Occurrence and Mortality in the Older Population: A Population-Based Cohort Study. Endoscopy 2025. [CrossRef]
- Castells, A.; Quintero, E.; Bujanda, L.; Castán-Cameo, S.; Cubiella, J.; Díaz-Tasende, J.; Lanas, Á.; Ono, A.; Serra-Burriel, M.; Frías-Arrocha, E.; et al. Articles Effect of Invitation to Colonoscopy versus Faecal Immunochemical Test Screening on Colorectal Cancer Mortality (COLONPREV): A Pragmatic, Randomised, Controlled, Non-Inferiority Trial. 2025. [CrossRef]
- Rasmussen, S.L.; Pedersen, L.; Torp-Pedersen, C.; Rasmussen, M.; Bernstein, I.; Thorlacius-Ussing, O. Post-Colonoscopy Colorectal Cancer and the Association with Endoscopic Findings in the Danish Colorectal Cancer Screening Programme. BMJ Open Gastroenterol 2025, 12, e001692. [CrossRef]
- Bretthauer, M.; Løberg, M.; Wieszczy, P.; Kalager, M.; Emilsson, L.; Garborg, K.; Rupinski, M.; Dekker, E.; Spaander, M.; Bugajski, M.; et al. Effect of Colonoscopy Screening on Risks of Colorectal Cancer and Related Death. New England Journal of Medicine 2022, 387, 1547–1556. [CrossRef]
- Wang, P.; Berzin, T.M.; Glissen Brown, J.R.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-Time Automatic Detection System Increases Colonoscopic Polyp and Adenoma Detection Rates: A Prospective Randomised Controlled Study. Gut 2019, 68, 1813–1819. [CrossRef]
- Jahn, B.; Bundo, M.; Arvandi, M.; Schaffner, M.; Todorovic, J.; Sroczynski, G.; Knudsen, A.; Fischer, T.; Schiller-Fruehwirth, I.; Öfner, D.; et al. One in Three Adenomas Could Be Missed by White-Light Colonoscopy – Findings from a Systematic Review and Meta-Analysis. BMC Gastroenterol 2025, 25. [CrossRef]
- Van Rijn, J.C.; Reitsma, J.B.; Stoker, J.; Bossuyt, P.M.; Van Deventer, S.J.; Dekker, E. Polyp Miss Rate Determined by Tandem Colonoscopy: A Systematic Review. American Journal of Gastroenterology 2006, 101, 343–350.
- Zhao, S.; Wang, S.; Pan, P.; Xia, T.; Chang, X.; Yang, X.; Guo, L.; Meng, Q.; Yang, F.; Qian, W.; et al. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-Analysis. Gastroenterology 2019, 156, 1661-1674.e11. [CrossRef]
- Misawa, M.; Kudo, S.E. Current Status of Artificial Intelligence Use in Colonoscopy. Digestion 2024.
- Aung, Y.Y.M.; Wong, D.C.S.; Ting, D.S.W. The Promise of Artificial Intelligence: A Review of the Opportunities and Challenges of Artificial Intelligence in Healthcare. Br Med Bull 2021, 139, 4–15.
- Gao, J.; Jiang, Q.; Zhou, B.; Chen, D. Convolutional Neural Networks for Computer-Aided Detection or Diagnosis in Medical Image Analysis: An Overview. Mathematical Biosciences and Engineering 2019, 16, 6536–6561.
- Shah, S.; Park, N.; Chehade, N.E.H.; Chahine, A.; Monachese, M.; Tiritilli, A.; Moosvi, Z.; Ortizo, R.; Samarasena, J. Effect of Computer-Aided Colonoscopy on Adenoma Miss Rates and Polyp Detection: A Systematic Review and Meta-Analysis. Journal of Gastroenterology and Hepatology (Australia) 2023, 38, 162–176. [CrossRef]
- Glissen Brown, J.R.; Mansour, N.M.; Wang, P.; Chuchuca, M.A.; Minchenberg, S.B.; Chandnani, M.; Liu, L.; Gross, S.A.; Sengupta, N.; Berzin, T.M. Deep Learning Computer-Aided Polyp Detection Reduces Adenoma Miss Rate: A United States Multi-Center Randomized Tandem Colonoscopy Study (CADeT-CS Trial). Clinical Gastroenterology and Hepatology 2022, 20, 1499-1507.e4. [CrossRef]
- Hiratsuka, Y.; Hisabe, T.; Ohtsu, K.; Yasaka, T.; Takeda, K.; Miyaoka, M.; Ono, Y.; Kanemitsu, T.; Imamura, K.; Takeda, T.; et al. Evaluation of Artificial Intelligence: Computer-Aided Detection of Colorectal Polyps. J Anus Rectum Colon 2025, 9, 79–87. [CrossRef]
- Biscaglia, G.; Cocomazzi, F.; Gentile, M.; Loconte, I.; Mileti, A.; Paolillo, R.; Marra, A.; Castellana, S.; Mazza, T.; Di Leo, A.; et al. Real-Time, Computer-Aided, Detection-Assisted Colonoscopy Eliminates Differences in Adenoma Detection Rate between Trainee and Experienced Endoscopists. Endosc Int Open 2022, 10, E616–E621. [CrossRef]
- Yamaguchi, D.; Shimoda, R.; Miyahara, K.; Yukimoto, T.; Sakata, Y.; Takamori, A.; Mizuta, Y.; Fujimura, Y.; Inoue, S.; Tomonaga, M.; et al. Impact of an Artificial Intelligence-Aided Endoscopic Diagnosis System on Improving Endoscopy Quality for Trainees in Colonoscopy: Prospective, Randomized, Multicenter Study. Digestive Endoscopy 2024, 36, 40–48. [CrossRef]
- Lou, S.; Du, F.; Song, W.; Xia, Y.; Yue, X.; Yang, D.; Cui, B.; Liu, Y.; Han, P. Artificial Intelligence for Colorectal Neoplasia Detection during Colonoscopy: A Systematic Review and Meta-Analysis of Randomized Clinical Trials; 2023;
- Wang, P.; Liu, P.; Glissen Brown, J.R.; Berzin, T.M.; Zhou, G.; Lei, S.; Liu, X.; Li, L.; Xiao, X. Lower Adenoma Miss Rate of Computer-Aided Detection-Assisted Colonoscopy vs Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology 2020, 159, 1252-1261.e5. [CrossRef]
- Maida, M.; Marasco, G.; Maas, M.H.J.; Ramai, D.; Spadaccini, M.; Sinagra, E.; Facciorusso, A.; Siersema, P.D.; Hassan, C. Effectiveness of Artificial Intelligence Assisted Colonoscopy on Adenoma and Polyp Miss Rate: A Meta-Analysis of Tandem RCTs. Digestive and Liver Disease 2025, 57, 169–175. [CrossRef]
- M Lee, M.C.; Parker, C.H.; C Liu, L.W.; Farahvash, A.; Jeyalingam, T. SYSTEMATIC REVIEW AND META-ANALYSIS Impact of Study Design on Adenoma Detection in the Evaluation of Artificial Intelligence-Aided Colonoscopy: A Systematic Review and Meta-Analysis;
- Maas, M.H.J.; Rath, T.; Spada, C.; Soons, E.; Forbes, N.; Kashin, S.; Cesaro, P.; Eickhoff, A.; Vanbiervliet, G.; Salvi, D.; et al. A Computer-Aided Detection System in the Everyday Setting of Diagnostic, Screening and Surveillance Colonoscopy: An International, Randomized Trial. Endoscopy 2024. [CrossRef]
- Bretthauer, M.; Ahmed, J.; Antonelli, G.; Beaumont, H.; Beg, S.; Benson, A.; Bisschops, R.; De Cristofaro, E.; Gibbons, E.; Häfner, M.; et al. Use of Computer-Assisted Detection (CADe) Colonoscopy in Colorectal Cancer Screening and Surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy 2025. [CrossRef]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; Campos-Tato, F.; Herrero, J.; Puga, M.; Remedios, D.; Rivas, L.; Sánchez, E.; Iglesias, Á.; Cubiella, J.; et al. Real-Time Polyp Detection Model Using Convolutional Neural Networks. Neural Comput Appl 2022, 34, 10375–10396. [CrossRef]
- Nogueira Rodríguez, A.; Daniel González, D.; Hugo López Fernández, P.D. Escola Internacional de Doutoramento DEEP LEARNING TECHNIQUES FOR COMPUTER-AIDED DIAGNOSIS IN COLORECTAL CANCER Dirixida Polos Doutores;
- Nogueira-Rodríguez, A.; Glez-Peña, D.; Reboiro-Jato, M.; López-Fernández, H. Negative Samples for Improving Object Detection—A Case Study in AI-Assisted Colonoscopy for Polyp Detection. Diagnostics 2023, 13. [CrossRef]
- Nogueira-Rodríguez, A.; Reboiro-Jato, M.; Glez-Peña, D.; López-Fernández, H. Performance of Convolutional Neural Networks for Polyp Localization on Public Colonoscopy Image Datasets. Diagnostics 2022, 12. [CrossRef]
- Davila-Piñón, P.; Nogueira-Rodríguez, A.; Díez-Martín, A.I.; Codesido, L.; Herrero, J.; Puga, M.; Rivas, L.; Sánchez, E.; Fdez-Riverola, F.; Glez-Peña, D.; et al. Optical Diagnosis in Still Images of Colorectal Polyps: Comparison between Expert Endoscopists and PolyDeep, a Computer-Aided Diagnosis System. Front Oncol 2024, 14. [CrossRef]
- Davila-Piñón, P.; Pedrido, T.; Díez-Martín, A.I.; Herrero, J.; Puga, M.; Rivas, L.; Sánchez, E.; Zarraquiños, S.; Pin, N.; Vega, P.; et al. PolyDeep Advance 1: Clinical Validation of a Computer-Aided Detection System for Colorectal Polyp Detection with a Second Observer Design. Diagnostics 2025, 15. [CrossRef]
- Maida, M.; Marasco, G.; Maas, M.H.J.; Ramai, D.; Spadaccini, M.; Sinagra, E.; Facciorusso, A.; Siersema, P.D.; Hassan, C. Effectiveness of Artificial Intelligence Assisted Colonoscopy on Adenoma and Polyp Miss Rate: A Meta-Analysis of Tandem RCTs. Digestive and Liver Disease 2024. [CrossRef]
- Makar, J.; Abdelmalak, J.; Con, D.; Hafeez, B.; Garg, M. Use of Artificial Intelligence Improves Colonoscopy Performance in Adenoma Detection: A Systematic Review and Meta-Analysis. Gastrointest Endosc 2024. [CrossRef]
- Mangas-Sanjuan, C.; de-Castro, L.; Cubiella, J.; Díez-Redondo, P.; Suárez, A.; Pellisé, M.; Fernández, N.; Zarraquiños, S.; Núñez-Rodríguez, H.; Álvarez-García, V.; et al. Role of Artificial Intelligence in Colonoscopy Detection of Advanced Neoplasias : A Randomized Trial. Ann Intern Med 2023. [CrossRef]
- Cubiella, J.; González, A.; Almazán, R.; Rodríguez-Camacho, E.; Rodiles, J.F.; Ferreiro, C.D.; Sandoval, C.T.; Gómez, C.S.; Bielza, N. de V.; Lorenzo, I.P.R.; et al. Pt1 Colorectal Cancer Detected in a Colorectal Cancer Mass Screening Program: Treatment and Factors Associated with Residual and Extraluminal Disease. Cancers (Basel) 2020, 12, 1–19. [CrossRef]
- Ali, M.L.; Zhang, Z. The YOLO Framework: A Comprehensive Review of Evolution, Applications, and Benchmarks in Object Detection. Computers 2024, 13.
- Lalinia, M.; Sahafi, A. Colorectal Polyp Detection in Colonoscopy Images Using YOLO-V8 Network. Signal Image Video Process 2024, 18, 2047–2058. [CrossRef]
|
Conventional group1 (N = 117) |
PolyDeep Group2 (N = 123) |
P4 | |
| Age (years) | 63.0 ± 6.8 | 61.6 ± 6.2 | 0.1 |
| Sex (male) | 69 (50.4%) | 80 (65.0%) | 0.4 |
| Indication (FIT) | 75 (64.1%) | 83 (67.5%) | 0.7 |
| Boston Bowel cleansing | 7.59 ± 1.28 | 7.42 ± 1.31 | 0.3 |
| First withdrawal time (minutes: seconds) | 13:34 ± 8:39 | 13:41 ± 07:37 | 0.9 |
| Second withdrawal time (minutes: seconds) | 07:58 ± 3:17 | 07:42 ± 03:57 | 0.6 |
| Detection of lesions (yes) | 90 (76.9%) | 94 (76.4%) | 0.7 |
| Number of polyps | 3.4 ± 3.3 | 3.4 ± 2.9 | 1.0 |
| Polyp size (millimetres) | 4.5 ± 4.7 | 4.9 ± 4.7 | 0.4 |
| Conventional group1 | PolyDeep group2 | ||||
| 1ª withdrawal | 2ª withdrawal3 | 1ª withdrawal | 2ª withdrawal3 | p4 | |
| Adenoma | 172 (81.9%) |
38 (18.1%) |
185 (78.7%) |
50 (21.3%) |
0.5 |
| Serrated lesion | 67 (74.4%) |
23 (25.6%) |
59 (76.6%) |
18 (23.4%) |
0.9 |
| Polyp5 | 239 (79.7%) |
61 (20.3%) |
244 (78.2%) |
68 (21.8%) |
0.7 |
| Other polyp | 12 (75.0%) |
4 (25.0%) |
16 (84.2%) |
3 (15.8%) |
- |
| Not histology | 12 (66.7%) |
6 (33.3%) |
6 (66.6%) |
3 (33.3%) |
- |
| Advanced adenomas6 | 40 (95.2%) |
2 (4.8%) |
37 (94.9%) |
2 (5.1%) |
1.0 |
| Advanced serrated lesion7 | 9 (64.3%) |
5 (35.7%) |
19 (86.4%) |
3 (13.6%) |
0.2 |
| Advanced polyp8 | 47 (88.7%) |
6 (11.3%) |
51 (92.7%) |
4 (7.3%) |
0.5 |
| Proximal polyp9 | 141 (81.5%) |
32 (18.5%) |
134 (80.7%) |
32 (19.3%) |
0.8 |
| Distal polyp10 | 98 (77.2%) |
29 (22.8%) |
110 (75.3%) |
36 (24.7%) |
0.8 |
| < 5 mm polyp | 161 (75.9%) |
51 (24.1%) |
149 (74.5%) |
51 (25.5%) |
0.8 |
| < 10 mm polyp | 203 (77.2%) |
60 (22.8%) |
209 (76.6%) |
64 (23.4%) |
0.9 |
| ≥ 5 mm polyp | 78 (88.6%) |
10 (11.4%) |
95 (84.8%) |
17 (15.2%) |
0.6 |
| Screening1 | p4 | Surveillance1 | p4 | |||
| Conventional group2 | PolyDeep Group3 | Conventional group2 | PolyDeep group3 | |||
| Adenoma miss rate | 14.9%5 | 20.4% | 0.2 | 25.8% | 24.1% | 1.0 |
| Serrated lesion miss rate | 29.4% | 25.0% | 0.7 | 20.5% | 15.4% | 1.0 |
| Polyp miss rate6 | 18.6% | 21.6% | 0.5 | 23.8% | 22.4% | 1.0 |
| Advanced polyp miss rate7 | 6.8% | 8.2% | 1.0 | 33.3% | 0.0% | 0.2 |
| Proximal polyp miss rate8 | 18.3% | 17.2% | 0.9 | 18.8% | 26.3% | 0.5 |
| Distal polyp miss rate9 | 18.9% | 26.5% | 0.3 | 34.4% | 17.2% | 0.2 |
| < 5 mm polyp miss rate | 21.3% | 25.0% | 0.6 | 28.2% | 26.9% | 1.0 |
| ≥ 5 mm polyp miss rate | 13.9% | 16.5% | 0.8 | 0.0% | 6.7% | 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





