Submitted:
10 September 2025
Posted:
12 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Fungal Culture
2.3. Production of Hybridomas and Screening by ELISA
2.4. Determination of Ig Class and Sub-Cloning Procedure
2.5. Antibody Purification and Enzyme Conjugation
2.6. Determination of Antibody Specificity
2.7. Epitope Characterisation by Heat
2.8. Polyacrylamide Gel Electrophoresis and Western Blotting
2.9. Antigen Production In Vitro
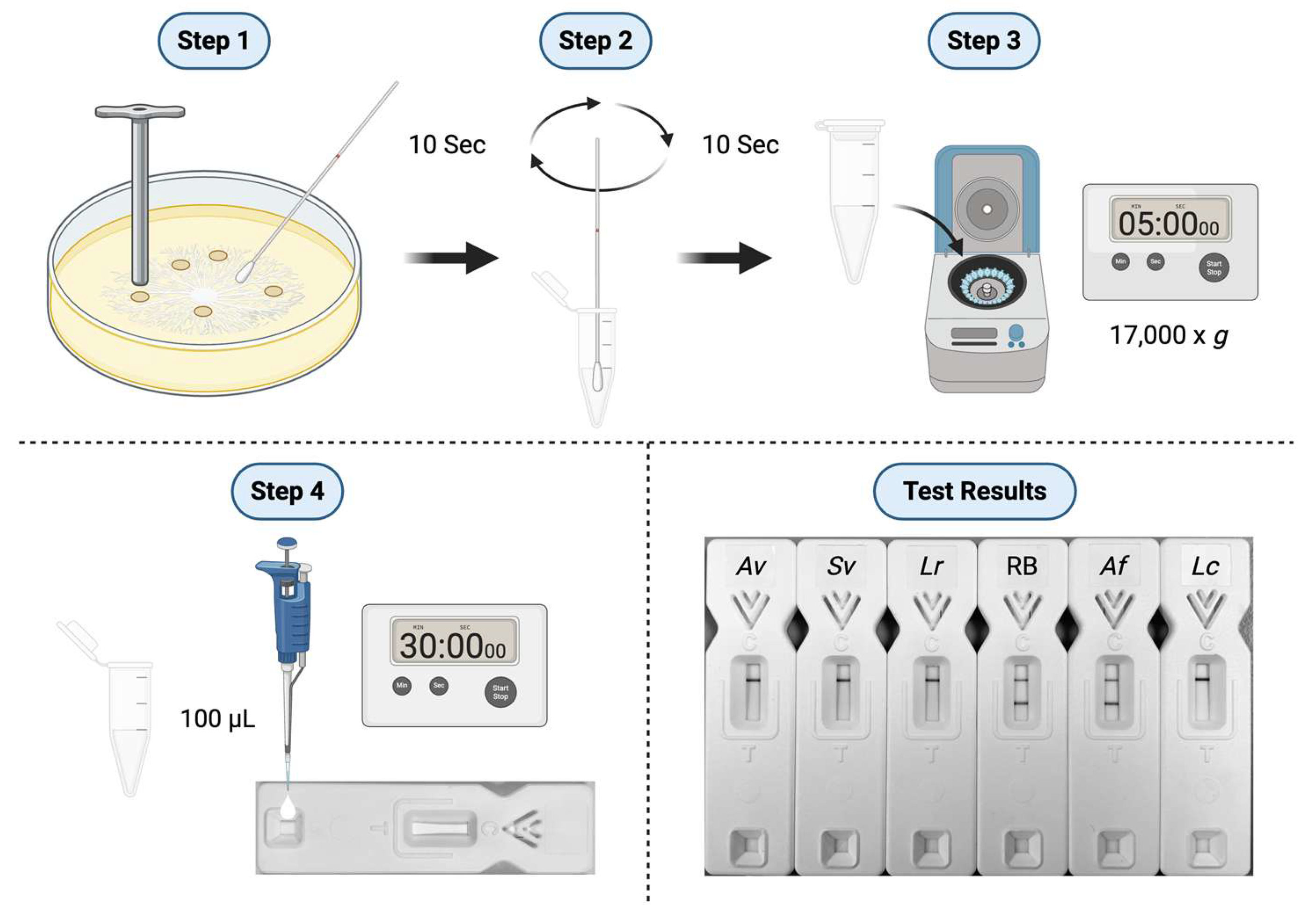
2.10. Culture-Based Detection and Differentiation of Mucorales Fungi
2.11. Statistical Analysis
3. Results
3.1. Production of Hybridomas and mAb Isotyping
3.2. Specificity of mAb JD4 and Epitope Characterisation
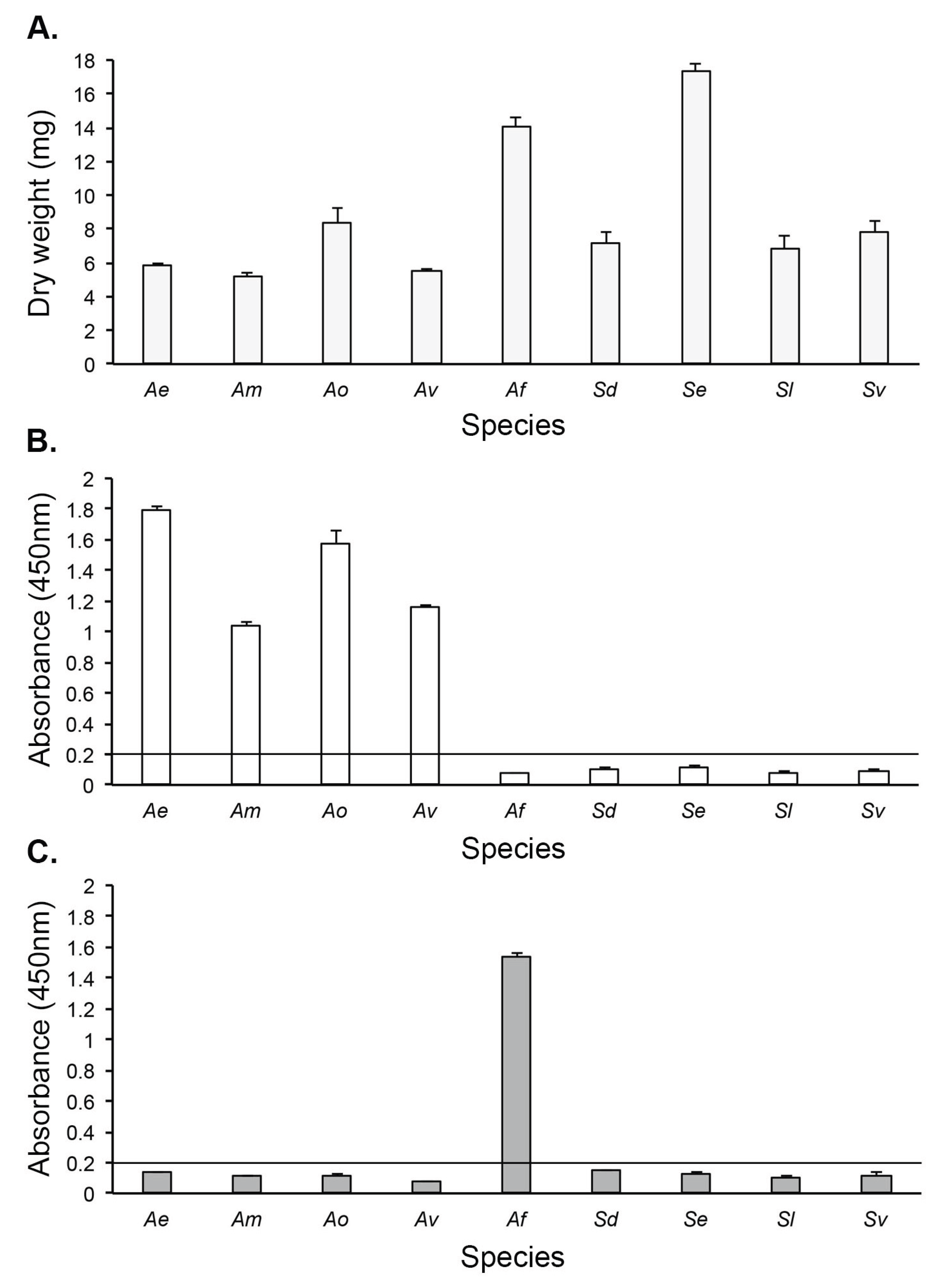
3.3. Antigen Production In Vitro
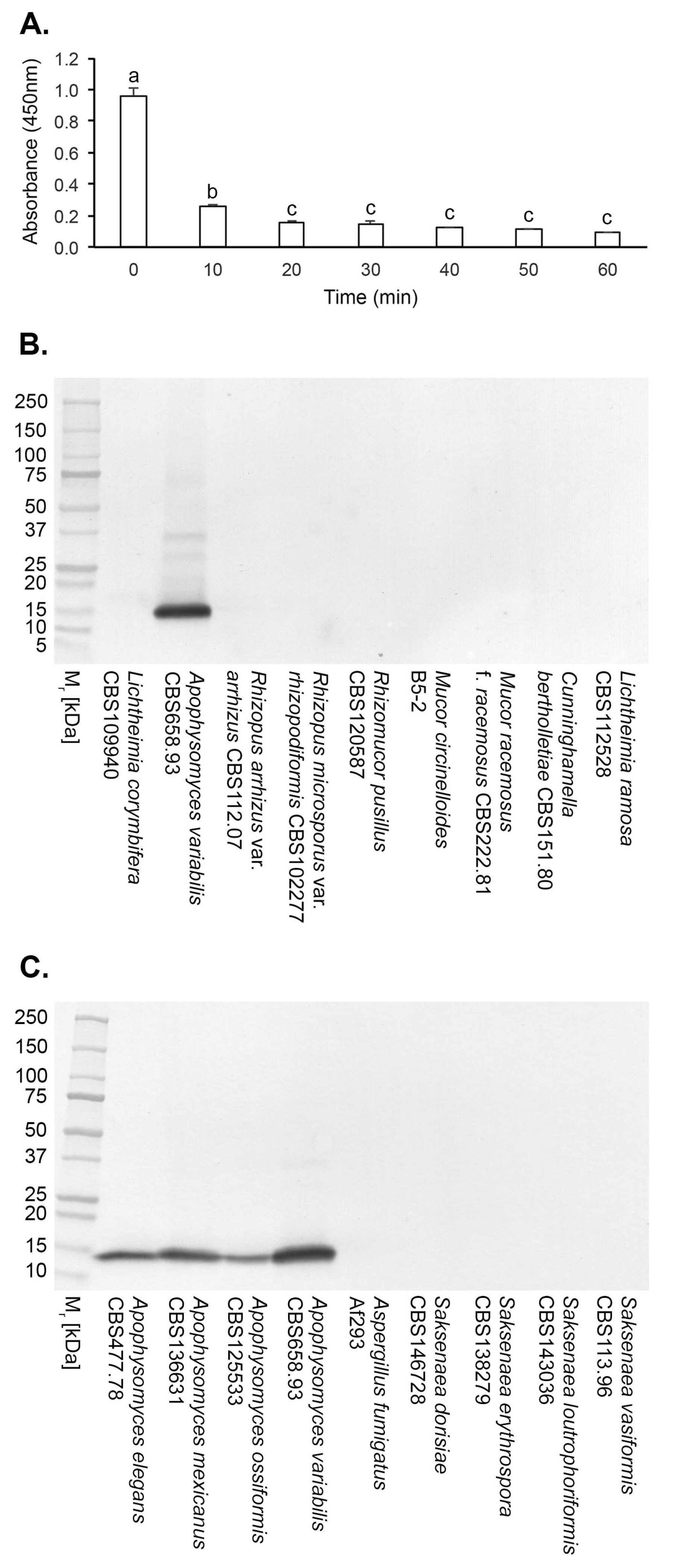
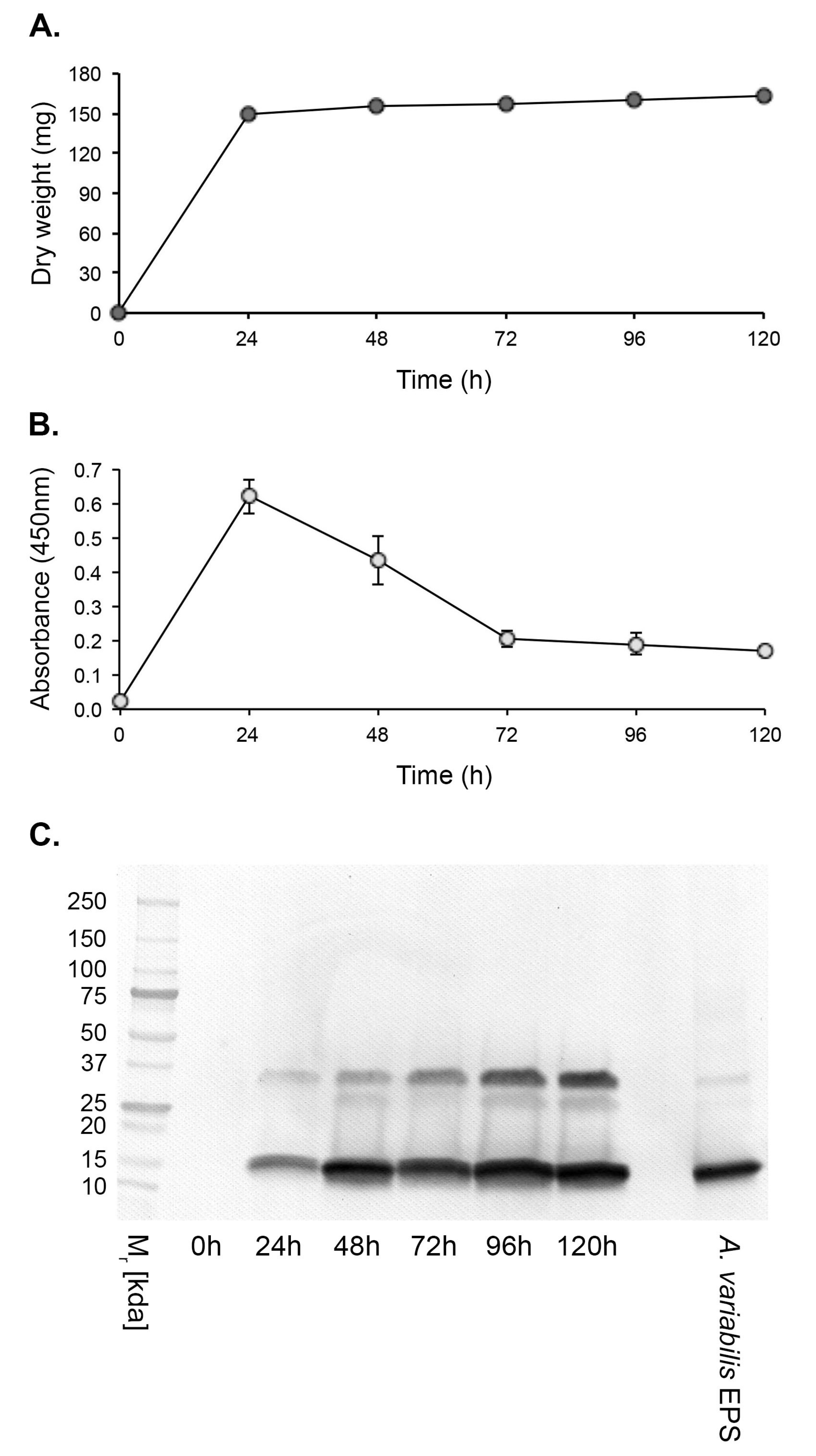
3.4. Culture-Based Detection and Differentiation of Mucorales Fungi
3.4.1. Lateral-flow immunoassay
3.4.2. Direct-ELISA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO (2022). WHO fungal priority pathogens list to guide research, development and public health action (Geneva: World Health Organization). License: CC BY-NC-SA 3.0 IGO.
- Brown, G.D.; Ballou, E.R.; Bates, S.; Bignell, E.M.; Borman, A.M.; Brand, A.C.; Brown, A.J.P.; Coelho, C.; Cook, P.C.; Farrer, R.A.; et al. The pathobiology of human fungal infections. Nat. Rev. Microbiol. 2024, 22, 687–704. [Google Scholar] [CrossRef]
- Morrissey, C.O.; Kim, H.Y.; Granham, K.; Dao, A.; Chakrabarti, A.; Perfect, J.R.; Alastruey-Izquierdo, A.; Harrison, T.S.; Bongomin, F.; Galas, M.; et al. Mucorales: a systematic review to inform the World Health Organization priority list of fungal pathogens. Med. Mycol. 2024, 62, myad130. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Alastruey-Izquierdo, A.; Arenz, D.; Chen, S.C.A.; Dannaoui, E.; Hochhegger, B.; Hoenigl, M.; Jensen, H.E.; Lagrou, K.; Lewis, R.E.; et al. Global guidelines for the diagnosis and management of mucormycosis: an initiative of the European Confederation of Medical Mycology in cooperation with the Mycoses Study Group Education and Research Consortium. Lancet Infect. Dis. 2019, 19, e405–e421. [Google Scholar] [CrossRef] [PubMed]
- Muthu, V.; Rudramurthy, S.; Chakrabarti, A.; Agarwal, R. Epidemiology and pathophysiology of COVID-19-associated mucormycosis: India versus the rest of the world. Mycopathologia 2021, 186, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Singh, R. Mucormycosis in India: unique features. Mycoses 2014, 57, 85–90. [Google Scholar] [CrossRef]
- Patel, A.; Kaur, H.; Xess, I.; Michael, J.S.; Savio, J.; Rudramurthy, S.; Singh, R.; Shastri, P.; Umabala, P.; Sardana, R.; et al. A multicentre observational study on the epidemiology, risk factors, management and outcomes of mucormycosis in India. Clin. Microbiol. Infect. 2020, 26, 944e9–944e15. [Google Scholar] [CrossRef]
- Erami, M.; Mirhendi, H.; Momen-Heravi, M.; Hezaveh, S.J.H.; Ahsaniarani, A.H.; Sabet, A.S.; Aboutalebian, S. A case of COVID-19-associated rhino-orbito-cerebral mucormycosis caused by Apophysomyces variabilis with a review of the literature. Front. Cell. Infect. Microbiol. 2022, 12, 898477. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Kumar, P.; Padhye, A.A.; Chatha, L.; Singh, S.K.; Das, A.; Wig, J.D.; Kataria, R.N. Primary cutaneous zygomycosis due to Saksenaea vasiformis and Apophysomyces elegans. Clin. Infect. Dis. 1997, 24, 580–582. [Google Scholar] [CrossRef]
- Pamidimukkala, U.; Sudhaharan, S.; Kancharia, A.; Vemu, L.; Challa, S.; Karanam, S.D.; Chavali, P.; Prakash, H.; Ghosh, A.K.; Gupta, S.; et al. Mucormycosis due to Apophysomyces species complex – 25 years’ experience at a tertiary care hospital in southern India. Med. Mycol. 2020, 58, 425–433. [Google Scholar] [CrossRef]
- Singh, S.; Kanaujia, R.; Kumar, M.B.; Naga Santhosh Irrinki, R.N.; Satish, S.N.; Choudhary, H.; Kaur, H.; Rudramurthy, S.M. Saksenaea vasiformis infection: extensive abdominal necrotizing fasciitis with systemic review and analysis of 65 cases. Mycoses 2023, 66, 697–704. [Google Scholar] [CrossRef]
- Sykes, B.A.; Krause, H.; Lamparelli, M.; Austin, D. Saksenaea mucormycosis: a rare and dangerous cause of necotising fasciitis. BMJ Case Rep. 2023, 16, e254183. [Google Scholar] [CrossRef] [PubMed]
- Landré, V.; Klingebiel, F.K-L.; van Niftrik, C.H.B.; Goetze, E.; Speck, R.F.; Hübner, C.T.; Pape, H-C.; Schäfer, F.P. Mucormycosis caused by Apophysomyces elegans—a case report and systematic review of the literature of rhino-orbital-cerebral cases of the genus Apophysomyces. J. Fungi 2025, 11, 368.
- Skiada, A.; Drogari-Apiranthitou, M.; Roilides, E.; Chander, J.; Khostelidi, S.; Klimko, N.; Hamal, P.; Chrenkova, V.; Kanj, S.S.; El Zein, S.; et al. A global analysis of cases of mucormycosis recorded in the European Confederation of Medical Mycology / International Society for Human and Animal Mycology (ECMM / ISHAM) Zygomyco.net registry from 2009 to 2022. Mycopathologia 2025, 190, 53. [Google Scholar] [CrossRef]
- Prakash, H.; Ghosh, A.K.; Rudramurthy, S.M.; Paul, R.A.; Guota, S.; Negi, V.; Chakrabarti, A. The environmental source of emerging Apophysomyces variabilis infection in India. Med. Mycol. 2016, 54, 567–575. [Google Scholar] [CrossRef]
- Li, X-Y.; Wang, S-J.; Huang, G-M.; Zheng, D-Y.; Al-Odaini, N.; Pan, K-S.; Zheng, Y-Q.; Cao, C-W. Apophysomyces variabilis as an emerging pathogen in mainland China. Mycoses 2023, 66, 795-800.
- Liang, K.P.; Tleyjeh, I.M.; Wilson, W.R.; Roberts, G.D.; Temesgen, Z. Rhino-orbitocerebral mucormycosis caused by Apophysomyces elegans. J. Clin. Microbiol. 2006, 44, 892–898. [Google Scholar] [CrossRef]
- Vitrat-Hincky, V.; Lebeau, B.; Bozonnet, E.; Falcon, D.; Pradel, P.; Faure, O.; Aubert, A.; Piolat, C.; Grillot, R.; Pelloux, H. Severe filamentous fungal infections after widespread tissue damage due to traumatic injury: six cases and review of the literature. Scand. J. Infect. Dis. 2009, 41, 491–500. [Google Scholar] [CrossRef]
- DeLa Cruz, W.P.; Calvano, T.P.; Griffith, M.E.; White, C.E.; Kim, S.H.; Sutton, D.A.; Thompson, E.H.; Fu, J.; Wickes, B.L.; Guarro, J.; et al. Invasive Apophysomyces variabilis infection in a burn patient. J. Clin. Microbiol. 2012, 50, 2814–2817. [Google Scholar] [CrossRef]
- Lelievre, L.; Garcia-Hermosa, D.; Abdoul, H.; Hivelin, M.; Chouaki, T.; Toubas, D.; Mamez, A-C.; Lantieri, L.; Lortholary, O.; Lanternier, F. Posttraumatic mucormycosis. Medicine 2014, 93, 395-404.
- Narayanan, S.; Narayanan, C.D.; Kindon, A.J.; Arora, A.; Haridas, P.A. Fatal fungal infection: the living dead. J. Surg. Case Rep. 2014, 2014, rju104. [Google Scholar] [CrossRef]
- Shankarappa, V.K.G.; Shivappa, S.G.; Puttaswamy, M.C.; Ramakrishna, R.; Ramalingaiah, R.; Puranik, V. Apophysomyces variabilis a flesh-eating fungus – a case report. IOSR J. Dent. Med. Sci. 2014, 13, 86–90. [Google Scholar] [CrossRef]
- Coronel-Pérez, I.; Rodríguez-Rey, E.; Castilla-Guerra, L.; Dominguez, M. Primary cutaneous mucormycosis due to Saksenaea vasiformis in an immunocompetent patient. Actas Dermosifiliogr. 2015, 106, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.Y.; Morales-López, S.E.; Rodríguez, G.J.; Álvarez-Moreno, C.A.; Ocampo, W.; Cepeda, M.L.; Mora-Valderrama, M.A. Necrotizing fasciitis caused by Apophysomyces variabilis in an immunocompetent patient. Med. Mycol. Case Rep. 2018, 20, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Samaddar, A.; Sharma, A.; Maurya, V.K.; Tak, V. Necrotizing fasciitis caused by Apophysomyces variabilis. IDCases 2019, 18, e00660. [Google Scholar] [CrossRef]
- Etienne, K.A.; Gillece, J.; Hilsabeck, R.; Schupp, J.M.; Colman, R.; Lockhart, S.R.; Gade, L.; Thompson, E.H.; Sutton, D.A.; Neblett-Fanfair, R.; et al. Whole genome sequence typing to investigate the Apophysomyces outbreak following a tornado in Joplin, Missouri, 2011. PLoS ONE 2012, 7, e49989. [Google Scholar] [CrossRef] [PubMed]
- Warketien, T.; Rodriguez, C.; Lloyd, B.; Wells, J.; Weintrob, A.; Dunne, J.R.; Ganesan, A.; Li, P.; Bradley, W.; Gaskins, L.J.; et al. Invasive mold infections following combat-related injuries. Clin. Infect. Dis. 2012, 55, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.L.; Finley, P.J.; Mikkleson, D.R.; Tibbs, B. Mucormycosis: a rare fungal infection in tornado victims. J. Burn Care Res. 2014, 35, e164–e171. [Google Scholar] [CrossRef] [PubMed]
- Kronen, R.; Liang, S.Y.; Bochicchio, G.; Bochicchio, K.; Powderly, W.G.; Spec, A. Invasive fungal infections secondary to traumatic injury. Int. J. Infect. Dis. 2017, 62, 102–111. [Google Scholar] [CrossRef]
- Walsh, T.J.; Hospenthal, D.R.; Petraitis, V.; Kontoyiannis, D.P. Necrotizing mucormycosis of wounds following combat injuries, natural disasters, burns, and other trauma. J. Fungi 2019, 5, 57. [Google Scholar] [CrossRef]
- Echaiz, J.F.; Burnham, C-A.; Bailey, T.C. A case of Apophysomyces trapeziformis nectrotizing soft tissue infection. Int. J. Infect. Dis. 2013, 17, e1240-e1242.
- Bertumen, J.B.; Schell, W.A.; Joyce, M.; Alley, C.; Woods, C.W. Diagnostic difficulty identifying Apophysomyces trapeziformis septic arthritis in a patient with multiple myeloma. JMM Case Rep. 2016, 3, e005075. [Google Scholar] [CrossRef]
- Ellis, J.J.; Ajello, L. An unusual source for Apophysomyces elegans and a method for stimulating sporulation of Saksenaea vasiformis. Mycologia 74, 144-145.
- Padhye, A.A.; Ajello, L. Simple method of inducing sporulation by Apophysomyces elegans and Saksenaea vasiformis. J. Clin. Microbiol. 1988, 26, 1861–1863. [Google Scholar] [CrossRef]
- Chander, J.; Stchigel, A.M.; Alastruey-Izquierdo, A.; Jayant, M.; Bala, K.; Rani, H.; Handa, U.; Punia, R.S.; Dalal, U.; Attri, A.K.; et al. Fungal necrotizing fasciitis, an emerging infectious disease caused by Apophysomyces (Mucorales). Rev. Iberoam. Micol. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Labuda, R.; Bernreiter, A.; Hochenauer, D.; Schüller, C.; Kubátová, A.; Strauss, J.; Wagner, M. Saksenaeae dorisiae sp. nov., a new opportunistic pathogenic fungus from Europe. Int. J. Microbiol. 2019, 6253829.
- Planegger, A.; Uyulmaz, S.; Poskevicius, A.; Zbinden, A.; Muller, N.J.; Clacagni, M. Cutaneous invasive fungal infections with Saksenaea species in immunocompetent patients in Europe: a systematic review and case report. Plast. Reconstr. Surg. Glob. Open. 2022, 10, e4230. [Google Scholar] [CrossRef]
- Shao, J.; Wan, Z.; Li, R.; Yu, J. Species identification and delineation of pathogenic Mucorales by Matrix-Assisted Laser Desorption Ionisation-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2018, 56, e01886–17. [Google Scholar] [CrossRef]
- Chander, J.; Singla, N.; Kaur, M.; Punia, R.P.S.; Attri, A.K.; Alastruey-Izquierdo, A.; Stchigel, A.; Cano-Lira, J.F.; Guarro, J. Apophysomyces variabilis, an emerging and worrisome cause of primary cutaneous necrotizing infections in India. J. Med. Mycol. 2021, 31, 101197. [Google Scholar] [CrossRef]
- Jain, D.; Kumar, Y.; Vasishta, R.K.; Rajesh, L.; Pattari, S.K.; Chakrabarti, A. Zygomycotic necrotizing fasciitis in immunocompetent patients: a series of 18. Mod. Pathol. 2006, 19, 1221–1226. [Google Scholar] [CrossRef]
- Thornton, C.R. The potential for rapid antigen testing for mucormycosis in the context of COVID-19. Expert Rev. Mol. Diagn. 2024, 3, 161–167. [Google Scholar] [CrossRef]
- Davies, G.E.; Thornton, C.R. Development of a monoclonal antibody and a serodiagnostic lateral-flow device specific to Rhizopus arrhizus (Syn. R. oryzae), the principal global agent of mucormycosis in humans. J. Fungi 2022, 8, 756. [Google Scholar]
- Thornton, C.R.; Davies, G.E.; Dougherty, L. Development of a monoclonal antibody and a lateral-flow device for the rapid detection of a Mucorales-specific biomarker. Front. Cell. Infect. Microbiol. 2023, 13, 1305662. [Google Scholar] [CrossRef] [PubMed]
- Hudson, A.C.; Corzo-Léon, D.E.; Kalinina, I.; Wilson, D.; Thornton, C.R.; Warris, A.; Ballou, E.R. Characterization of the spatiotemporal localization of a pan-Mucorales-specific antigen during germination and immunohistochemistry. J. Infect. Dis. 2025, 231, e244–e253. [Google Scholar] [CrossRef]
- Rousselot, J.; Millon, L.; Scherer, E.; Bourgeois, N.; Imbert, S.; Dupont, D.; Debourgogne, A.; Maubon, D.; Bellanger, A.P.; Thornton, C.R. Detection of Mucorales antigen in bronchoalveolar lavage samples using a newly developed lateral-flow device. J. Clin. Microbiol. 2025, 63, e00226–25. [Google Scholar] [CrossRef]
- Thornton, C.R. Immunological Methods for Fungi. In Molecular and Cellular Biology of Filamentous Fungi. In Molecular and Cellular Biology of Filamentous Fungi: A Practical Approach, 1st ed.; Talbot, N.J., Ed.; Oxford University Press: Oxford, UK, 2001; pp. 227–256. [Google Scholar]
- Mah, J.-E.; Yu, J.-H. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Euk. Cell 2006, 5, 1585–1595. [Google Scholar] [CrossRef]
- Thornton, C.R. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin. Vaccine Immunol. 2008, 15, 1095–1105. [Google Scholar] [CrossRef]
- Thornton, C.R. Detection of the ‘big five’ mold killers of humans: Aspergillus, Fusarium, Lomentospora, Scedosporium and Mucormycetes. Adv. Appl. Microbiol. 2020, 110, 1–61. [Google Scholar]
- Roden, M.M.; Zaoutis, T.E.; Buchanan, W.I.; Knudsen, T.A.; Sarkisova, T.A.; Schaufer, R.L.; et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 2005, 41, 634–653. [Google Scholar] [CrossRef]
- Graves, B.; Morrissey, C.O.; Wei, A.; Coutsouvelis, J.; Ellis, S.; Pham, A.; Gooi, J.; Ananda-Rajah, M. Isavuconazole as salvage therapy for mucormycosis. Med. Mycol. Case Rep. 2016, 11, 36–39. [Google Scholar] [CrossRef]
- Pham, D.; Howard-Jones, A.R.; Sparks, R.; Stefani, M.; Sivalingham, V.; Halliday, C.L.; Beardsley, J.; Chen, C.-A. Epidemiology, modern diagnostics, and the management of Mucorales infections. J. Fungi 2023, 9, 659. [Google Scholar] [CrossRef] [PubMed]
- Caramalho, R.; Tyndall, J.D.A.; Monk, B.C.; Larentis, T.; Lass-Flörl, C.; Lackner, M. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci. Rep. 2017, 7, 15898. [Google Scholar] [CrossRef] [PubMed]
- Skiada, A.; Lanternier, F.; Groll, A.H.; Pagano, L.; Zimmerli, S.; Herbrecht, R.; Lortholary, O.; Petrikkos, G.L. Diagnosis and treatment of mucormycosis in patients with hematological malignancies: Guidelines from the 3rd European Conference on Infections in Leukemia (ECIL 3). Haematologica 2013, 98, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Alastruey-Izquierdo, A.; Castelli, M.V.; Cuesta, I.; Monzon, A.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Activity of Posaconazole and other antifungal agents against Mucorales strains identified by sequencing of internal transcribed spacers. Antimicrob. Agents Chemotherap. 2009, 53, 1686–1689. [Google Scholar] [CrossRef]
- Skiada, A.; Pavleas, I.; Drogari-Apiranthitou, M. Rare fungal infectious agents: a lurking enemy. F1000Res. 2017, 6, 1917. [Google Scholar] [CrossRef]
- Bonifaz, A.; Tirado-Sánchez, A.; Hernández-Medel, M.L.; Kassak, J.J.; Araiza, J.; González, G.M. Mucormycosis with cutaneous involvement. A retrospective study of 115 cases at a tertiary care hospital in Mexico. Australas. J. Dermatol. 2021, 62, 162–167. [Google Scholar] [CrossRef]
- Pedreira-Rincón, J.; Rivas, L.; Comenge, J.; Skouridou, V.; Camprubí-Ferrer, D.; Muñoz, J.; O’Sullivan, C.K.; Chamorro-Garcia, A.; Parolo, C. A comprehensive review of competitive lateral flow assays over the past decade. Lab Chip 2025, 25, 2578–2608. [Google Scholar] [CrossRef]
- Castrejón-Pérez, A.D.; Welsh, E.C.; Miranda, I.; Ocampo-Candiani, J.; Welsh, O. Cutaneous mucormycosis. An. Bras. Dermatol. 2017, 92, 304–311. [Google Scholar] [CrossRef]
- Murray, J.; Lu, Z.A.; Miller, K.; Meadows, A.; Totten, M.; Zhang, S.X. Disseminated aspergillosis and mucormycosis diagnosed at autopsy: a report of two cases of coinfection and a review of the literature. J. Fungi 2023, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Keighley, C.; Wolfe, R.; Lee, W.L.; Slavin, M.A.; Kong, D.C.M.; Chen, S.C.-A. The Epidemiology and clinical manifestations of Mucormycosis: A systematic review and meta-analysis of case reports. Clin. Microbiol. Infect. 2019, 25, 26–34. [Google Scholar] [CrossRef] [PubMed]
- William, A.; Kaur, R.; Rawat, D.; Kandir, N.S.S.; Sharma, A. Necrotizing fasciitis in neonate by Lichtheimia ramosa: a case study. Access Microbiol. 2022, 4, 000327. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S. ; Ramsay, A.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Millon, L.; Herbrecht, R.; Grenouillet, F.; Morio, F.; Alanio, A.; Letscher-Bru, V.; Cassaing, S.; Chouaki, T.; Kauffman-Lacroix, C.; Poirier, P.; et al. Early diagnosis and monitoring of mucormycosis by detection of circulating DNA in serum: retrospective analysis of 44 cases collected through the French Surveillence Network of Invasive Fungal Infections (RESSIF). Clin. Microbiol. Infect. 2016, 22, 810. [Google Scholar] [CrossRef]
- Shen, H.; Cai, X.; Liu, J.; Yan, G.; Ye, Y.; Ding, R.; Wu, J.; Li, L.; Shen, Q.; Ma, Y.; et al. Case report: the clinical utility of metagenomic next-generation sequencing in mucormycosis diagnosis caused by fatal Lichtheimia ramosa infection in pediatric neuroblastoma. Front. Pediatr. 2023, 11, 1130775. [Google Scholar] [CrossRef]
- Gomes, M.Z.R.; Lewis, R.E.; Kontoyiannis, D.P. Mucormycosis caused by unusual Mucormycetes, non-Rhizopus, -Mucor, and -Lichtheimia species. Clin. Microbiol. Rev. 2011, 24, 411–445. [Google Scholar] [CrossRef]
- Alvarez, E.; Stchigel, A.M.; Cano, J.; Sutton, D.A.; Fothergill, A.W.; Chander, J.; Salas, V.; Rinaldi, M.G.; Guarro, J. Molecular phylogenetic diversity of the emerging mucoralean fungus Apophysomyces: proposal of three new species. Rev. Iberoam. Micol. 2010, 27, 80–89. [Google Scholar] [CrossRef]
- Snell, B.; Tavakoli, K. Necrotizing fasciitis caused by Apophysomyces elegans complicating soft tissue and pelvic injuries in a tsunami survivor from Thailand. Plast. Reconst. Surg. 2007, 119, 448–449. [Google Scholar] [CrossRef]
- Bonifaz, A.; Stchigel, A.M.; Guarro, J.; Guevara, E.; Pintos, L.; Sanchis, M.; Cano-Lira, J.F. Primary cutaneous mucormycosis produced by the new species Apophysomyces mexicanus. J. Clin. Microbiol. 2014, 52, 4428–4431. [Google Scholar] [CrossRef]
- Miller, T.J.; Scheckter, C.C.; Watt, A.J. Humeral osteomyelitis with Apophysomyces ossiformis in an immunocompetent patient: a case report. Surg. Case Rep. 2019, 2, 3–3. [Google Scholar]
- Martínez-Herrera, E.; Frías-De-León, M.G.; Julián-Castrejón, A.; Cruz-Benítez, L.; Xicohtencatl-Cortes, J.; Hernández-Castro, R. Rhino-orbital mucormycosis due to Apophysomyces ossiformis in a patient with diabetes mellitus: a case report. BMC Infect. Dis. 2020, 20, 614. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.; Laínez-Arteaga, I.; Galindo-Morales, C.; Acevedo-Almendárez, L.; Aguilar, K.; Valladares, D.; López, M.; Fontecha, G. First molecular identification of three clinical isolates of fungi causing mucormycosis in Honduras. Infect. Dis. Rep. 2022, 14, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Wolkow, N.; Jakobiec, F.A.; Stagner, A.M.; Piantadosi, A.L.; Basgoz, N.; Lefebvre, D. Chronic orbital and calvarial fungal infection with Apophysomyces variabilis in an immunocompetent patient. Clin. Pathol. Rev. 2017, 62, P70–82. [Google Scholar] [CrossRef] [PubMed]
- Pamidimukkala, U.; Sudhaharan, S.; Kancharia, A.; Vemu, L.; Challa, S.; Karanam, S.D.; Chavali, P.; Prakash, H.; Ghosh, A.K.; Gupta, S.; et al. Mucormycosis due to Apophysomyces species complex – 25 years’ experience at a tertiary care hospital in southern India. Med. Mycol. 2020, 58, 425–433. [Google Scholar] [CrossRef]
- Hospental, D.R.; Chung, K.K.; Lairet, K.; Thompson, E.H.; Guarro, J.; Renz, E.M.; Sutton, D.A. Saksenaea erythrospora infection following combat trauma. J. Clin. Microbiol. 2011, 49, 3707–3709. [Google Scholar] [CrossRef]
- Relloso, S.; Romano, V.; Landaburu, M.F.; Herrera, F.; Smayevsky, J.; Veciño, C. , Mujica, M.T. Saksenaea erythrospora infection following a serious sailing accident. J. Med. Microbiol. 2014, 63, 317–321. [Google Scholar]
- Rodríguez, J.Y.; Rodríguez, G.J.; Morales-López, S.E.; Cantillo, C.E.; Le Pape, P.; Álvarez-Moreno, C.A. Saksenaea erythropora infection after medical tourism for esthetic breast augmentation surgery. Int. J. Infect. Dis. 2016, 49, 107–110. [Google Scholar] [CrossRef]
- Chander, J.; Singla, N.; Kaur, M. , Punia, R.S.; Attri, A.; Alastruey-Izquierdo, A.; Cano-Lira, J.F.; Stchigel, A.M.; Guarro, J. Saksenaea erythrospora, an emerging mucoralean fungus causing severe necrotizing skin and soft tissue infections – a study from a tertiary care hospital in north India. Infect. Dis. 2017, 49, 170–177. [Google Scholar]
- Tendolkar, U.; Baradkar, V.; Baveja, S.; Gore, M.; Bankar.; Gore, S. Cutaneous zygomycosis due to Saksenaea vasiformis in a patient with paraparesis, burns and pressure ulcer. Int. J. Inf. Dis. 2011, 10, 6.
- Neblett-Fanfair, R.; Benedict, K.; Bos, J.; Bennett, S.D.; Lo, Y-C.; Adebanjo, T.; Etienne, K.; Deak, E.; Derado, G.; Shieh, W-J.; et al. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N. Engl. J. Med. 2012, 367, 2214-2225.
| Species | Isolate Number | Source1 |
JD4 Direct-ELISA Absorbance (450nm)2 |
|||
| Apophysomyces elegans | 477.78 | CBS | 1.466±0.005 | |||
| Apophysomyces mexicanus | 136361 | CBS | 1 1.224±0.006 | |||
| Apophysomyces ossiformis | 125533 | CBS | 0.674±0.018 | |||
| Apophysomyces variabilis | 658.93 | CBS | 1.646±0.022 | |||
| Aspergillus fumigatus | Af293 | FGSC | 0.082±0.003 | |||
| Cunninghamella bertholletiae | 151.80 | CBS | 0.084±0.011 | |||
| Fusarium solani | 224.34 | CBS | 0.049±0.000 | |||
| Lichtheimia corymbifera | 109940 | CBS | 0.093±0.002 | |||
| Lichtheimia ornata | 142195 | CBS | 0.083±0.000 | |||
| Lichtheimia hyalospora | 146576 | CBS | 0.073±0.002 | |||
| Lichtheimia ramosa | 112528 | CBS | 0.059±0.003 | |||
| Lichtheimia ramosa | 124197 | CBS | 0.071±0.005 | |||
| Lichtheimia ramosa | 2845 | NCPF | 0.067±0.002 | |||
| Lomentospora prolificans | 3.1 | CRT | 0.056±0.001 | |||
| Mucor circinelloides | B-52 | CRT | 0.050±0.001 | |||
| Mucor racemosus f. racemosus | 222.81 | CBS | 0.076±0.003 | |||
| Rhizomucor pusillus | 120587 | CBS | 0.073±0.002 | |||
| Rhizopus arrhizus | 111233 | CBS | 0.050±0.001 | |||
| Rhizopus arrhizus var. arrhizus | 112.07 | CBS | 0.044±0.002 | |||
| Rhizopus microsporus var. rhizopodiformis | 102277 | CBS | 0.039±0.005 | |||
| Saksenaea dorisiae | 146728 | CBS | 0.044±0.001 | |||
| Saksenaea erythrospora | 138279 | CBS | 0.044±0.002 | |||
| Saksenaea loutrophoriformis | 143036 | CBS | 0.045±0.006 | |||
| Saksenaea vasiformis | 113.96 | CBS | 0.056±0.001 | |||
| Scedosporium apiospermum | 8353 | CBS | 0.045±0.001 | |||
| Scedosporium aurantiacum | 121926 | CBS | 0.062±0.003 | |||
| Scedosporium boydii | 835.96 | CBS | 0.057±0.003 | |||
| Species | Isolate Number | Source1 |
TG11-LFD (a.u.)2 |
| Apophysomyces elegans | 477.78 | CBS | 34.5±1.5 |
| Apophysomyces mexicanus | 136361 | CBS | 27.8±0.1 |
| Apophysomyces ossiformis | 125533 | CBS | 30.8±0.3 |
| Apophysomyces variabilis | 658.93 | CBS | 15.5±4.5 |
| Aspergillus fumigatus ΔAfbrlA7 | A1176 | FGSC | 474.0±19.0 |
| Lichtheimia ramosa3 | 226533346 | DRH | 15.7±1.5 |
| Saksenaea dorisiae | 146728 | CBS | 38.9±0.5 |
| Saksenaea erythrospora | 138279 | CBS | 34.4±1.7 |
| Saksenaea loutrophoriformis | 143036 | CBS | 21.0±2.8 |
| Saksenaea vasiformis | 113.96 | CBS | 22.5±1.0 |
| Negative Control4 | Af293 | FGSC | 591.7±79.9 |
| Positive Control5 | 109940 | CBS | 29.5±2.2 |
| RB only | - | - | 506.9±20.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





