Submitted:
07 September 2025
Posted:
08 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
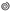 m2), with a limited number of molecular species (<10) and copy numbers (typically, <100K). Monte-Carlo driven simulations of significantly larger scale, such as MCell3 [14] and MCell4 [15], have been developed and can potentially serve for broader, and more detailed description of the IS. Still, these simulations have not been designed so far to dynamically modify the simulated surfaces in response to molecular interactions. Thus, such simulations cannot readily capture the complex dynamics of signaling and cell reorganization that occur at the immune synapse and enable T cell decision-making.
m2), with a limited number of molecular species (<10) and copy numbers (typically, <100K). Monte-Carlo driven simulations of significantly larger scale, such as MCell3 [14] and MCell4 [15], have been developed and can potentially serve for broader, and more detailed description of the IS. Still, these simulations have not been designed so far to dynamically modify the simulated surfaces in response to molecular interactions. Thus, such simulations cannot readily capture the complex dynamics of signaling and cell reorganization that occur at the immune synapse and enable T cell decision-making.2. Results
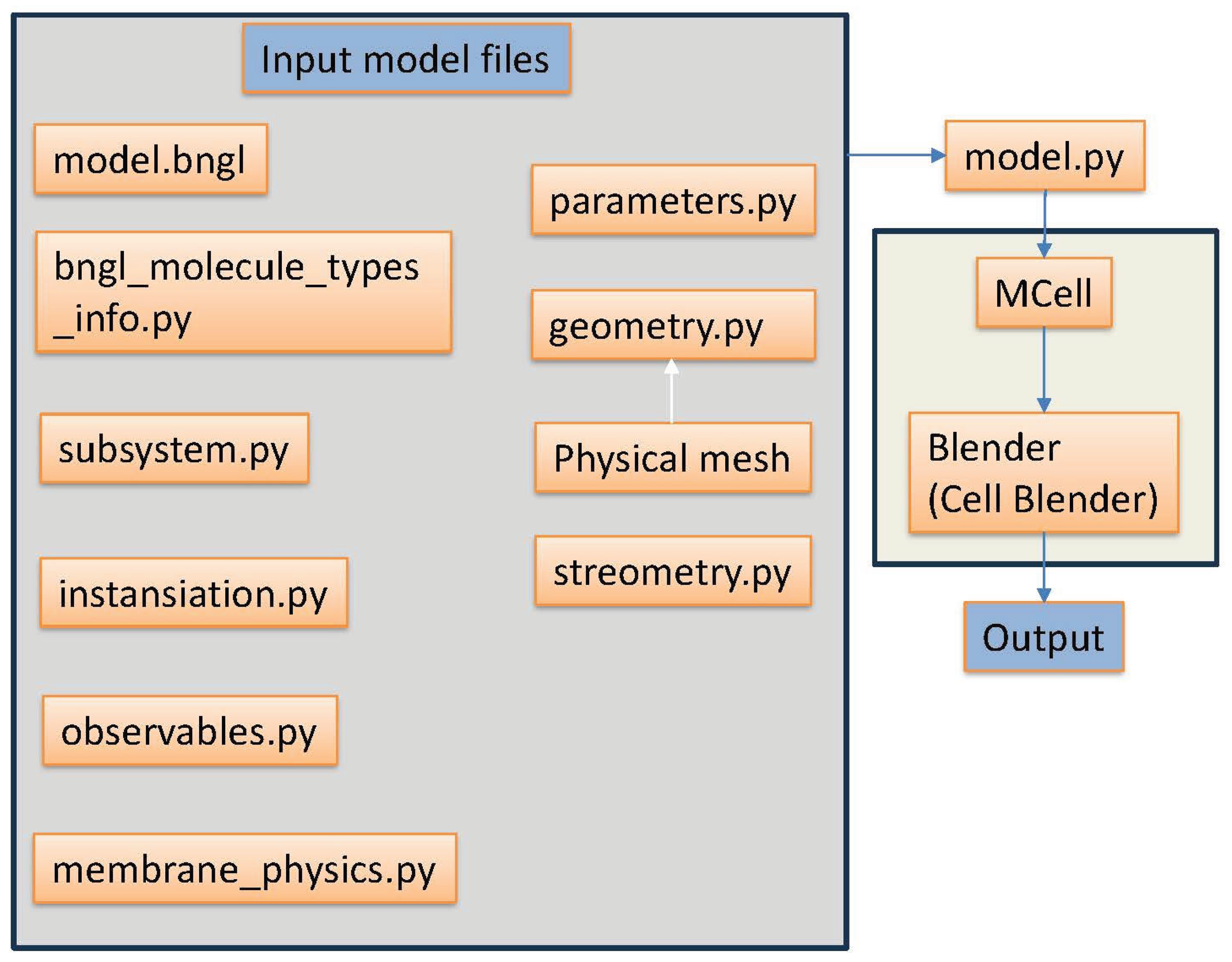
2.1. Our Approach for Large-Scale Monte-Carlo Simulations with Added Forces
2.2. Integration of Multiple Force Types into MCell4
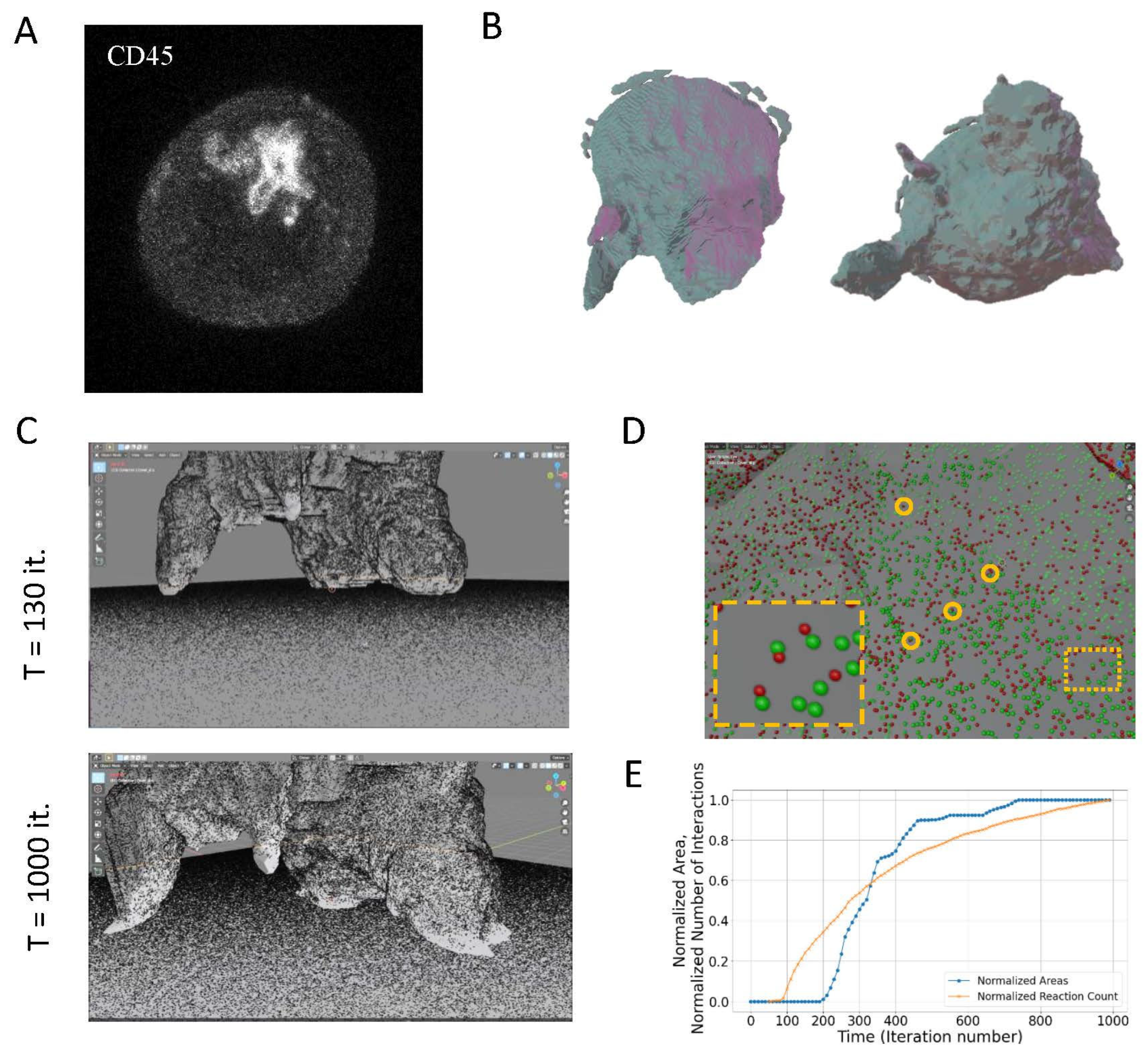
2.3. Simulations of Interacting Cells
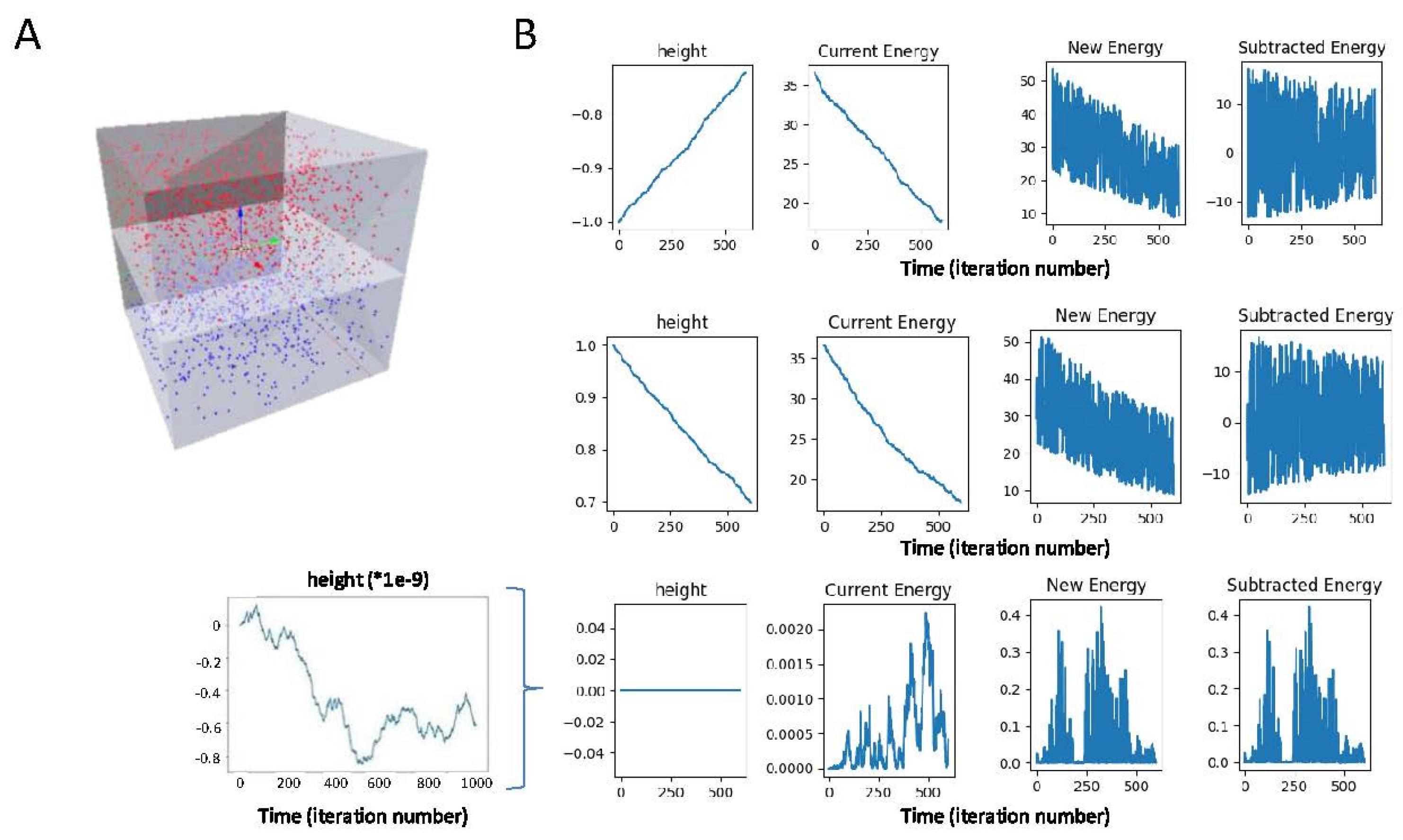
2.4. T Cell Spreading: Towards Realistic Simulation of the Immune Synapse
3. Discussion
4. Methods
4.1. Detailed Molecular Simulation
4.2. Sample Preparation and Confocal Microscopy
Supplementary Materials
Data Availability Statement
Institutional Review Board Statement
Funding
Author Contributions
Acknowledgments
References
- Wilson, J.; Hunt, T. Molecular Biology of the Cell: The Problems Book; Molecular biology of the cell; Garland Pub., 1994. ISBN 9780815316213.
- Cemerski, S.; Shaw, A. Immune Synapses in T-Cell Activation. Curr Opin Immunol 2006, 18, 298–304. [Google Scholar] [CrossRef]
- Monks, C.R.F.; Freiberg, B.A.; Kupfer, H.; Sciaky, N.; Kupfer, A. Three-Dimensional Segregation of Supramolecular Activation Clusters in T Cells. Nature 1998, 395, 82–86. [Google Scholar] [CrossRef]
- Freiberg, B.A.; Kupfer, H.; Maslanik, W.; Delli, J.; Kappler, J.; Zaller, D.M.; Kupfer, A. Staging and Resetting T Cell Activation in SMACs. Nat Immunol 2002, 3, 911–917. [Google Scholar] [CrossRef]
- Hammer, J.A.; Wang, J.C.; Saeed, M.; Pedrosa, A.T. Origin, Organization, Dynamics, and Function of Actin and Actomyosin Networks at the T Cell Immunological Synapse. Annu Rev Immunol 2019, 37, 201–224. [Google Scholar] [CrossRef]
- Dustin, M.L.; Springer, T.A. T-Cell Receptor Cross-Linking Transiently Stimulates Adhesiveness through Lfa-1. Nature 1989, 341, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Springer, T.A.; Dustin, M.L.; Kishimoto, T.K.; Marlin, S.D. The Lymphocyte Function-Associated LFA-1, CD2, and LFA-3 Molecules: Cell Adhesion Receptors of the Immune System. Annu Rev Immunol 1987, 5, 223–252. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Riven, I.; Feigelson, S.W.; Kartvelishvily, E.; Tohya, K.; Miyasaka, M.; Alon, R.; Haran, G. Three-Dimensional Localization of T-Cell Receptors in Relation to Microvilli Using a Combination of Superresolution Microscopies. Proc Natl Acad Sci U S A 2016, 113. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.T.; Fernandes, R.A.; Ganzinger, K.A.; Lee, S.F.; Siebold, C.; McColl, J.; Jönsson, P.; Palayret, M.; Harlos, K.; Coles, C.H.; et al. Initiation of T Cell Signaling by CD45 Segregation at “Close Contacts.”. Nat Immunol 2016, 17. [CrossRef]
- Neve-Oz, Y.; Sajman, J.; Razvag, Y.; Sherman, E. InterCells: A Generic Monte-Carlo Simulation of Intercellular Interfaces Captures Nanoscale Patterning at the Immune Synapse. Front Immunol 2018, 9. [Google Scholar] [CrossRef]
- Weikl, T.R.; Lipowsky, R. Pattern Formation during T-Cell Adhesion. Biophys J 2004, 87, 3665–3678. [Google Scholar] [CrossRef]
- Dharan, N.; Farago, O. Interplay between Membrane Elasticity and Active Cytoskeleton Forces Regulates the Aggregation Dynamics of the Immunological Synapse. Soft Matter 2017, 13, 6938–6946. [Google Scholar] [CrossRef]
- Razvag, Y.; Neve-Oz, Y.; Sajman, J.; Reches, M.; Sherman, E. Nanoscale Kinetic Segregation of TCR and CD45 in Engaged Microvilli Facilitates Early T Cell Activation. Nat Commun 2018, 9, 732. [Google Scholar] [CrossRef]
- Kerr, R.A.; Bartol, T.M.; Kaminsky, B.; Dittrich, M.; Chang, J.C.J.; Baden, S.B.; Sejnowski, T.J.; Stiles, J.R. Fast Monte Carlo Simulation Methods for Biological Reaction-Diffusion Systems in Solution and on Surfaces. Siam Journal on Scientific Computing 2008, 30, 3126–3149. [Google Scholar] [CrossRef]
- Husar, A.; Ordyan, M.; Garcia, G.C.; Yancey, J.G.; Saglam, A.S.; Faeder, J.R.; Bartol, T.M.; Sejnowski, T.J. MCell4 with BioNetGen: A Monte Carlo Simulator of Rule-Based Reaction-Diffusion Systems with Python Interface. bioRxiv 2022, 2022.05.17.492333. [CrossRef]
- Harris, L.A.; Hogg, J.S.; Tapia, J.J.; Sekar, J.A.P.; Gupta, S.; Korsunsky, I.; Arora, A.; Barua, D.; Sheehan, R.P.; Faeder, J.R. BioNetGen 2.2: Advances in Rule-Based Modeling. Bioinformatics 2016, 32, 3366–3368. [Google Scholar] [CrossRef]
- Sajman, J.; Razvag, Y.; Schidorsky, S.; Kinrot, S.; Hermon, K.; Yakovian, O.; Sherman, E. Adhering Interacting Cells to Two Opposing Coverslips Allows Super-Resolution Imaging of Cell-Cell Interfaces. Commun Biol 2021, 4. [Google Scholar] [CrossRef] [PubMed]
- Bunnell, S.C.; Barr, V.A.; Fuller, C.L.; Samelson, L.E. High-Resolution Multicolor Imaging of Dynamic Signaling Complexes in T Cells Stimulated by Planar Substrates. Sci. STKE 2003, 2003, pl8. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Dinet, C.; Dillard, P.; Nassereddine, A.; Puech, P.H.; Limozin, L.; Sengupta, K. Biphasic Mechanosensitivity of T Cell Receptor-Mediated Spreading of Lymphocytes. Proc Natl Acad Sci U S A 2019, 116, 5908–5913. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.; Barr, V.; Manley, S.; Patterson, G.; Balagopalan, L.; Akpan, I.; Regan, C.K.; Merrill, R.K.; Sommers, C.L.; Lippincott-Schwartz, J.; et al. Functional Nanoscale Organization of Signaling Molecules Downstream of the T Cell Antigen Receptor. Immunity 2011, 35, 705–720. [Google Scholar] [CrossRef]
- Lillemeier, B.F.; Mörtelmaier, M.A.; Forstner, M.B.; Huppa, J.B.; Groves, J.T.; Davis, M.M. TCR and Lat Are Expressed on Separate Protein Islands on T Cell Membranes and Concatenate during Activation. Nat Immunol 2010, 11, 90–96. [Google Scholar] [CrossRef]
- Balagopalan, L. *; Sherman, E.*; Barr, V.A.A.; Samelson, L.E.E. Imaging Techniques for Assaying Lymphocyte Activation in Action. Nat Rev Immunol 2011, 11, 21–33. [Google Scholar] [CrossRef]
- Sherman, E. Resolving Protein Interactions and Organization Downstream the T Cell Antigen Receptor Using Single-Molecule Localization Microscopy: A Review. Methods Appl Fluoresc 2016, 4. [Google Scholar] [CrossRef]
- Schamel, W.W.A.A.; Arechaga, I.; Risueño, R.M.; Van Santen, H.M.; Cabezas, P.; Risco, C.; Valpuesta, J.M.; Alarcón, B. Coexistence of Multivalent and Monovalent TCRs Explains High Sensitivity and Wide Range of Response. J Exp Med 2005, 202, 493–503. [Google Scholar] [CrossRef]
- Xu, C.; Gagnon, E.; Call, M.E.; Schnell, J.R.; Schwieters, C.D.; Carman, C. V; Chou, J.J.; Wucherpfennig, K.W. Regulation of T Cell Receptor Activation by Dynamic Membrane Binding of the CD3epsilon Cytoplasmic Tyrosine-Based Motif. Cell 2008, 135, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Alarcón, B.; Alarcon, B. TCR Nanoclusters as the Framework for Transmission of Conformational Changes and Cooperativity. Front Immunol 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Minguet, S.; Swamy, M.; Alarcón, B.; Luescher, I.F.; Schamel, W.W.A.A.; Alarcon, B.; Luescher, I.F.; Schamel, W.W.A.A. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Neve-Oz, Y.; Razvag, Y.; Sajman, J.; Sherman, E. Mechanisms of Localized Activation of the T Cell Antigen Receptor inside Clusters. Biochim Biophys Acta Mol Cell Res 2015, 1853, 810–821. [Google Scholar] [CrossRef]
- Pageon, S. V.; Tabarin, T.; Yamamoto, Y.; Ma, Y.; Bridgeman, J.S.; Cohnen, A.; Benzing, C.; Gao, Y.; Crowther, M.D.; Tungatt, K.; et al. Functional Role of T-Cell Receptor Nanoclusters in Signal Initiation and Antigen Discrimination. Proc Natl Acad Sci U S A 2016, 113, E5454–E5463. [Google Scholar] [CrossRef]
- Razvag, Y.; Neve-Oz, Y.; Sajman, J.; Yakovian, O.; Reches, M.; Sherman, E. T Cell Activation through Isolated Tight Contacts. Cell Rep 2019, 29, 3506-+. [Google Scholar] [CrossRef] [PubMed]
- Razvag, Y.; Neve-Oz, Y.; Sherman, E.; Reches, M. Nanoscale Topography-Rigidity Correlation at the Surface of T Cells. ACS Nano 2019, 13, 346–356. [Google Scholar] [CrossRef]
- van der Merwe, P.A.; Dushek, O. Mechanisms for T Cell Receptor Triggering. Nat Rev Immunol 2011, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.A.; Grünewald, F.; van Tilburg, P.A.M.; König, M.; Gilbert, B.R.; Brier, T.A.; Thornburg, Z.R.; Luthey-Schulten, Z.; Marrink, S.J. Molecular Dynamics Simulation of an Entire Cell. Front Chem 2023, 11, 1–9. [Google Scholar] [CrossRef]
- Raveh, B.; Sun, L.; White, K.L.; Sanyal, T.; Tempkin, J.; Zheng, D.; Bharath, K.; Singla, J.; Wang, C.; Zhao, J.; et al. Bayesian Metamodeling of Complex Biological Systems across Varying Representations. Proc Natl Acad Sci U S A 2021, 118, 1–12. [Google Scholar] [CrossRef]
- Neve-Oz, Y.; Sherman, E.; Raveh, B. Bayesian Metamodeling of Early T-Cell Antigen Receptor Signaling Accounts for Its Nanoscale Activation Patterns. Front Immunol 2024, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Maritan, M.; Autin, L.; Karr, J.; Covert, M.W.; Olson, A.J.; Goodsell, D.S. Building Structural Models of a Whole Mycoplasma Cell. J Mol Biol 2022, 434, 167351. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Litvinov, S.; Koumoutsakos, P. Bending Models of Lipid Bilayer Membranes: Spontaneous Curvature and Area-Difference Elasticity. Comput Methods Appl Mech Eng 2020, 359, 112758. [Google Scholar] [CrossRef]
- Maksimova, N.A.; Garrison, L.H.; Eisenstein, D.J.; Hadzhiyska, B.; Bose, S.; Satterthwaite, T.P. A BACUS S UMMIT : A Massive Set of High-Accuracy, High-Resolution N -Body Simulations 1 I N T RO D U C T I O N 2 A BAC U S S U M M I T : S U I T E S P E C I F I C AT I O N S. 2021, 4037, 4017–4037.
- Helfrich, W. Elastic Properties of Lipid Bilayers: Theory and Possible Experiments. Zeitschrift fur Naturforschung - Section C Journal of Biosciences 1973, 28, 693–703. [Google Scholar] [CrossRef]
| Total num. of molecules* | Set time | Simulation speed [iterations/sec] |
|---|---|---|
| 4K | 126.6 seconds | 0.9 |
| 100K | 2.35 hours | 0.161 |
| 500K | 65 hours | 0.038 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




