Submitted:
26 August 2025
Posted:
28 August 2025
You are already at the latest version
Abstract
Taxonomic clarity within the genus Antithamnion is important in the molecular phylogeny of red algae. Antithamnion hubbsii is reported for the first time from the Korean coast, previously misidentified as A. nipponicum due to morphological similarities. Specimens collected from Gangneung were analyzed using plastid-encoded rbcL and psaA genes, confirming their identity as A. hubbsii. Korean specimens exhibit indeterminate lateral axes, oppositely arranged pinnae and pinnules, and distinctive gland cells on the adaxial surface of pinnules. Molecular analyses revealed minimal genetic divergence between the two species (1-3 base pair differences in rbcL, none in psaA). While PTP analysis differentiated them as distinct species, ASAP and ABGD analyses grouped them as a single species. Despite these results, our phylogenetic analyses showed Korean A. hubbsii forming a distinct clade with strong bootstrap support. However, further research is needed to clarify the taxonomic boundaries between A. hubbsii and A. nipponicum. Our ecological observations revealed that this North American invasive species competes with both native red algal communities and established non-natives, apparently outcompeting A. nipponicum in shared habitats. This suggests potential shifts in community composition and ecological functions along the Korean coast as this invader spreads.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling, Culture and Microscopy
2.2. Molecular Analysis
3. Results
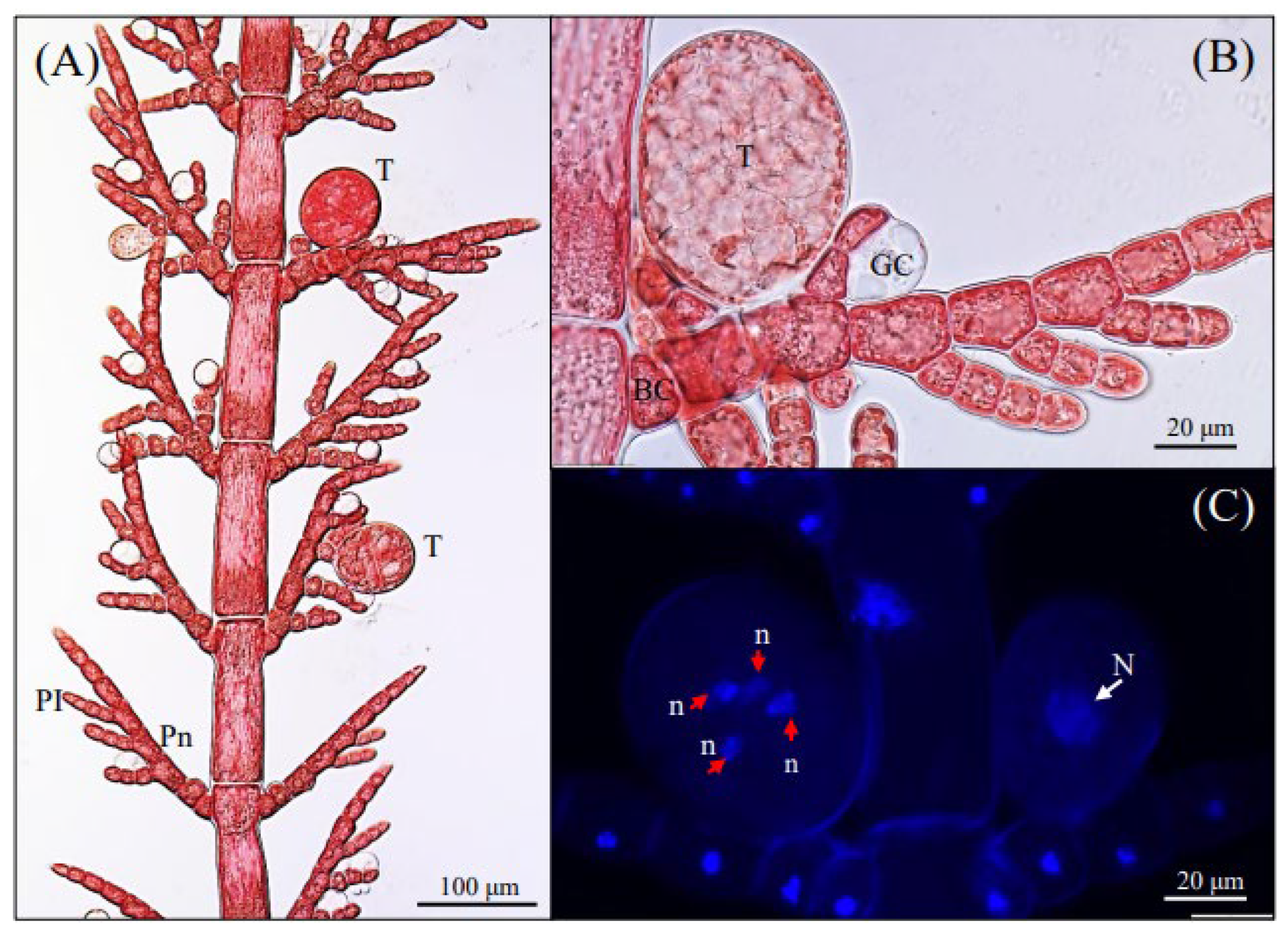
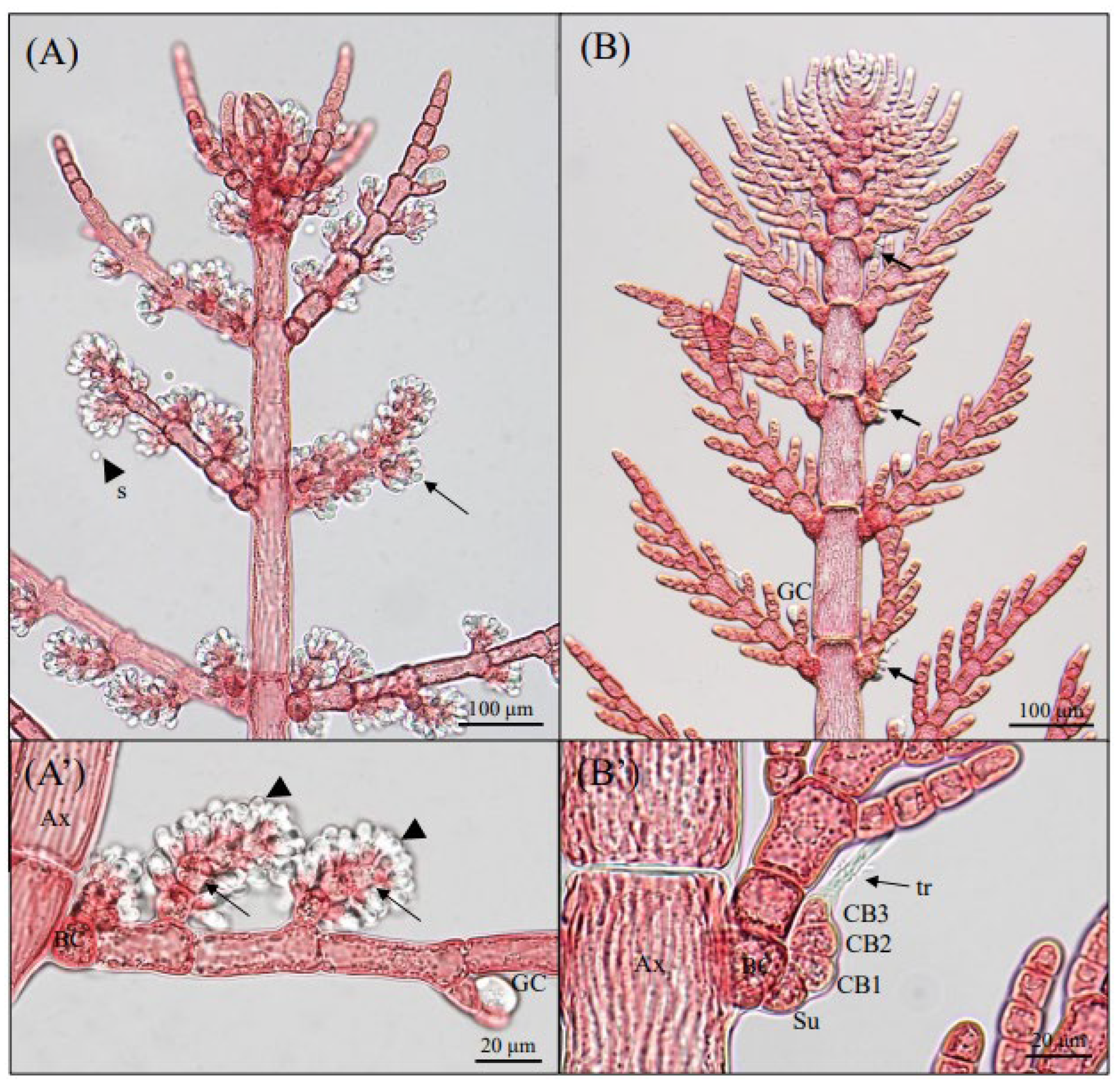
3.1. Morphological Observations
| Species | Gland cells | Indeterminae laterals | Pinnae | Branching plane | Tetrasporangia |
|---|---|---|---|---|---|
| New collection, Antithamnion hubsii Shim & Kim, 2024 |
Borne on pinnae, adaxial, covering 2 cells | Paired | Pinnate (diploid) Adaxially pectinate (haploid) |
Distichous | 1-cell pedicel |
| Antithamnion hubsii Dawson, 1963 | Borne on pinnae, adaxial, covering 2 cells | Paired | Pinnate | Distichous | NA |
|
Antithamnion nipponicum Yamada et Inagaki |
Borne on pinnae, adaxial, covering 2 cells | Paired | Pinnate | Distichous | 1-cell pedicel |
| Antithamnion cruciatum Nägeli, 1847 | On pinnae, covering 2 cells | Paired w/ determinate lateral | Branched decussate | Decussate | 1-cell pedicel or sessile, tetrahedral |
| Antithamnion defectum Kylin, 1925 | On pinnae, covering 2 cells | Unpaired | Branched, pectinate | Distichous | Pedicellate, undefined pedicel length |
| Antithamnion densum Howe, 1914 | On pinnae, covering 2 cells | Paired w/ determinate lateral | Adaxially pectinate | Distichous | 1-cell pedicel |
| Antithamnion sparsum Tokida, 1932 | On pinnae, covering 2 cells | Unpaired | Adaxially pectinate | Distichous | 1-cell pedicel |
| Antithamnionella spirographidis Wollaston, 1968 | On primary branches, partly covering 1 cell | Unpaired | Simple | Distichous | Sessile |
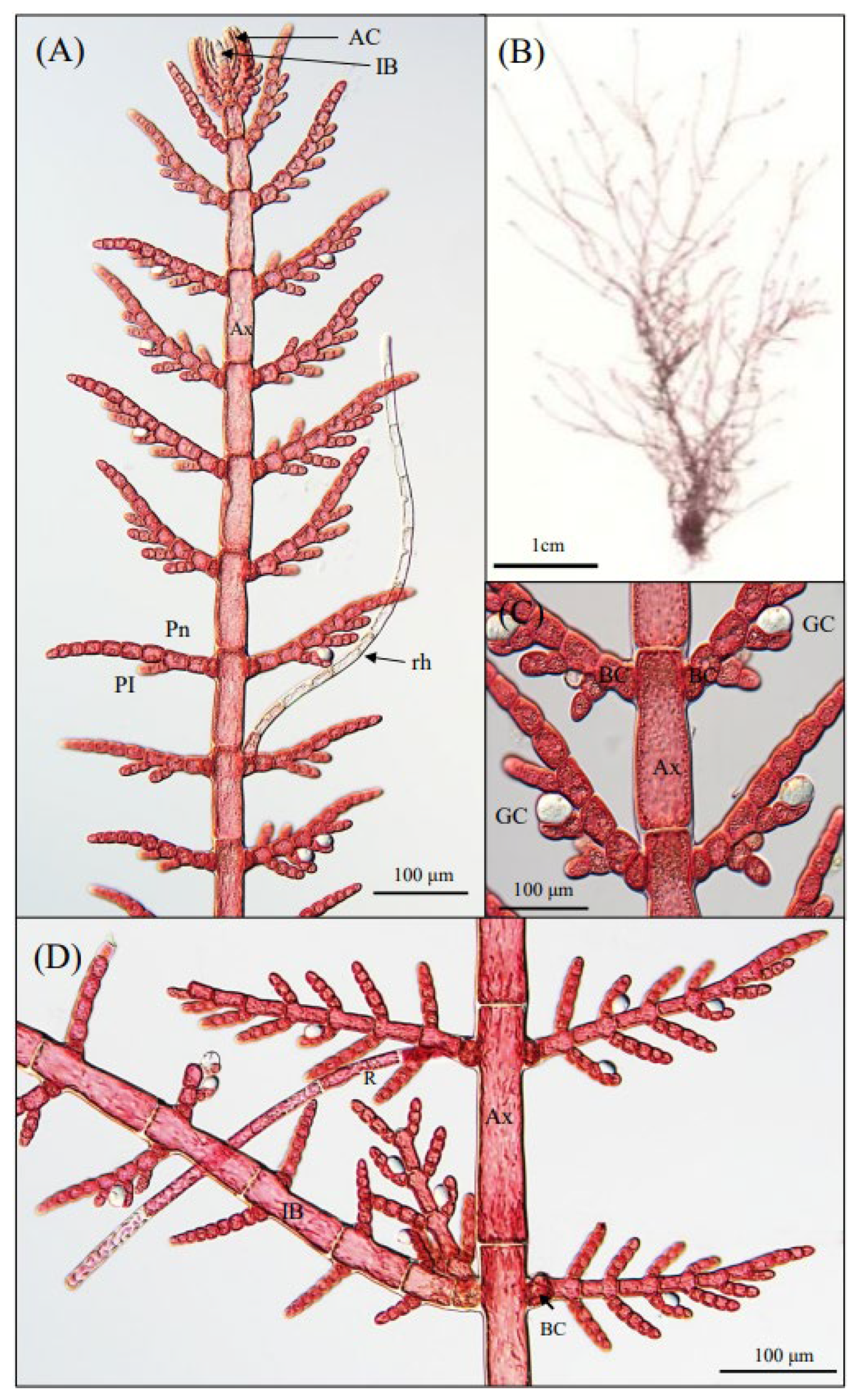
3.2. Phylogenetic Analyses
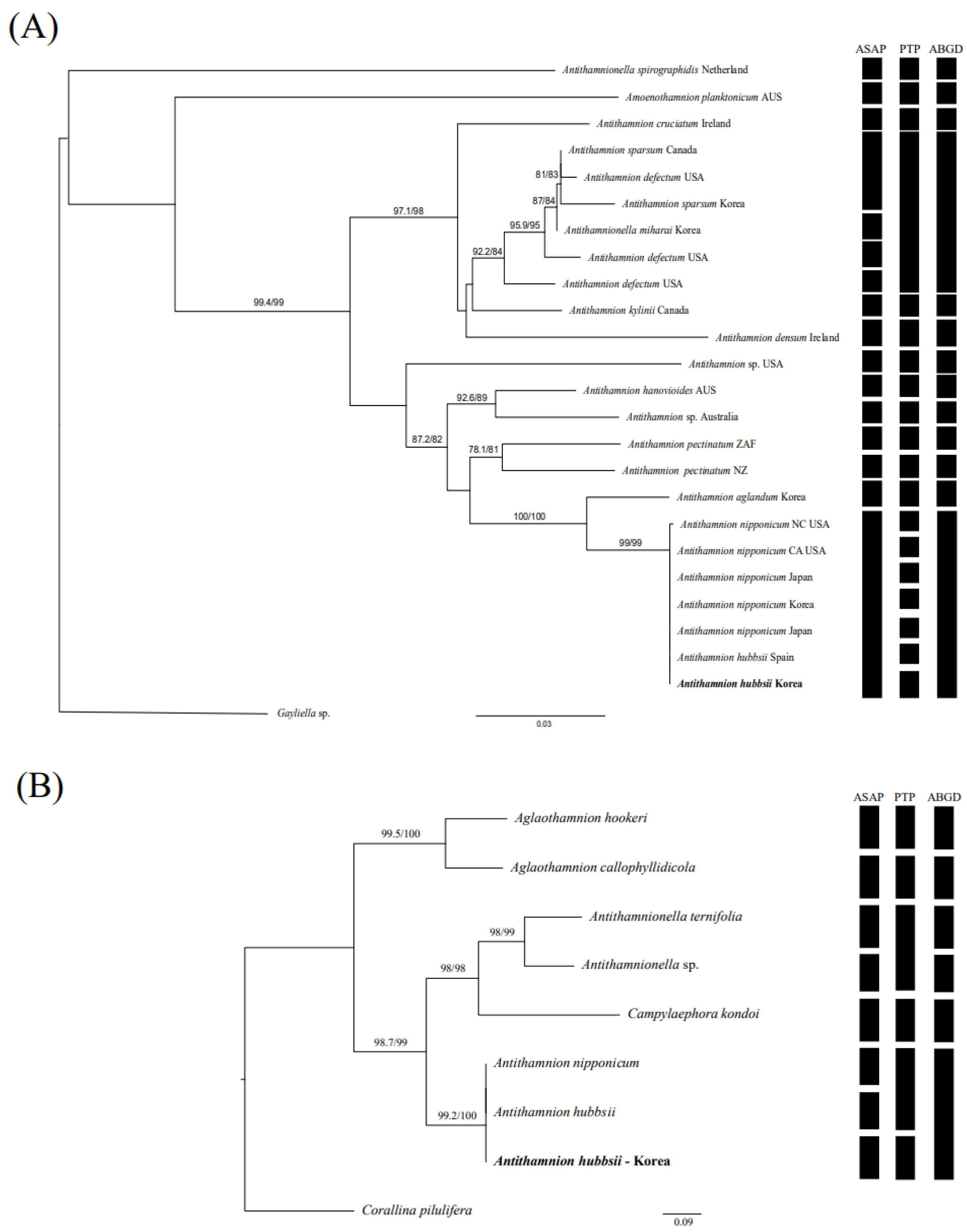
4. Discussion
Acknowledgement
References
- Athanasiadis A and Tittley I. 1994. Antithamnioid algae (Rhodophyta, Ceramiaceae) newly recorded from the Azores. Phycologia 33, 77-80. [CrossRef]
- Brooks CM and Krumhansl K. 2023. First record of the Asian Antithamnion sparsum Tokida, 1932 (Ceramiales, Rhodophyta) in Nova Scotia, Canada. BioInvasions Records 12:745-752.
- Carlton JT and Geller JB. 1993. Ecological roulette: The global transport of nonindigenous marine organisms. Science 261:78-82.
- Chernomor O, von Haeseler A and Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65:997-1008. [CrossRef]
- Cho TO, Boo SM, Hommersand MH, Maggs CA, McIvor L and Fredericq S. 2005. Gayliella gen. nov. in the tribe Ceramieae (Ceramiaceae, Rhodophyta) based on molecular and morphological evidence. Journal of Phycology 41:726-738. [CrossRef]
- Cho TO, Won BY and Fredericq S. 2007. Antithamnion nipponicum (Ceramiaceae, Rhodophyta), incorrectly known as A. pectinatum in western Europe, is a recent introduction along the North Carolina and Pacific coasts of North America. European Journal of Phycology 40:323-335. [CrossRef]
- Dawson EY. 1925. Studies of intertidal algae at Pacific Grove, California. Bulletin of the California Academy of Sciences 24:31-59.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791.
- Freshwater DW, Tudor K, O’Shaughnessy K and Wysor B. 2010. DNA barcoding in the red algal order Gelidiales: Comparison of COI with rbcL and verification of the “barcoding gap”. Cryptogamie, Algologie 31:435-449.
- Hommersand MH, Freshwater DW, Lopez-Bautista JM and Fredericq S. 2009. Proposal of the Euptiloteae Hommersand et Fredericq, trib. nov. and transfer of some southern hemisphere Ptiloteae to the Callithamnieae (Ceramiaceae, Rhodophyta). Journal of Phycology 45:1065-1080. [CrossRef]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A and Jermiin LS. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14:587-589. [CrossRef]
- Kang JH, Jang JE, Kim JH and Kim MS. 2020. DNA barcoding of the genus Grateloupia (Rhodophyta) from Korea: An assessment of DNA barcoding markers for species identification. Diversity 12:3.
- Klochkova TA, Kang SH, Cho GY, Pueschel CM, West JA and Kim GH. 2006. Biology of a terrestrial green alga, Chlorococcum sp. (Chlorococcales, Chlorophyta), collected from the Miruksazi stupa in Korea. Phycologia 45:349-358. [CrossRef]
- Leliaert F, Verbruggen H, Vanormelingen P, Steen F, López-Bautista JM, Zuccarello GC and De Clerck O. 2014. DNA-based species delimitation in algae. European Journal of Phycology 49:179-196. [CrossRef]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A and Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37:1530-1534. [CrossRef]
- Nägeli C. 1847. Die neueren Algensysteme und Versuch zur Begründung eines eigenen Systems der Algen und Florideen. Neue Denkschriften der Allgemeinen Schweizerischen Gesellschaft für die Gesammten Naturwissenschaften 9:1-275.
- Olenin S, Alemany F, Cardoso AC, Gollasch S, Goulletquer P, Lehtiniemi M, McCollin T, Minchin D, Miossec L, Occhipinti-Ambrogi A, Ojaveer H, Jensen KR, Stankiewicz M, Wallentinus I and Aleksandrov B. 2011. Marine Strategy Framework Directive – Task Group 2 Report: Non-indigenous species. Luxembourg: Office for Official Publications of the European Communities.
- Payo DA, Leliaert F, Verbruggen H, D’hondt S, Calumpong HP and De Clerck O. 2013. Extensive cryptic species diversity and fine-scale endemism in the marine red alga Portieria in the Philippines. Proceedings of the Royal Society B: Biological Sciences 280:20122660. [CrossRef]
- Puillandre N, Lambert A, Brouillet S and Achaz G. 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21:1864-1877. [CrossRef]
- Rueness J, Heggøy E, Husa V and Sjøtun K. 2007. First report of the Japanese red alga Antithamnion nipponicum on the west coast of Norway. Aquatic Invasions 2:431-434.
- Williams SL and Smith JE. 2007. A global review of the distribution, taxonomy, and impacts of introduced seaweeds. Annual Review of Ecology, Evolution, and Systematics 38:327-359. [CrossRef]
- Yoon HS, Hackett JD and Bhattacharya D. 2002. A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proceedings of the National Academy of Sciences 99:11724-11729. [CrossRef]
- Zhang J, Kapli P, Pavlidis P and Stamatakis A. 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869-2876. [CrossRef]
- Zuccarello GC, Burger G, West JA and King RJ. 1999. A mitochondrial marker for red algal intraspecific relationships. Molecular Ecology 8:1443-1447. [CrossRef]
| Species | Location of collection and/or source of culture; collector or depositor | Date | GeneBank Accession NO. |
|---|---|---|---|
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene | |||
|
Antithamnion aglandum Kim et Lee |
Jeoongdori, Wando, South Korea; S.M. Boo, T.O. Cho & H.G. Choi |
29.01.1999 | AY594700 |
| Antithamnion cruciatum | Mulroy Bay, Co. Donegal, Ireland; C.A. Maggs |
16.02.1993 | JN089394 |
| Antithamnion defectum | Sitka, AK, USA | 21.06.2005 | GO252484 |
| Antithamnion defectum | California, La Jolla, USA; C.A. Maggs |
27.07.1995 | JN089391 |
| Antithamnion defectum | Skellig Rocks, Co. Kerry Ireland; C.A. Maggs |
01.06.1992 | JN089395 |
|
Antithamnion hanovioides (Sonder) De Toni |
Pennington Bay, Kangaroo Island, S. Australia; T.O. Cho & B.Y. Won |
07.09.1995 | AY591927 |
| Antithamnion hubbsii | Spain | 07.05.2019 | MK814610 |
|
Antithamnion hubbsii Shim et Kim |
Sungeut beach, Gangwon-do, South Korea; E. Shim, S.Y. Kim & G.H. Kim |
06.01.2024 | PV453999 |
| Antithamnion kylinii | Canada | JN089393 | |
|
Antithamnion nipponicum Yamada et Inagaki |
Hachinohe, Aomori, Japan; M. Kamiya |
21.05.1995 | AY594699 |
|
Antithamnion nipponicum NC-1 |
In front of Duke University Marine Laboratory, Pivers I, Beaufort, Carteret Co., North Carolina, USA; T.O. Cho & B.Y. Won | 29.10.2003 | AY591928 |
|
Antithamnion nipponicum |
Maengbang, Samcheok, South Korea; E.C. Yang & S.M. Boo |
2016 | AY295174 |
|
Antithamnion nipponicum CA |
Halfmoon Bay, California, USA; T.O. Cho & B.Y. Won |
AY591930 | |
| Antithamnion nipponicum | Aragami, Funakoshi, Yamada, Japan; Masahiro Suzuki |
29.08.2014 | LC821146 |
|
Antithamnion pectinatum (Montagne) Brauner |
Lee Bay, Stewart Island, New Zealand; W.A. Nelson |
03.10.2004 | DQ023481 |
| Antithamnion pectinatum | Kenton On Sea, South Africa; Faith Mshiywa |
24.11.2017 | OR939842 |
| Antithamnion sp. | Mallacoota, VIC, Australia; H. Verbruggen & K. Dixon |
2019 | MK125356 |
| Antithamnion sp. | Ewing Bank, Offshore Louisiana, USA; J.Richards |
29.05.2011 | KY994130 |
|
Antithamnion sparsum Tokida |
Nova Sotia, Canada | 2021 | OP600459 |
| Antithamnion sparsum | Daechon, South Korea; H.G. Cho |
23.04.1992 | JN089392 |
|
Antithamnion ternifolia (Hooker et Harvey) Lyle |
Williamston, Australia; J. West |
28.05.2002 | AY591926 |
| Amoenothamnion planktonicum | WestAP, Australia | OR359632 | |
| Antithamnionella miharai | Youngeumjun, Sokcho, South Korea | 09.05.2006 | GQ252485 |
| Antithamnionella spirographidis | Monterey Bay, California, USA; T.O. Cho |
11.12.1999 | AY591923 |
| Photosystem I P700 apoprotein (psaA) | |||
| Aglaothamnion callophyllidicola | Kamo Bay, Oki Island, Japan; E.C. Yang & S.M. Boo |
07.05.2003 | DQ787601 |
| Aglaothamnion hookeri | Castilo San Cristobal, Canary, Spain | 24.04.2004 | EU195020 |
| Antithamnion hubbsii | MK814610 | ||
|
Antithamnion hubbsii Shim et Kim |
Sungeut beach, Gangwon-do, South Korea; E. Shim, S.Y. Kim & G.H. Kim |
06.01.2024 | PV454032 |
| Antithamnion nipponicum | - | 2019 | AY295136 |
| Antithamnionella sp. | Choshi, Chiba, Japan; E.C. Yang & S.M. Boo |
27.07.2002 | DQ787600 |
| Antithamnionella ternifolia | - | 2019 | MK814608 |
| Campylaephora kondoi | Hakampo, Taean, South Korea; E.C. Yang & S.M. Boo |
2003 | AY295138 |
| Corallina pilulifera | Nagasaki, Choshi, Chiba, Japan; E.C. Yang & S.M. Boo |
01.08.2004 | DQ787594 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





