1. Introduction
Articular hyaline cartilage is an avascular and aneural tissue that covers the bone surfaces of synovial joints, providing resistance to compression and reducing friction during movement. It consists of a single cell population, chondrocytes, embedded in an extracellular matrix (ECM) rich in type II collagen, aggrecan, and other structural proteins essential for its biomechanical function [
1,
2,
3]. However, due to its limited endogenous regenerative capacity, articular cartilage injuries tend to progress toward degenerative diseases such as osteoarthritis, affecting the quality of life of millions of people worldwide [
4,
5]. Currently available treatments, such as microfracture, autologous chondrocyte implantation (ACI), and allografts, provide symptomatic relief but fail to achieve complete cartilage tissue regeneration, thereby limiting their long-term efficacy [
6].
In this context, tissue bioengineering has explored the development of 3D cultures or cartilage organoids derived from stem cells as a promising strategy for articular cartilage repair. Cartilage organoids are three-dimensional structures derived from stem cells that mimic the cellular organization and ECM composition of native cartilage, providing a physiologically relevant model for its study and clinical application [
7,
8]. Nevertheless, questions remain regarding the optimal stem cell type for generating organoids that closely resemble native cartilage and possess in vivo regenerative potential [
9]. In this regard, although both mesenchymal stem cells (MSCs) [
10] and skeletal stem cells (SSCs) [
11] have demonstrated chondrogenic potential. MSCs have been shown to generate hypertrophic chondrocytes or fibrocartilage-producing cells, which are more associated with inflammation and tissue damage than with the formation of native hyaline cartilage [
12,
13,
14]. This observation raises questions about their suitability as a cellular source for cartilage regeneration strategies. Moreover, it remains unclear whether the interaction between these two chondrogenic populations could yield optimized organoids for tissue repair, particularly in the context of the stem cell niche hypothesis [
15,
16]. According to this hypothesis, MSCs primarily function as support cells for a more restricted and tissue-specific stem cell population, in this case, SSCs, which would be directly responsible for cartilage regeneration. Therefore, further characterization of the functional roles of each cell type within the organoid microenvironment is necessary, along with additional studies to determine whether MSC–SSC co-culture enhances the structural stability of the resulting organoids and reduces the risk of hypertrophy and fibrosis in cartilage tissue.
Another critical factor in the formation of functional cartilage is oxygen tension. It has been demonstrated that physiological oxygen levels in articular cartilage (1–5% O
2) promotes chondrogenic differentiation and ECM production characteristic of hyaline cartilage, whereas in vitro normoxia (21% O
2) induces a hypertrophic phenotype and the production of type X collagen, which is associated with endochondral ossification [
17,
18,
19]. However, there is limited evidence regarding the effect of cartilage physiological oxygen levels or cartilage physioxia on cartilage organoid generation.
In cartilage organoid development, in addition to identifying the optimal stem cell populations for their generation and defining appropriate physical conditions, such as oxygen levels, ECM protein production within these structures is a key factor in their potential clinical application. In cartilage organoid models, ECM characterization has primarily focused on analyzing classical structural proteins, such as type II collagen and aggrecan, due to their essential roles in cartilage formation and functionality. However, other cartilage-associated proteins with key functions during specific developmental stages and in determining tissue quality have been scarcely explored in these models. Among them, podoplanin (PDPN), involved in prenatal chondrogenesis, and proteoglycan 4 (PRG4), associated with joint lubrication and considered a potential prognostic marker of success in cartilage regeneration strategies, stand out [
20,
21]. PDPN is a transmembrane glycoprotein expressed in chondrocytes during embryonic development and in the growth plate, playing a role in cellular organization and cartilage stability [
22,
23]. Its expression has been associated with cytoskeletal regulation and cell-ECM communication [
24], although its function in vitro-generated cartilage organoids remains unclear. On the other hand, PRG4, also known as lubricin, is a key protein for cartilage lubrication and the prevention of joint friction [
25]. Its expression has been linked to optimal tribological properties following cartilage regeneration processes [
26]. Moreover, PRG4 expression increases after the successful implantation of human embryonic stem cell (hESC)-derived chondrocytes [
27] and induced pluripotent stem cell (iPSC)-derived cartilage organoids [
20], suggesting its potential as a biomarker for successful organoid transplantation in cartilage regeneration therapies. Despite advancements in organoid development, it remains undetermined whether PDPN and PRG4 expression in these structures is constitutive or dependent on specific conditions such as physiological cartilage physioxia. Furthermore, it is unknown whether their expression correlates with the production of chondrocytes or ECM proteins characteristic of hyaline cartilage.
Since cartilage regeneration represents a significant clinical challenge and current treatments fail to fully restore its structure and function, it is essential to deepen our understanding of cartilage organoids and identify key biomarkers associated with their development and quality. In this context, the present study contributes to this knowledge by generating and characterizing the structural and functional properties of cartilage organoids derived from mesenchymal and skeletal stem cells, cultured under cartilage physioxia conditions that replicate the articular cartilage microenvironment. Additionally, we evaluated classical ECM proteins, such as type II collagen and aggrecan, as well as other proteins relevant to chondrogenesis or tissue quality indicators, including podoplanin (PDPN) and proteoglycan 4 (PRG4), which have been scarcely studied in cartilage organoids.
2. Materials and Methods
2.1. Donor Selection and Bone Marrow Sample Collection
Bone marrow (BM) samples were obtained from patients undergoing hip prosthetic replacement at the Department of Orthopedics and Traumatology, Hospital Universitario San Ignacio (Bogotá, Colombia), who voluntarily agreed to participate in the study. Written informed consent was obtained in accordance with approvals from the Research and Ethics Committees of the Faculty of Sciences at Pontificia Universidad Javeriana (Acta No. 10, June 17, 2021) and Hospital Universitario San Ignacio (Acta No. 11, June 24, 2021). Samples were collected by orthopedic surgeons during surgery and subsequently transported to the Faculty of Sciences at Pontificia Universidad Javeriana for processing and analysis. Donor selection was conducted according to previously published inclusion criteria [
28,
29]. Eligible patients were those over 18 years of age, with no prior diagnosis of hematopoietic system neoplasms, seronegative for hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus (HIV), and not receiving immunosuppressive treatments.
2.2. Isolation and Culture of Skeletal Stem Cells (SSCs) and Mesenchymal Stem Cells (MSCs)
Mononuclear cells (MNCs) were isolated from BM samples using a density gradient with Histopaque 1077 (d = 1.077 g/cm
3, Sigma-Aldrich®). Specific cell populations were then separated to obtain MSCs and SSCs. For MSC isolation, MNCs were directly cultured at a density of 160×10
3 cells/cm
2 in IMDM medium (Gibco, Thermo Scientific®) supplemented with 10% fetal bovine serum (FBS, Gibco, Thermo Scientific®), 1% sodium pyruvate (Sigma-Aldrich®), 1% non-essential amino acids (Gibco, Thermo Scientific®), and 1% penicillin/streptomycin (Eurobio®). Cultures were maintained at 37°C and 5% CO
2 for 72 hours, after which non-adherent cells were removed. Adherent cells with fibroblast-like morphology were cultured until reaching 70% confluence [
30,
31,
32]. For SSC isolation, a negative immunomagnetic separation was performed from 10×10
6 MNCs using anti-CD45 MicroBeads (Miltenyi Biotec®), followed by a second negative immunomagnetic separation using anti-CD146-PE and anti-PE MicroBeads (Miltenyi Biotec®), yielding CD45-/CD146- cells. A subsequent positive immunomagnetic selection was performed using anti-CD73-PE, anti-CD164-APC, anti-PE MicroBeads, and anti-APC MicroBeads (Miltenyi Biotec®) to obtain CD45-/CD146-/CD73+/CD164+ cells. These cells were cultured at a density of 2×10
4/cm
2 in MEM-alpha medium (Gibco, Thermo Scientific®) supplemented with 10% human platelet lysate (STEMCELL Technologies®), 1% sodium pyruvate (Sigma-Aldrich®), and 1% penicillin/streptomycin (Eurobio®). Cultures were maintained at 37°C and 5% CO
2 for 72 hours, after which non-adherent cells were removed. Adherent cells with fibroblast-like morphology were cultured until reaching over 70% confluence [
33,
34]. For both MSCs and SSCs, three passages of the primary culture were performed, followed by individual morphological and phenotypic characterization (data not shown). Subsequently, MSCs and SSCs from each donor were pooled to create a homogeneous population for each cell type, which was then subjected to morphological, phenotypic, and functional characterization. Pooling of each cell population and their co-culture was conducted to reduce donor-dependent biological variability in the isolated cells, as previously demonstrated [
35,
36].
2.3. Morphological, Phenotypic, and Functional Characterization of MSC, SSC, and MSC-SSC Pools
Three cell pools were generated from the characterized primary cultures: SSCs, MSCs, and an MSC-SSC co-culture at a 1:1 ratio. Morphological, phenotypic, and functional characterization of the pools was performed in accordance with ISO 24651 standards for MSCs and experimental background for SSCs [
31,
34]. Morphological characterization was conducted using inverted microscopy with an OLYMPUS CKX3 microscope. For a more detailed morphological evaluation, cells were subjected to cytocentrifugation in an Aerospray® Hematology Pro system. Cells were then stained with Wright’s stain (Sigma-Aldrich®) and evaluated under a conventional optical microscope. The phenotype of the cell populations was determined using qRT-PCR to assess the expression of pluripotency-associated genes [
37,
38,
39,
40] and spectral flow cytometry (Aurora, Cytek® Biosciences). Flow cytometry data were analyzed using FlowJo v10.10.0 to establish the antigenic profile reported for each cell population [
11,
31]. The gating strategy included doublet and debris exclusion, selection of highly viable cells, and generation of fluorescence intensity histograms to determine the median fluorescence intensity (MFI) of each antigen.
Functional characterization of the three cell pools (SSCs, MSCs, and MSC-SSC co-culture) was performed through colony formation and multipotent differentiation assays. Colony formation assays involved culturing 0.5×10
2 cells from each pool in their respective culture medium. After 14 days at 37°C and 5% CO
2, colonies were fixed with 4% paraformaldehyde (Sigma-Aldrich®) and stained with 3% crystal violet (Sigma-Aldrich®) for counting. Multipotent differentiation capacity was assessed by culturing cells in lineage-specific induction media: osteogenesis (StemPro® Osteogenesis Differentiation Kit, Gibco, Thermo Scientific®), adipogenesis (StemPro® Adipogenesis Differentiation Kit, Gibco, Thermo Scientific®), and chondrogenesis (StemPro® Chondrogenesis Differentiation Kit, Gibco, Thermo Scientific®). Osteogenesis was confirmed by Von Kossa staining (Sigma-Aldrich®) for calcium deposits, adipogenesis by Oil Red O staining (Sigma-Aldrich®) for intracellular lipid vacuoles, and chondrogenesis by Alcian Blue staining for glycosaminoglycans (GAG) (Santa Cruz Biotechnology®). Multipotency was further evaluated through qRT-PCR expression analysis of lineage-specific transcription factors: RUNX2 (osteogenesis), PPARγ (adipogenesis), and SOX9 (chondrogenesis) (
Table 1) [
41].
2.4. Maintenance of Control Cell Lines: NHAC-kn and HPdLF
To compare the phenotypic and functional characteristics of the obtained pools and, subsequently, the characteristics of the developed organoids, control cell lines were used, including cartilage-constituting cells (articular chondrocytes) and cells not associated with articular hyaline cartilage (gingival fibroblasts). The Human Knee Articular Chondrocytes (NHAC-kn) CC-2550 cell line (Lonza®) was cultured in CGM™ Chondrocyte Growth Medium BulletKit™ supplemented with 1% penicillin/streptomycin (Gibco, Thermo Scientific®). The Human Periodontal Ligament Fibroblasts (HPdLF) CC-7049 cell line (Lonza®) was cultured in RPMI (STEMCELL Technologies®) supplemented with 1% penicillin/streptomycin (Eurobio®). These cells were provided by Centro de Investigaciones Odontológicas (CIO) of Pontificia Universidad Javeriana.
2.5. Cartilage Organoid Generation from MSCs, SSCs, and MSC-SSC Co-Culture
Organoids were generated from the three previously described cell pools, which included skeletal stem cells (SSC), mesenchymal stem cells (MSC), and a 1:1 co-culture pool of SSC and MSC. Each of these cell conditions was seeded into ultra-low attachment 96-well plates (Corning®) at a density of 1 × 105 cells per well, using an appropriate volume of culture medium. Two culture conditions were established: without chondrogenic induction medium (-IM), using DMEM-F12 (Gibco, Thermo Scientific®) supplemented with 10% KnockOut™ serum (Gibco, Thermo Scientific®) and 1% penicillin/streptomycin (Eurobio®); and with chondrogenic induction medium (+IM), using the StemPro® Chondrogenesis Differentiation Kit (Gibco, Thermo Scientific®) under the same supplementation conditions. After seeding each pool, the plates were centrifuged at 500×g for 15 minutes to promote cell aggregation and then incubated under physioxic conditions (5% O2, 5% CO2, 37°C) for a period of 10 days. Organoid formation was based on spontaneous cell self-aggregation without the use of scaffolds, facilitated by the low-adhesion surface of the culture plates and the physioxic environment, which closely mimics the native cartilage microenvironment. Simultaneously, control cell lines (NHAC-kn and HPdLF) were cultured under the same conditions, starting with 1×103 HPdLF cells and 4×103 NHAC-kn cells. All experiments described below were performed in biological triplicates to ensure reproducibility and to allow for appropriate statistical analysis of the results.
2.6. Evaluation of Cartilage Organoid Morphology, Viability, and Physioxia Condition Validation
The evaluation of morphology, viability, and validation of cartilage physioxia (5% O2) conditions in the organoids was conducted on days 1, 5, and 10, with data acquisition performed using the Cytation 5 Cell Imaging Multimode Reader (BioTek®). Morphological evaluation was carried out by measuring the diameters of the 3D structures in the culture plate using the Line Tool in the Gen5 software on the multifunctional imaging system. Viability assessment of organoids and control cell lines was performed on days 1, 5, and 10 using the LIVE/DEAD Cell Imaging Kit (Invitrogen, Thermo Scientific®) following the manufacturer’s protocol. Image acquisition was conducted using the multifunctional imaging system, and data analysis was performed with ImageJ software. Validation of physioxia conditions in 3D structures was conducted using the BioTracker™ 520 Green Hypoxia Dye (Sigma-Aldrich®) [
42] and data analysis performed in ImageJ software.
2.7. Evaluation of Gene and Protein Expression Associated with Chondrogenesis in Cartilage Organoids Under Physioxia
After 10 days of culture, total RNA was extracted from the organoids using the RNeasy Plus Mini Kit (Qiagen®) following the manufacturer’s protocol and qRT-PCR was performed according to the manufacturer’s instructions. Two-step quantitative real-time PCR (qRT-PCR) was performed. cDNA was synthesized from 1 ng/µl of total RNA using QuantiTect Reverse Transcription Kit” (Qiagen®). Quantitative PCR (qPCR) was carried out using PowerUpTM SYBRTM Green Master Mix” (Biosystems®). Each target and housekeeping gene was analyzed in three technical replicates [
43]. The expression levels of SOX9, ACAN, PDPN, PGR4, RUNX2, and COL10 mRNA were determined. Expression levels were normalized using the housekeeping genes B2M, GAPDH, and ACTB (
Table 1) following the ΔΔCt method [
44]. Protein expression associated with chondrocyte generation in organoids was assessed through enzymatic digestion (1% trypsin, Eurobio®) and mechanical dissociation by repeated pipetting. The resulting cells were transferred to flow cytometry tubes with a 35 μm cell strainer (Falcon™), incubated with Zombie Aqua Fixable Viability dye (Biolegend®) and subsequently stained with the following monoclonal antibodies: CD44-PerCy7 (Clone G44-26, BD Pharmingen®), CD105 BV421 (Clone SN6h, Biolegend®), CD146 BV605 (Clone P1H12, Biolegend®) and PDPN APC (Clone NC-08, Biolegend®). Data acquisition was performed using the Cytek Aurora® spectral flow cytometer and analysis was conducted with FlowJo 10.10.0 software. The immunophenotype was represented using t-SNE dimensionality reduction algorithms and heatmaps based on the mean fluorescence intensity of each antigen in viable cells [
45].
Table S1 shows the primers used for the qRT-PCR assays.
2.8. Determination of Cartilage Extracellular Matrix Proteins in Organoids
After 10 days of culture, organoids were transferred to Eppendorf tubes with 0.5% gelatin (Sigma-Aldrich®) for transportation to the Pathology Department at Hospital Universitario San Ignacio. The 3D structures were then embedded in CryoMatrix (Feather Health®) and sectioned in a KD-3000 cryostat microtome at -20°C to obtain 5 µm thick slices. Sections were subsequently transferred to 98% ethanol for one minute and then incubated at 80°C. For immunohistochemistry (IHC), the Autostainer Link 48 automated system was used with the following antibodies: Aggrecan (Clone BC-3, Novus®), Collagen I alpha 1 (Clone COL-1, Novus®), Collagen II (Clone 5B2.5, Novus®), Collagen X alpha 1 (Clone SR3302, Novus®) and PRG4 (NBP1-19048, Novus®). IHC results were analyzed by double-blind microscopic evaluation performed by pathologists from the Pathology Department at Hospital Universitario San Ignacio. The results were then tabulated and assigned a score as follows: 0 (negative), 1 (mildly positive), 2 (moderately positive), 3 (intensely positive). Negative controls consisted of adipose tissue sections, while positive controls included sections of fetal trachea, skin, and intervertebral disc.
2.9. Statistical Analysis
The Shapiro-Wilk test was used to assess data normality. Based on these results, the Mann-Whitney U test and Kruskal-Wallis test, followed by Dunn’s multiple comparison test, were applied for non-parametric distributions. For parametric distributions, one-way ANOVA followed by Tukey’s multiple comparison test was performed. Statistical significance was set at p ≤ 0.05. Statistical analyses were conducted using GraphPad Prism 10.1.2.324.
4. Discussion
The results of this study provide valuable insights into the characterization and functional behavior of skeletal stem cells (SSCs) and mesenchymal stem cells (MSCs) derived from human bone marrow, as well as their capacity to form hyaline cartilage organoids under cartilage physioxia conditions. This discussion focuses on three main aspects: (i) initial characterization of the cell populations, (ii) formation and characterization of organoids, including molecular and functional assessments related to hyaline cartilage, and (iii) the potential role of PRG4 and PDPN as key markers for evaluating the chondrogenic quality of organoids.
The phenotypic and immunophenotypic characterization of SSCs was consistent with previous reports, showing expression of CD73, CD105, CD164, and PDPN, with absence of CD31, CD34, CD45, and CD146. These cells exhibited high colony-forming capacity (up to 22 CFUs) and demonstrated strong chondrogenic and osteogenic potential, but limited adipogenic differentiation, reflecting their preference for the osteochondral lineage. In contrast, MSCs displayed a classical mesenchymal phenotype, expressing CD146, a marker associated with greater plasticity and hypertrophic potential. Their colony-forming capacity was lower (17 CFUs), and they exhibited multipotent differentiation potential, including chondrogenesis, osteogenesis, and adipogenesis [
11,
30,
31,
33,
34].
The SSC-MSC co-culture allowed for the exploration of potential synergies between both populations, revealing a positive effect on chondrogenesis, with higher glycosaminoglycan production and increased SOX9 expression, surpassing the levels observed in individual cell pools. This interaction suggests possible paracrine signaling and partial recreation of a physiological niche. Additionally, a reduction in adipogenesis was observed, indicating preferential commitment toward chondrogenesis, with relevant implications for cartilage engineering applications. Organoids generated under cartilage physioxia conditions exhibited high viability (>90%), maintained diameters close to 600 μm, and underwent progressive compaction. This greater cohesion was more evident in SSC-derived and SSC-MSC co-culture organoids, likely due to increased extracellular matrix (ECM) protein production and differences in cell adhesion molecule expression, although this remains to be confirmed. Molecular evaluation of these organoids revealed a gene expression profile similar to that of chondrocytes, with SOX9, ACAN, PDPN, RUNX2, and COL10 expression, along with notable PRG4 expression. Protein detection further demonstrated that, particularly in SSC-derived and SSC-MSC co-culture organoids, high levels of aggrecan and PRG4 were produced, with low type X collagen expression. In contrast, MSC-derived organoids exhibited a more hypertrophic protein profile, with higher RUNX2 and type X collagen expression, suggesting a greater tendency toward calcification.
The results of this study suggest that PDPN expression may play a key role in the structural stability and maintenance of an immature chondrogenic phenotype in SSC-derived organoids and SSC-MSC co-culture organoids. Although chondrogenic induction did not significantly increase PDPN expression at the gene level, it did enhance PDPN protein expression, which may be related to better organoid compaction, higher aggrecan production, and elevated SOX9 expression. In contrast, MSC-derived organoids, which exhibited lower PDPN expression, showed reduced three-dimensional stability, absence of aggrecan, higher type X collagen production, and greater RUNX2 expression, reflecting a tendency toward hypertrophy. These findings highlight PDPN as a functional marker associated with the chondrogenic quality of organoids, with the potential to promote a phenotype compatible with immature hyaline cartilage, particularly in SSC and SSC-MSC co-culture organoids under conditions simulating the physiological articular microenvironment. Similarly, PRG4 emerged as a relevant marker for assessing chondrogenic organoid quality, given its role in cartilage lubrication and homeostasis, as well as its hypertrophy-inhibitory function [
54]. High PRG4 expression in SSC-derived and SSC-MSC co-culture organoids, combined with its low expression in MSC-derived organoids, reinforces its potential as a functional predictor for organoids with regenerative potential.
Future studies should evaluate these organoids in vivo models under relevant biomechanical conditions, allowing for a deeper investigation of the functional roles of PRG4 and PDPN during cartilage defect regeneration. Additionally, it will be essential to analyze the transcriptomic and proteomic profiles before and after implantation, as well as assess tribological properties (friction and lubrication), to further establish these organoids as a viable tissue engineering strategy. Finally, integrating in vitro mechanical stress simulations could provide key insights into cellular adaptation to the native articular environment, allowing for further optimization of the functional chondrogenic organoid generation process.
Author Contributions
CAMA: Writing - original draft, Data curation, Methodology. ANSD: Writing - original draft, Data curation, Methodology. TCCB: Writing - original draft, Data curation, Methodology. LFUG: Review and editing, Investigation, Methodology, Results Discussion. JMC: Review and editing, Investigation, Methodology, Results Discussion. RJQ: Review and editing, Investigation, Methodology, Results Discussion. LFJG; Review and editing, Investigation, Methodology, Results Discussion. JAFZ: Review and editing, Investigation, Methodology, Results Discussion. RCS: Investigation, Methodology, Results Discussion. JCUR: Review and editing, Investigation, Methodology, Results Discussion. IGR: Review and editing, Investigation, Methodology, Results Discussion. RS: Review and editing. CLCP: Review and editing, Methodology, Conceptualization, Supervision, Results Discussion. VMRP: Writing - original draft, Writing - review and editing, Data curation, Formal Analysis, Investigation, Methodology.
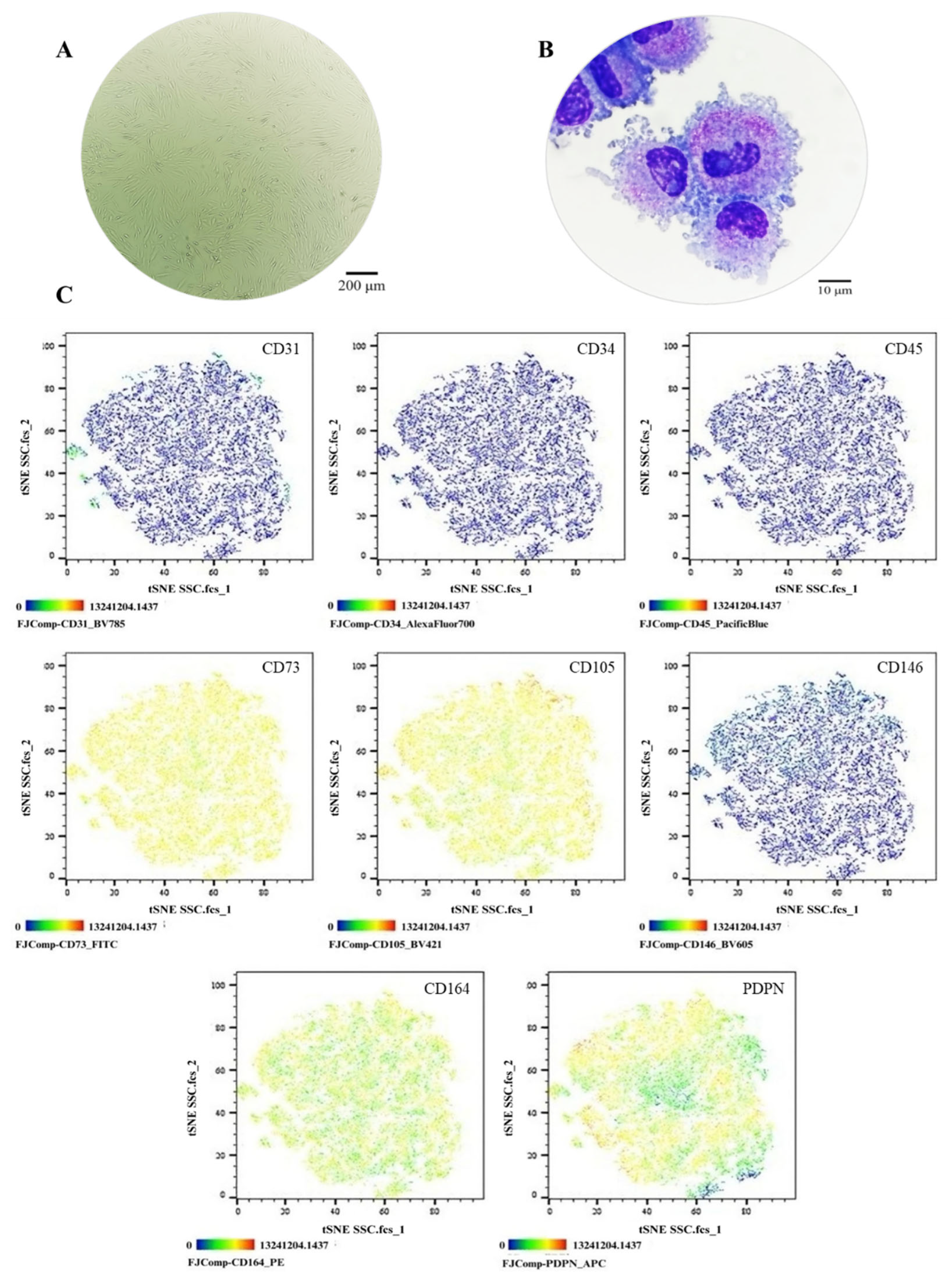
Figure 1.
Phenotypic Characterization of the SSC Pool. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 1.
Phenotypic Characterization of the SSC Pool. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 2.
Functional Characterization of the SSC Pool. (A) Colony-forming unit (CFU) assay. Representative images of SSC-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of SSC-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. SSC cultured without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.0082). (E) Osteogenic differentiation. SSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.1287). (G) Adipogenic differentiation. SSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.0082). Statistical significance was assessed using a two-tailed Student’s t-test.
Figure 2.
Functional Characterization of the SSC Pool. (A) Colony-forming unit (CFU) assay. Representative images of SSC-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of SSC-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. SSC cultured without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.0082). (E) Osteogenic differentiation. SSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.1287). (G) Adipogenic differentiation. SSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) SSC (p = 0.0082). Statistical significance was assessed using a two-tailed Student’s t-test.
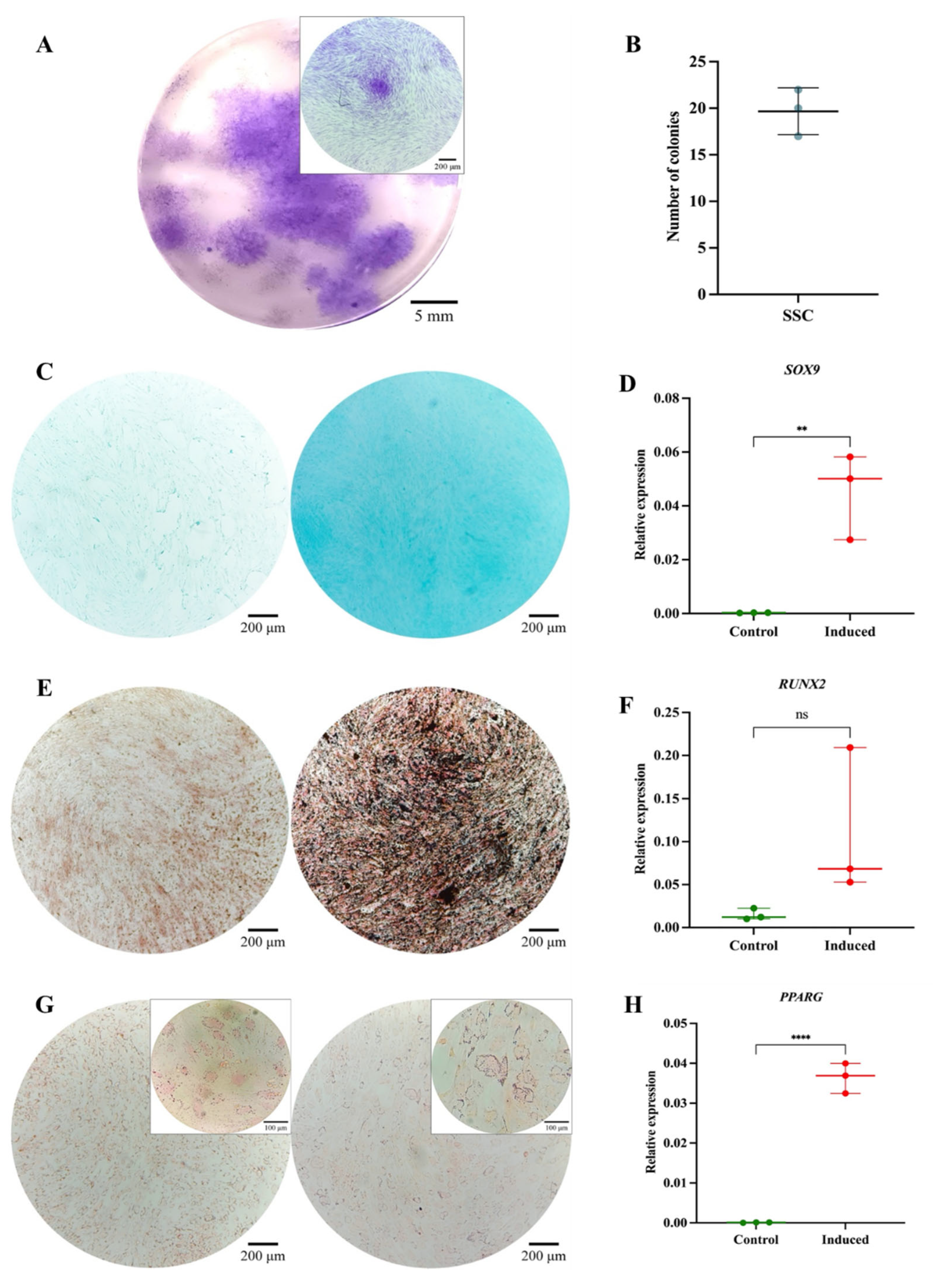
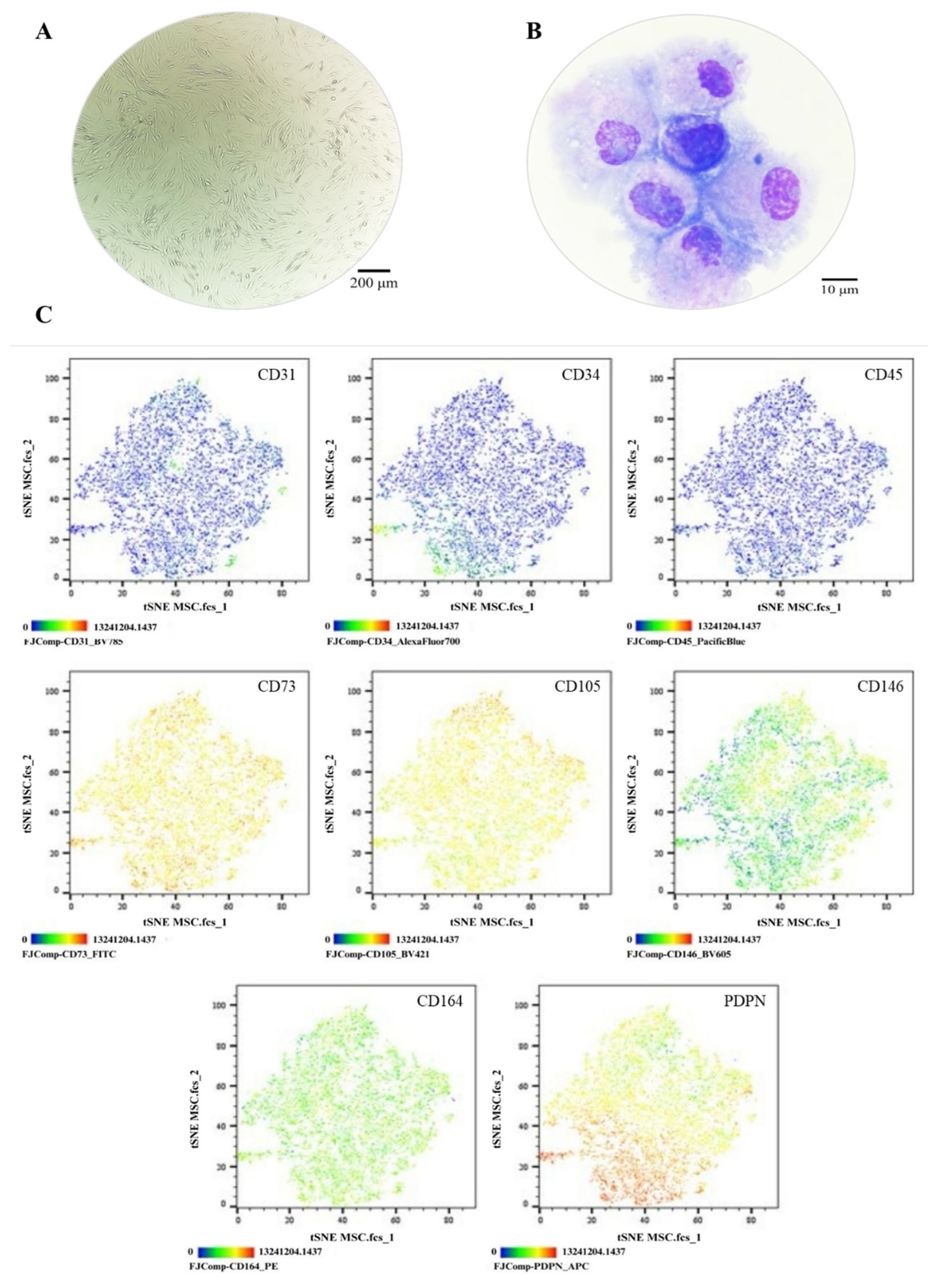
Figure 3.
Phenotypic Characterization of MSCs. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 3.
Phenotypic Characterization of MSCs. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 4.
Functional Characterization of the MSC Pool. (A) Colony-forming unit (CFU) assay. Representative images of MSC-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of MSC-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. MSC cultured without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). (E) Osteogenic differentiation. MSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.3744). (G) Adipogenic differentiation. MSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0010). Statistical significance was assessed using a two-tailed Student’s t-test.
Figure 4.
Functional Characterization of the MSC Pool. (A) Colony-forming unit (CFU) assay. Representative images of MSC-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of MSC-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. MSC cultured without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). (E) Osteogenic differentiation. MSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.3744). (G) Adipogenic differentiation. MSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0010). Statistical significance was assessed using a two-tailed Student’s t-test.
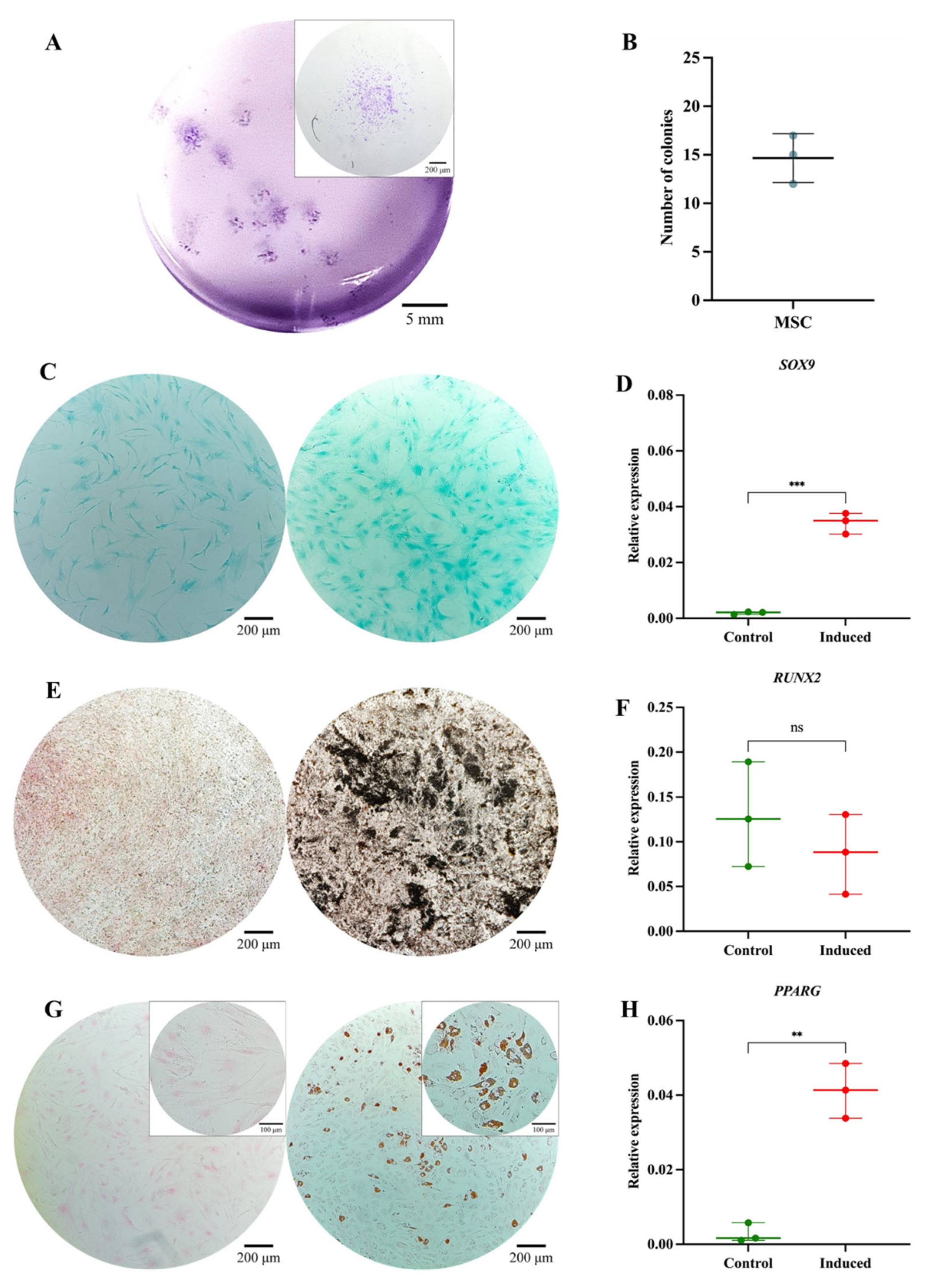
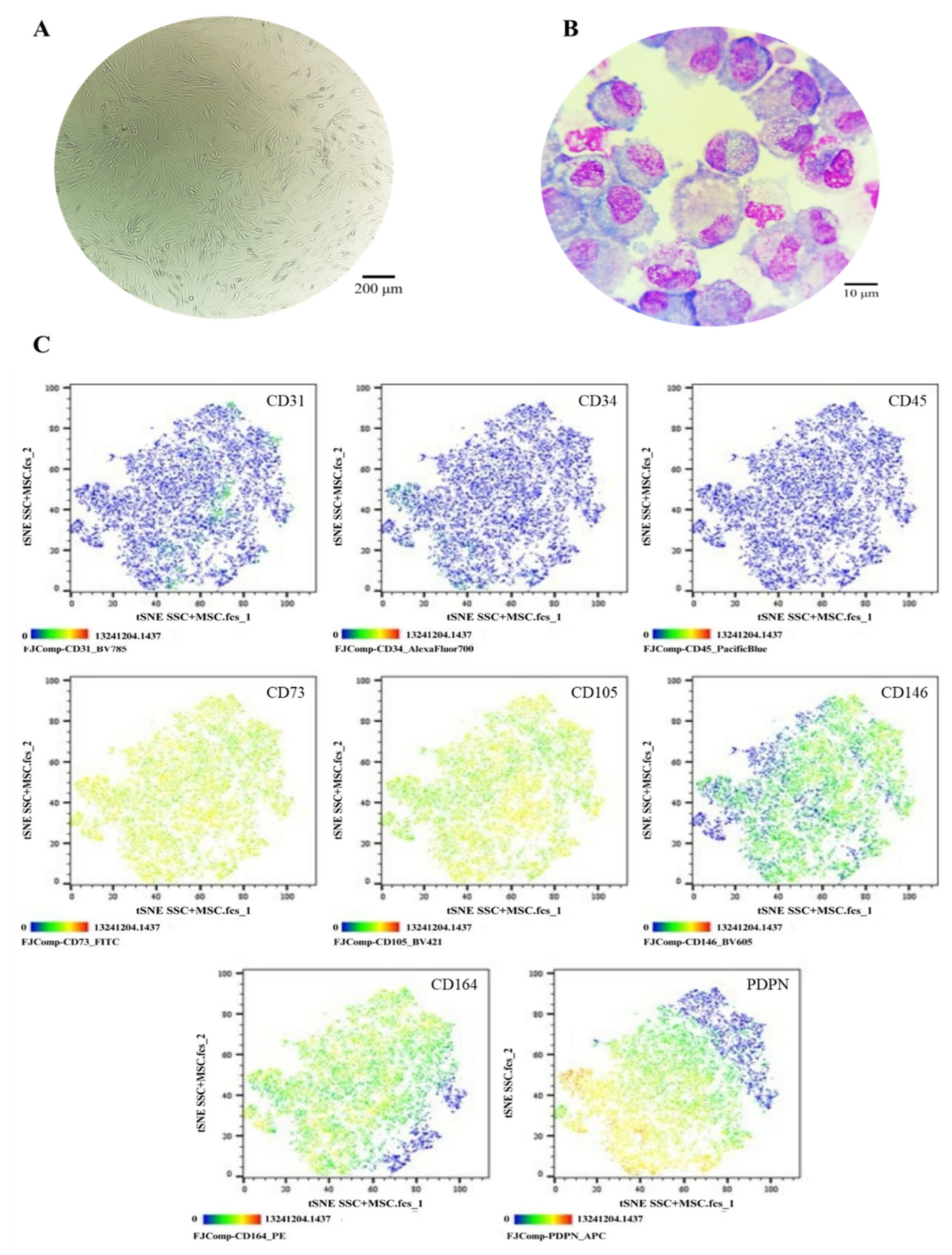
Figure 5.
Phenotypic Characterization of SSCs in Co-Culture with MSCs. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 5.
Phenotypic Characterization of SSCs in Co-Culture with MSCs. (A) Cell morphology in culture, Olympus inverted microscope (10X). (B) Cell morphology in cytospin, ZEISS Axiolab 5 microscope (100X). (C) Immunophenotype analysis by spectral flow cytometry is displayed as a heatmap using t-SNE dimensionality reduction. The antigen assessed is indicated in the top right corner of each panel, and the color scale in the bottom left corner denotes antigen expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 6.
Functional Characterization of the SSC Pool in Co-Culture with MSCs. (A) Colony-forming unit (CFU) assay. Representative images of SSC-MSC co-culture-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of SSC-MSC co-culture-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. SSC-MSC co-culture without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). (E) Osteogenic differentiation. MSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.5282). (G) Adipogenic differentiation. MSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). Statistical significance was assessed using a two-tailed Student’s t-test.
Figure 6.
Functional Characterization of the SSC Pool in Co-Culture with MSCs. (A) Colony-forming unit (CFU) assay. Representative images of SSC-MSC co-culture-derived colonies after 14 days of culture, stained with 3% crystal violet. Images were acquired using an Olympus inverted microscope at 10× magnification (inset, 40×). (B) Absolute quantification of SSC-MSC co-culture-derived colonies after 14 days of culture. (C) Chondrogenic differentiation. SSC-MSC co-culture without (left) or with (right) chondrogenic differentiation medium. Alcian Blue staining at 10× magnification. (D) SOX9 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). (E) Osteogenic differentiation. MSC cultured without (left) or with (right) osteogenic differentiation medium. Von Kossa staining at 10× magnification. (F) RUNX2 gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.5282). (G) Adipogenic differentiation. MSC cultured without (left) or with (right) adipogenic differentiation medium. Oil Red O staining at 10× magnification (inset, 40×). (H) PPARγ gene expression in undifferentiated (control) and differentiated (induced) MSC (p = 0.0001). Statistical significance was assessed using a two-tailed Student’s t-test.
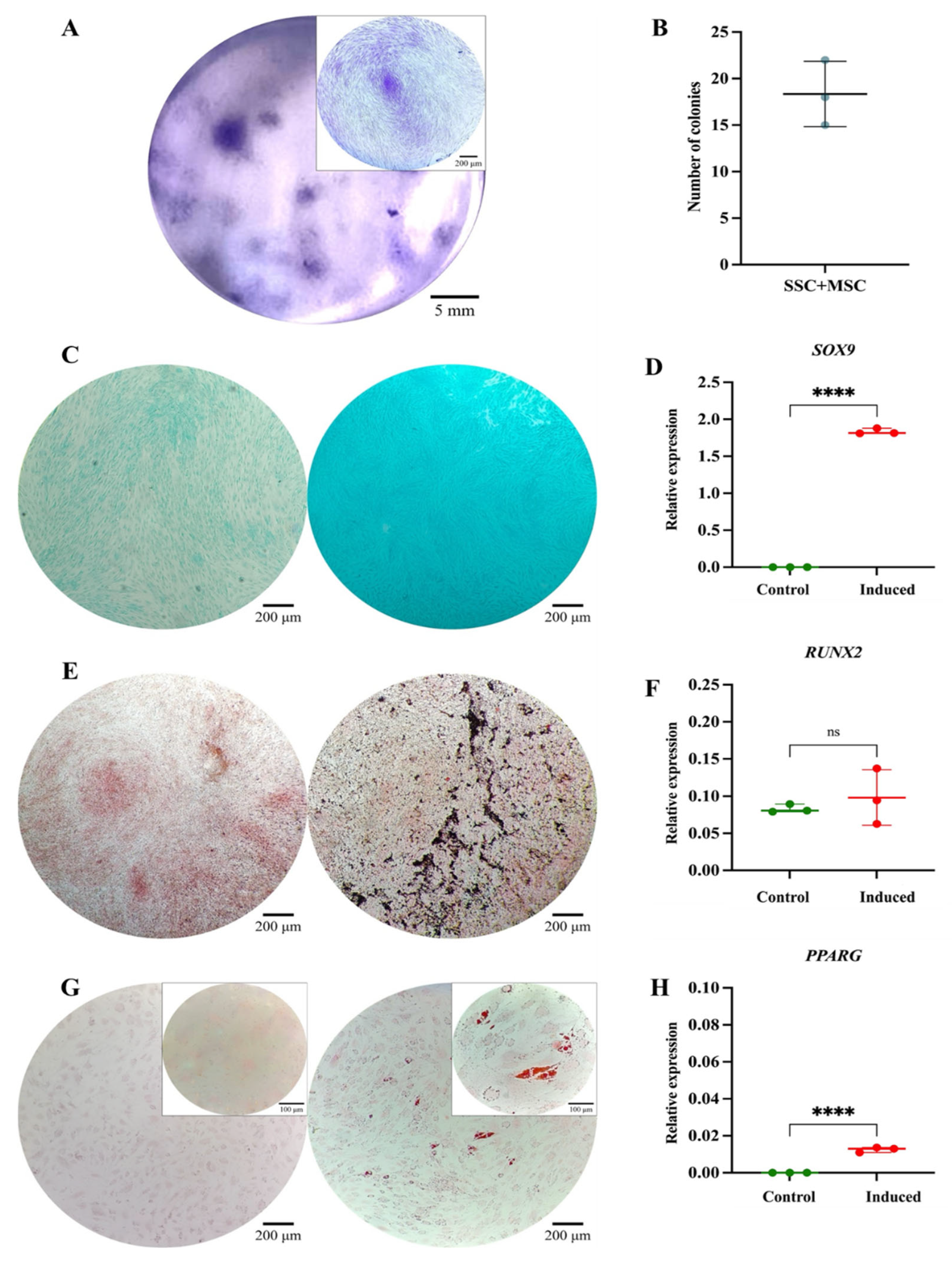
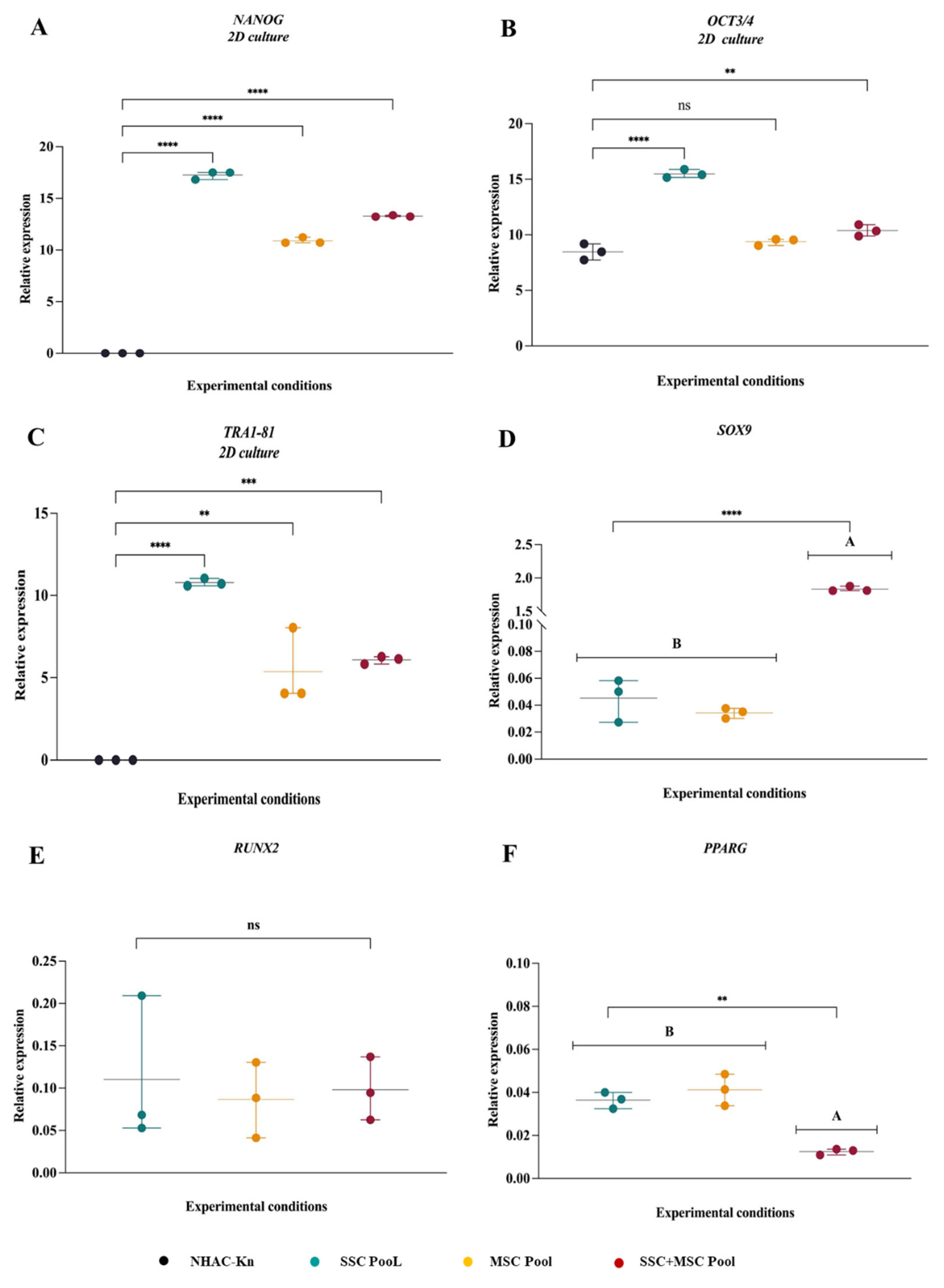
Figure 7.
Comparative gene expression profiles associated with pluripotency and multipotency in SSC, MSC, and SSC–MSC co-culture pools. (A–C) Relative expression levels of pluripotency-associated genes (NANOG, OCT3/4, and TRA1-81) in SSC, MSC, and SSC–MSC co-culture pools cultured in basal medium (i.e., without differentiation stimuli). (D–F) Expression of lineage-specific transcription factors related to multipotency: SOX9 (chondrogenesis), RUNX2 (osteogenesis), and PPARγ (adipogenesis) in cell pools subjected to lineage-specific differentiation media. Statistical significance was assessed using one-way ANOVA followed by post hoc multiple comparisons. **p < 0.01; ****p < 0.0001.
Figure 7.
Comparative gene expression profiles associated with pluripotency and multipotency in SSC, MSC, and SSC–MSC co-culture pools. (A–C) Relative expression levels of pluripotency-associated genes (NANOG, OCT3/4, and TRA1-81) in SSC, MSC, and SSC–MSC co-culture pools cultured in basal medium (i.e., without differentiation stimuli). (D–F) Expression of lineage-specific transcription factors related to multipotency: SOX9 (chondrogenesis), RUNX2 (osteogenesis), and PPARγ (adipogenesis) in cell pools subjected to lineage-specific differentiation media. Statistical significance was assessed using one-way ANOVA followed by post hoc multiple comparisons. **p < 0.01; ****p < 0.0001.
Figure 8.
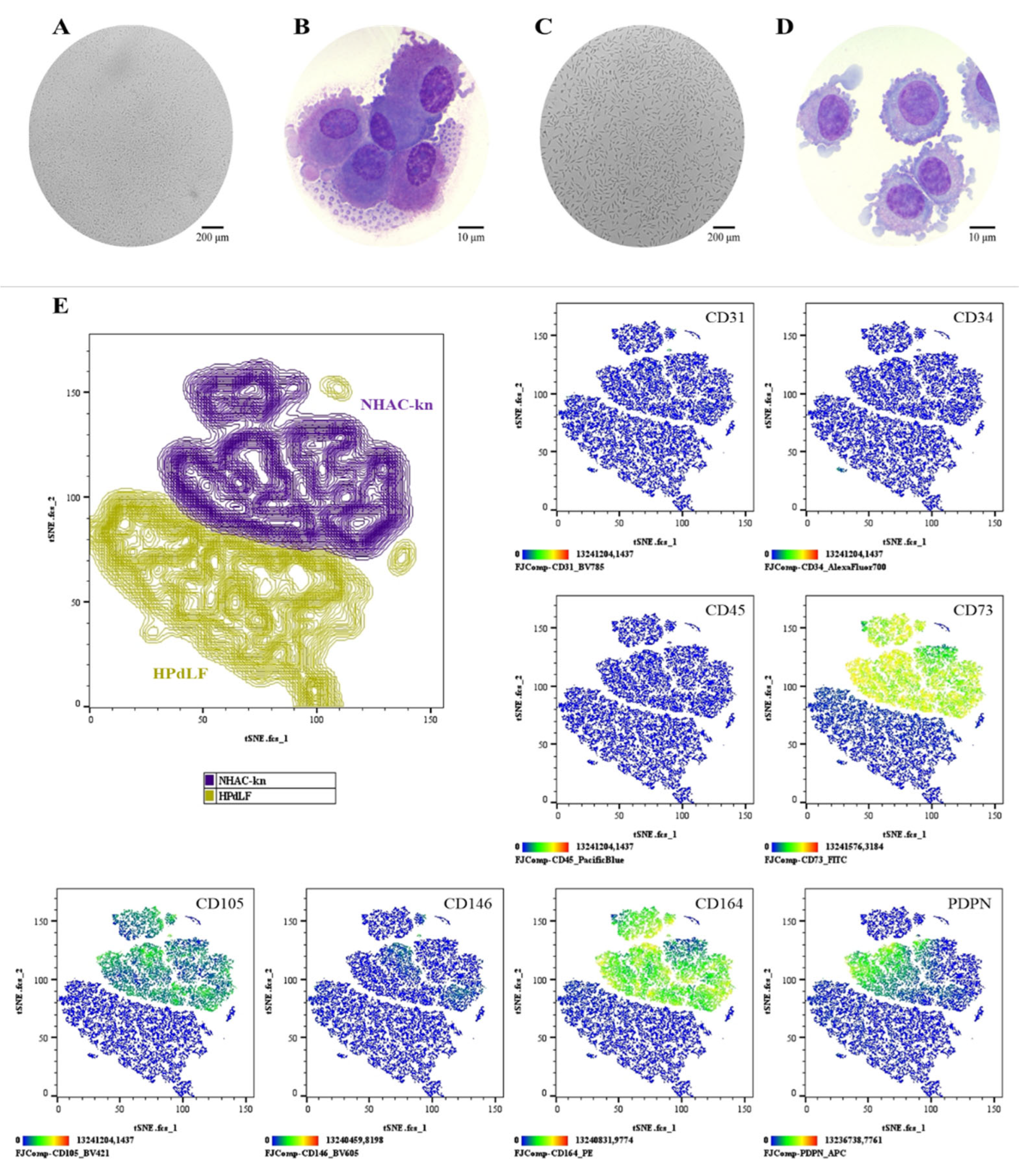
Comparative phenotypic characterization of control cell populations. (A–B) NHAC-kn: cell morphology in culture (Olympus inverted microscope, 10×) and in cytospin preparations (ZEISS Axiolab 5 microscope, 100×). (C–D) HPdLF: cell morphology in culture (Olympus inverted microscope, 10×) and in cytospin preparations (ZEISS Axiolab 5 microscope, 100×). (E) t-SNE analysis was performed to visualize the spatial segregation of the two control cell populations, providing a reference framework for the interpretation of subsequent immunophenotyping plots. Immunophenotypic profiling by spectral flow cytometry, displayed as heatmaps generated using t-SNE dimensionality reduction. The antigen analyzed is indicated in the top right corner of each panel. The color scale in the bottom left indicates expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 8.
Comparative phenotypic characterization of control cell populations. (A–B) NHAC-kn: cell morphology in culture (Olympus inverted microscope, 10×) and in cytospin preparations (ZEISS Axiolab 5 microscope, 100×). (C–D) HPdLF: cell morphology in culture (Olympus inverted microscope, 10×) and in cytospin preparations (ZEISS Axiolab 5 microscope, 100×). (E) t-SNE analysis was performed to visualize the spatial segregation of the two control cell populations, providing a reference framework for the interpretation of subsequent immunophenotyping plots. Immunophenotypic profiling by spectral flow cytometry, displayed as heatmaps generated using t-SNE dimensionality reduction. The antigen analyzed is indicated in the top right corner of each panel. The color scale in the bottom left indicates expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 9.
Diameter and viability assessment of cartilage organoids derived from co-cultured SSC, MSC, and SSC–MSC groups. (A) Representative images of cartilage organoids at days 1, 5, and 10 of culture, derived from SSC, MSC, and SSC–MSC groups, cultured with or without chondrogenic differentiation medium. Images were acquired using a Cytation 5 imaging system (BioTek) under 10× magnification. (B) Viability assessment of spheroids derived from control cell populations (NHAC-kn and HPdLF) and from SSC, MSC, and SSC–MSC groups, cultured with or without chondrogenic differentiation medium at days 1, 5, and 10. Cell viability was evaluated using the LIVE/DEAD™ Cell Imaging Kit (Invitrogen, Thermo Fisher Scientific®). Live cells fluoresce green (calcein AM), and dead cells fluoresce red (ethidium homodimer-1). Images were acquired with the Cytation 5 system. 10× magnification. (C) Quantification of spheroid diameters at days 1, 5, and 10 of culture. (D) Percentage viability of control cell populations and cartilage organoids at days 1, 5, and 10. The dashed line represents the 90% viability threshold, indicating that all populations maintained >90% viability under the experimental conditions throughout the culture period. Data on diameters and viability under the experimental conditions were acquired and analyzed using the Cytation 5 imaging system (BioTek).
Figure 9.
Diameter and viability assessment of cartilage organoids derived from co-cultured SSC, MSC, and SSC–MSC groups. (A) Representative images of cartilage organoids at days 1, 5, and 10 of culture, derived from SSC, MSC, and SSC–MSC groups, cultured with or without chondrogenic differentiation medium. Images were acquired using a Cytation 5 imaging system (BioTek) under 10× magnification. (B) Viability assessment of spheroids derived from control cell populations (NHAC-kn and HPdLF) and from SSC, MSC, and SSC–MSC groups, cultured with or without chondrogenic differentiation medium at days 1, 5, and 10. Cell viability was evaluated using the LIVE/DEAD™ Cell Imaging Kit (Invitrogen, Thermo Fisher Scientific®). Live cells fluoresce green (calcein AM), and dead cells fluoresce red (ethidium homodimer-1). Images were acquired with the Cytation 5 system. 10× magnification. (C) Quantification of spheroid diameters at days 1, 5, and 10 of culture. (D) Percentage viability of control cell populations and cartilage organoids at days 1, 5, and 10. The dashed line represents the 90% viability threshold, indicating that all populations maintained >90% viability under the experimental conditions throughout the culture period. Data on diameters and viability under the experimental conditions were acquired and analyzed using the Cytation 5 imaging system (BioTek).
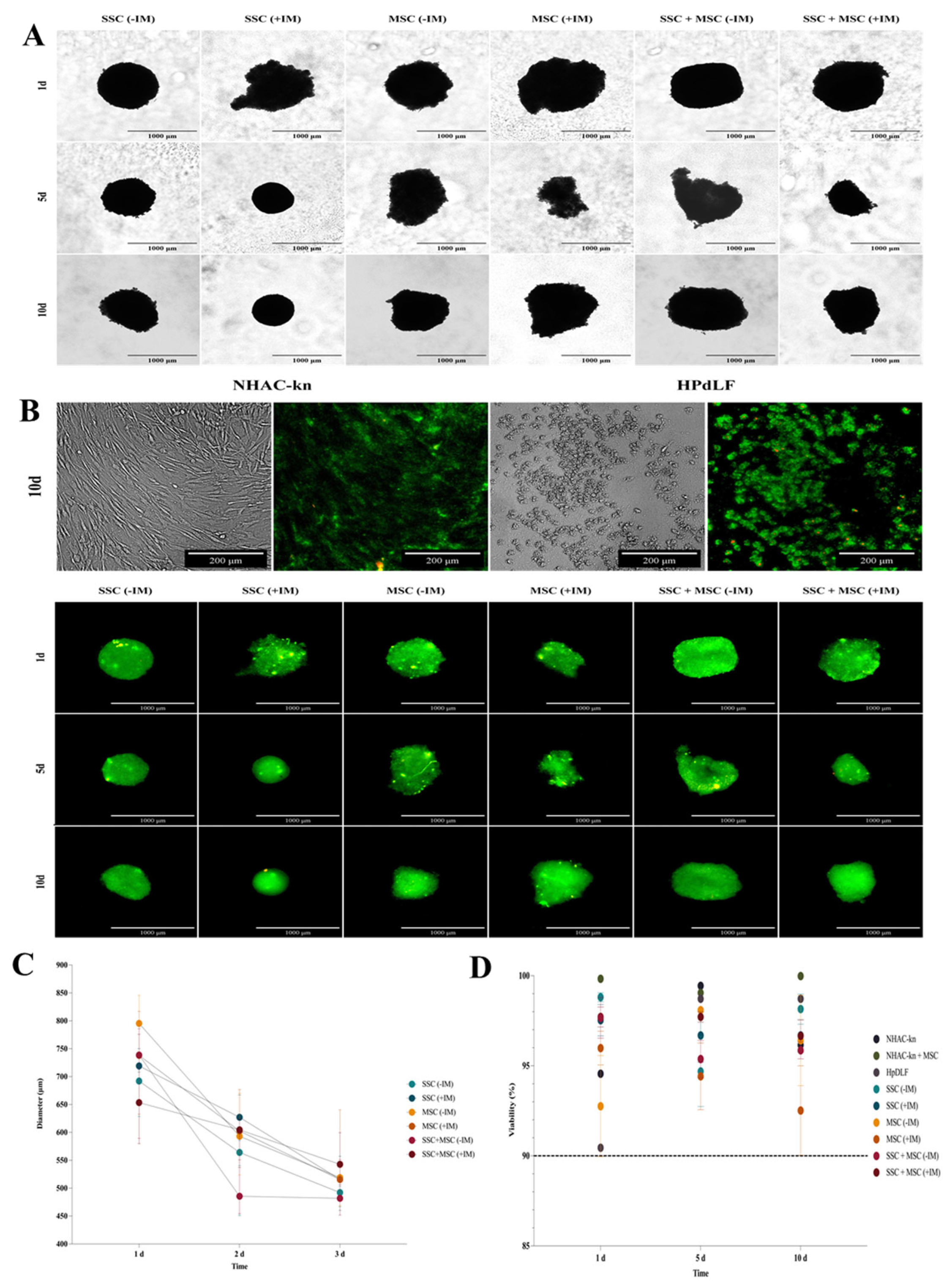
Figure 10.
Assessment of oxygen levels in cartilage organoids and control cell populations. (A) Representative images of control cell lines (NHAC-kn and HPdLF) after 10 days of culture under 5% O2. Brightfield and fluorescence images were acquired following staining with the hypoxia-sensitive probe BioTracker™ 520 Green Hypoxia Dye (Sigma-Aldrich®). Under normoxic conditions (21% O2), the dye remains inactive or exhibits low fluorescence. Under hypoxic conditions (5% O2), it is chemically reduced by intracellular enzymes, such as oxidoreductases, resulting in green fluorescence activation. (B) Quantification of fluorescence intensity in control cell populations cultured under 21% and 5% O2. Increased fluorescence intensity indicates lower oxygen availability. (C) Fluorescence and brightfield images of cartilage organoids derived from SSC, MSC, and SSC–MSC co-culture pools, cultured with or without chondrogenic differentiation medium. Organoids were cultured at 21% O2 (no fluorescence) or 5% O2 for 1, 5, and 10 days, showing increasing green fluorescence under hypoxic conditions. (D) Quantification of fluorescence intensity in cartilage organoids cultured under 21% and 5% O2. Significantly higher fluorescence was observed at 5% O2. Statistical significance was determined by one-way ANOVA with post hoc multiple comparisons. ***p < 0.001; ****p < 0.0001.
Figure 10.
Assessment of oxygen levels in cartilage organoids and control cell populations. (A) Representative images of control cell lines (NHAC-kn and HPdLF) after 10 days of culture under 5% O2. Brightfield and fluorescence images were acquired following staining with the hypoxia-sensitive probe BioTracker™ 520 Green Hypoxia Dye (Sigma-Aldrich®). Under normoxic conditions (21% O2), the dye remains inactive or exhibits low fluorescence. Under hypoxic conditions (5% O2), it is chemically reduced by intracellular enzymes, such as oxidoreductases, resulting in green fluorescence activation. (B) Quantification of fluorescence intensity in control cell populations cultured under 21% and 5% O2. Increased fluorescence intensity indicates lower oxygen availability. (C) Fluorescence and brightfield images of cartilage organoids derived from SSC, MSC, and SSC–MSC co-culture pools, cultured with or without chondrogenic differentiation medium. Organoids were cultured at 21% O2 (no fluorescence) or 5% O2 for 1, 5, and 10 days, showing increasing green fluorescence under hypoxic conditions. (D) Quantification of fluorescence intensity in cartilage organoids cultured under 21% and 5% O2. Significantly higher fluorescence was observed at 5% O2. Statistical significance was determined by one-way ANOVA with post hoc multiple comparisons. ***p < 0.001; ****p < 0.0001.
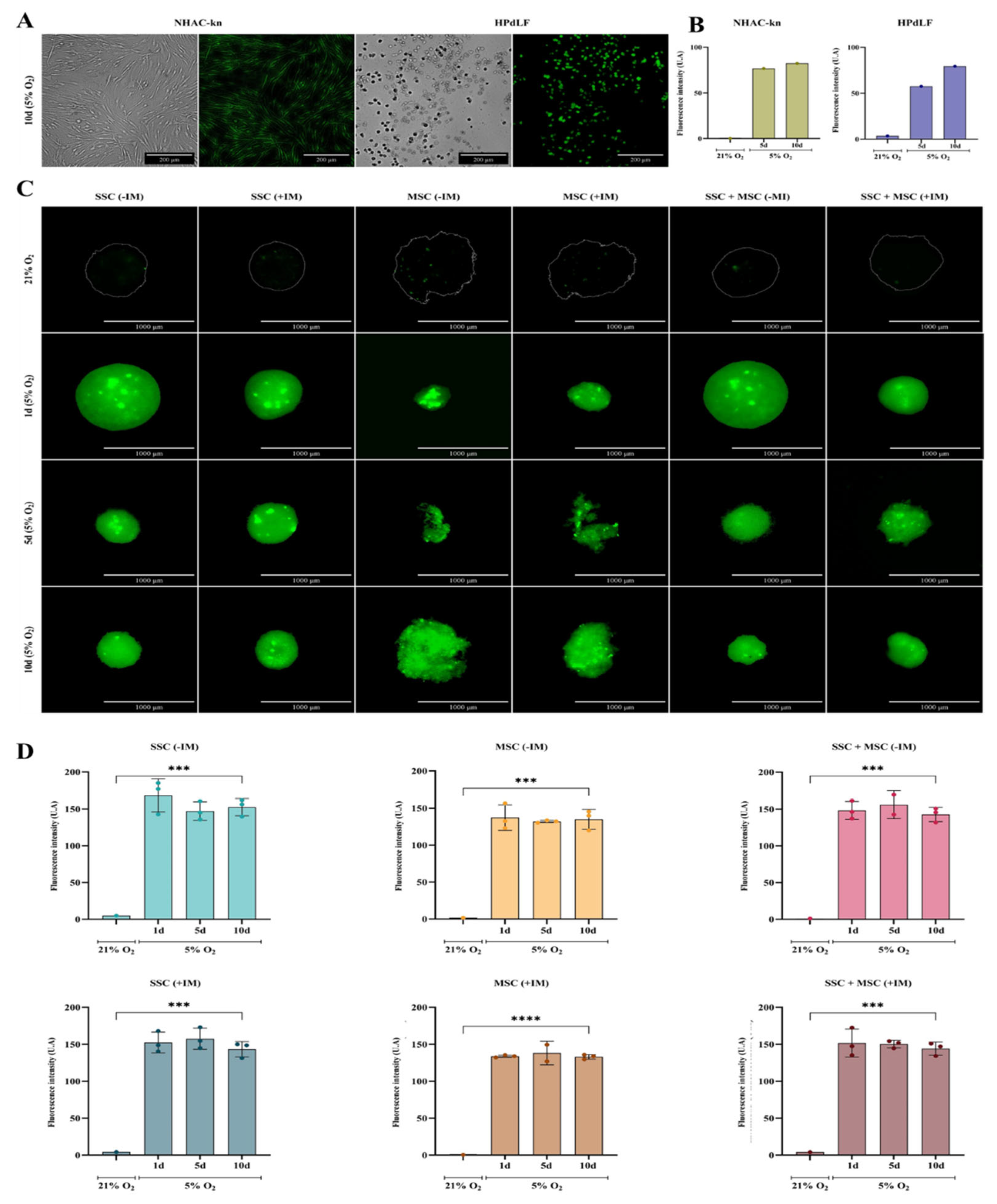
Figure 11.
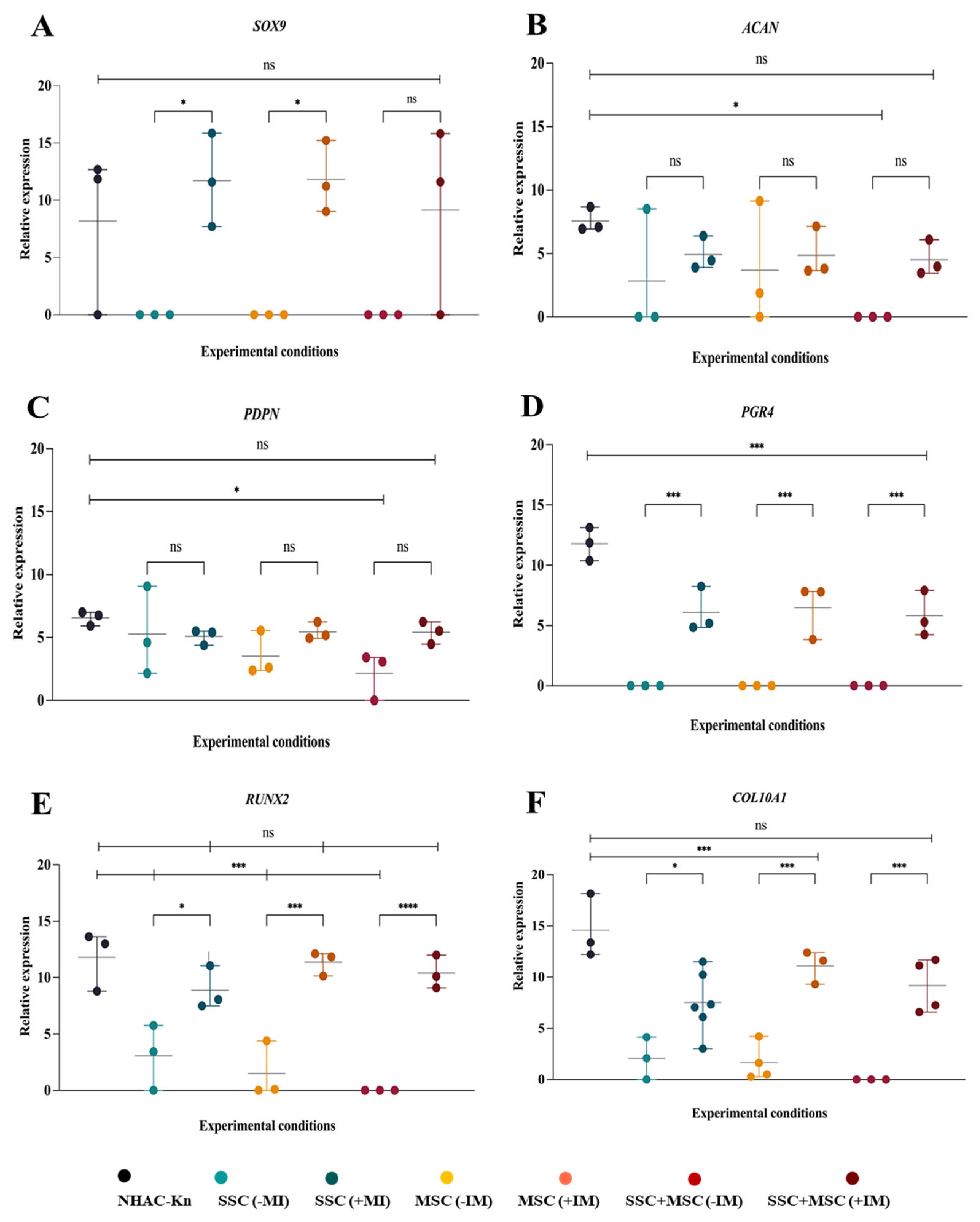
Expression of genes associated with chondrogenesis in cartilage organoids derived from SSC, MSC, and SSC–MSC co-culture pools after 10 days of culture. (A–D) Relative expression of SOX9, ACAN, PDPN, and PRG4 in chondrocytes from the NHAC-kn cell line and in cartilage organoids cultured with or without chondrogenic differentiation medium. (E–F) Relative expression of RUNX2 and COL10A1 in chondrocytes from the NHAC-kn cell line and in cartilage organoids under the same conditions. Statistical significance was assessed using a two-tailed Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Figure 11.
Expression of genes associated with chondrogenesis in cartilage organoids derived from SSC, MSC, and SSC–MSC co-culture pools after 10 days of culture. (A–D) Relative expression of SOX9, ACAN, PDPN, and PRG4 in chondrocytes from the NHAC-kn cell line and in cartilage organoids cultured with or without chondrogenic differentiation medium. (E–F) Relative expression of RUNX2 and COL10A1 in chondrocytes from the NHAC-kn cell line and in cartilage organoids under the same conditions. Statistical significance was assessed using a two-tailed Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Figure 12.
Generation of chondrocytes from cartilage organoids derived from SSCs, MSCs, and SSC–MSC co-cultures. After 10 days of culture in chondrogenic differentiation medium, cartilage organoids were enzymatically disaggregated, and the generation of chondrocytes exhibiting a CD44+/CD105+/CD146+/PDPN+ phenotype was assessed by spectral flow cytometry. The immunophenotypic profiles of organoid-derived cells were compared with those of control cell lines (NHAC-kn and HPdLF). (A) t-SNE analysis illustrating the spatial distribution of the two control populations and SSC-derived organoid cells. Heatmaps and t-SNE plots show the expression levels of CD44, CD105, CD146, and PDPN. (B) t-SNE analysis illustrating the spatial distribution of the control populations and MSC-derived organoid cells. Heatmaps and t-SNE plots show the expression of the same antigenic markers. (C) t-SNE analysis illustrating the spatial distribution of the control populations and cells derived from SSC–MSC co-culture organoids. Heatmaps and t-SNE plots indicate expression of CD44, CD105, CD146, and PDPN. Statistical significance was assessed using one-way ANOVA followed by multiple comparisons. ****p < 0.0001. The antigen analyzed is indicated in the top right corner of each panel. The color scale in the bottom left indicates expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
Figure 12.
Generation of chondrocytes from cartilage organoids derived from SSCs, MSCs, and SSC–MSC co-cultures. After 10 days of culture in chondrogenic differentiation medium, cartilage organoids were enzymatically disaggregated, and the generation of chondrocytes exhibiting a CD44+/CD105+/CD146+/PDPN+ phenotype was assessed by spectral flow cytometry. The immunophenotypic profiles of organoid-derived cells were compared with those of control cell lines (NHAC-kn and HPdLF). (A) t-SNE analysis illustrating the spatial distribution of the two control populations and SSC-derived organoid cells. Heatmaps and t-SNE plots show the expression levels of CD44, CD105, CD146, and PDPN. (B) t-SNE analysis illustrating the spatial distribution of the control populations and MSC-derived organoid cells. Heatmaps and t-SNE plots show the expression of the same antigenic markers. (C) t-SNE analysis illustrating the spatial distribution of the control populations and cells derived from SSC–MSC co-culture organoids. Heatmaps and t-SNE plots indicate expression of CD44, CD105, CD146, and PDPN. Statistical significance was assessed using one-way ANOVA followed by multiple comparisons. ****p < 0.0001. The antigen analyzed is indicated in the top right corner of each panel. The color scale in the bottom left indicates expression levels: negative (blue), weakly positive (green), moderately positive (yellow), and strongly positive (red).
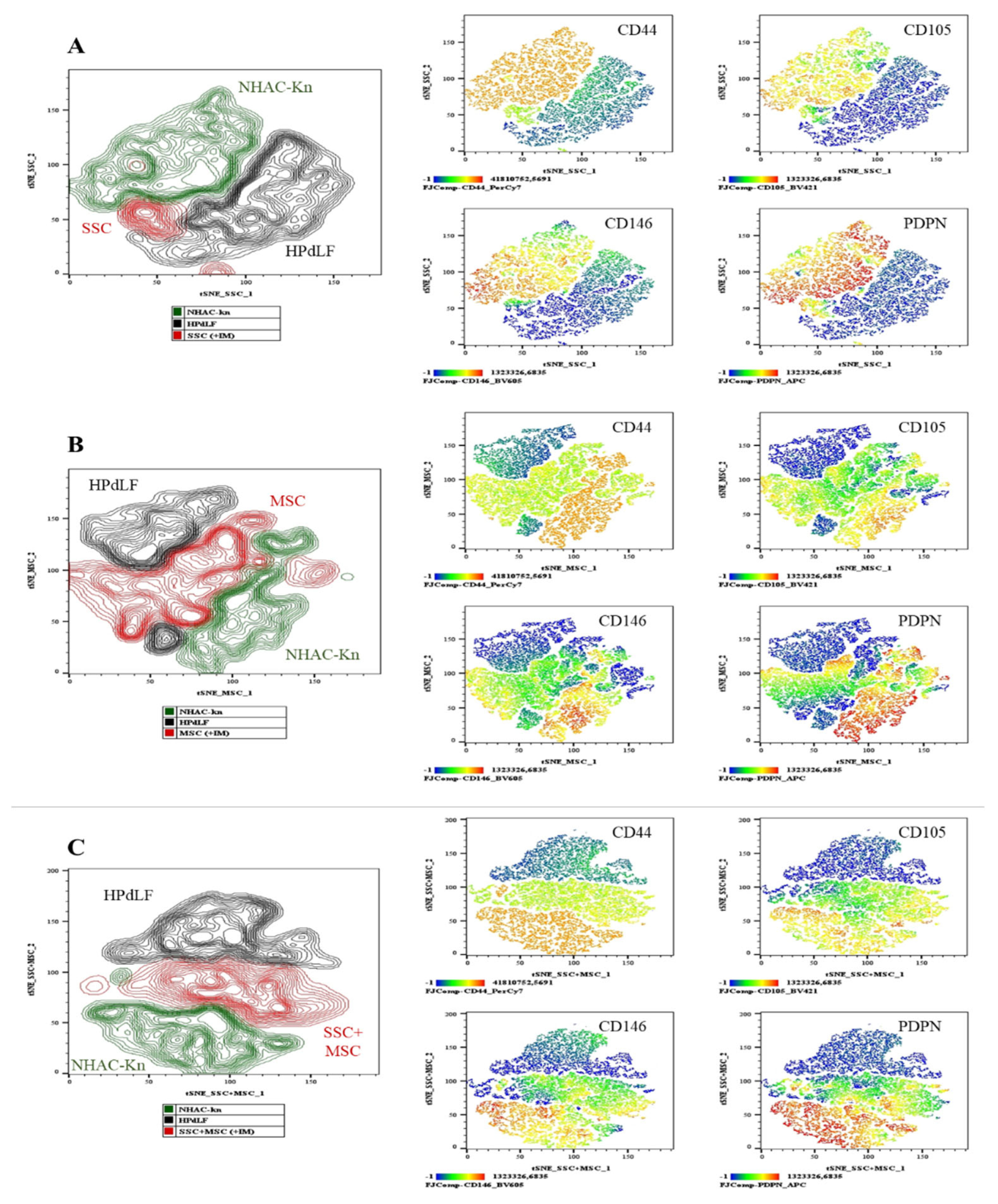
Figure 13.
Extracellular matrix (ECM) protein production in cartilage organoids. (A) Immunohistochemical detection of aggrecan, type II collagen, proteoglycan-4, type I collagen, and type X collagen in SSC-derived organoids cultured without (–MI) or with (+MI) chondrogenic induction medium. Images acquired with an Olympus microscope at 10× magnification. (B) Immunohistochemical detection of the same ECM proteins in MSC-derived organoids under the same conditions. (C) Immunohistochemical detection of ECM proteins in organoids derived from SSC–MSC co-cultures, with or without chondrogenic induction medium. Images acquired with an Olympus microscope at 10× magnification. (D–F) Semi-quantitative scoring of ECM protein expression (aggrecan, type II collagen, proteoglycan-4, type I collagen, and type X collagen) in SSC-derived organoids (D), MSC-derived organoids (E), and SSC–MSC co-culture organoids (F), cultured with or without chondrogenic induction medium. (G) Comparative scoring of aggrecan, type II collagen, and proteoglycan-4 expression between cartilage organoids and the NHAC-kn chondrocyte line. (H) Comparative scoring of type I and type X collagen expression between cartilage organoids and the NHAC-kn chondrocyte line. Statistical significance was determined using the Kruskal–Wallis test followed by multiple comparisons. ****p < 0.0001.
Figure 13.
Extracellular matrix (ECM) protein production in cartilage organoids. (A) Immunohistochemical detection of aggrecan, type II collagen, proteoglycan-4, type I collagen, and type X collagen in SSC-derived organoids cultured without (–MI) or with (+MI) chondrogenic induction medium. Images acquired with an Olympus microscope at 10× magnification. (B) Immunohistochemical detection of the same ECM proteins in MSC-derived organoids under the same conditions. (C) Immunohistochemical detection of ECM proteins in organoids derived from SSC–MSC co-cultures, with or without chondrogenic induction medium. Images acquired with an Olympus microscope at 10× magnification. (D–F) Semi-quantitative scoring of ECM protein expression (aggrecan, type II collagen, proteoglycan-4, type I collagen, and type X collagen) in SSC-derived organoids (D), MSC-derived organoids (E), and SSC–MSC co-culture organoids (F), cultured with or without chondrogenic induction medium. (G) Comparative scoring of aggrecan, type II collagen, and proteoglycan-4 expression between cartilage organoids and the NHAC-kn chondrocyte line. (H) Comparative scoring of type I and type X collagen expression between cartilage organoids and the NHAC-kn chondrocyte line. Statistical significance was determined using the Kruskal–Wallis test followed by multiple comparisons. ****p < 0.0001.
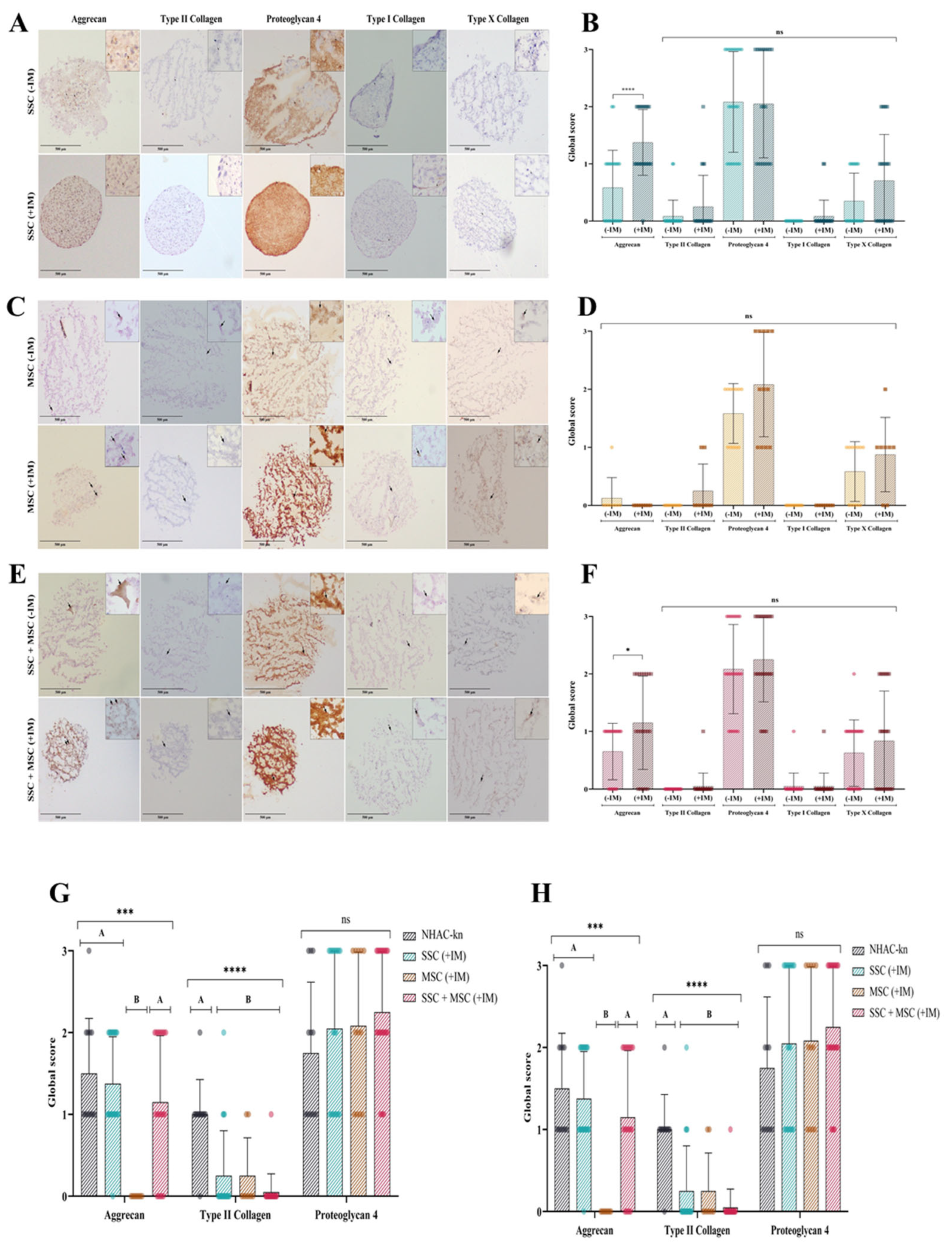
Table 1.
Donor Characteristics and Bone Marrow Samples for SSC and MSC Isolation.
Table 1.
Donor Characteristics and Bone Marrow Samples for SSC and MSC Isolation.
| |
Sample No. |
Age (years) |
Gender (M: male / F: female) |
BM Volume (mL) |
| SSC |
01 |
55 |
M |
90 |
| 02 |
61 |
M |
65 |
| 03 |
69 |
M |
42 |
| 04 |
59 |
M |
95 |
| 05 |
73 |
F |
75 |
| 06 |
68 |
F |
55 |
| MSC |
07 |
79 |
M |
90 |
| 08 |
61 |
M |
75 |
| 09 |
83 |
F |
80 |
| 10 |
73 |
M |
90 |
| 11 |
84 |
F |
80 |
| 12 |
60 |
M |
80 |