Submitted:
18 August 2025
Posted:
20 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Clinical Study Population and Tissue Sample Collection
2.2. DNA Methylation Analysis
2.2.1. Global Methylation Profiling Using Illumina EPIC Methylation Array
2.2.2. Bioinformatic Analysis
3. Results
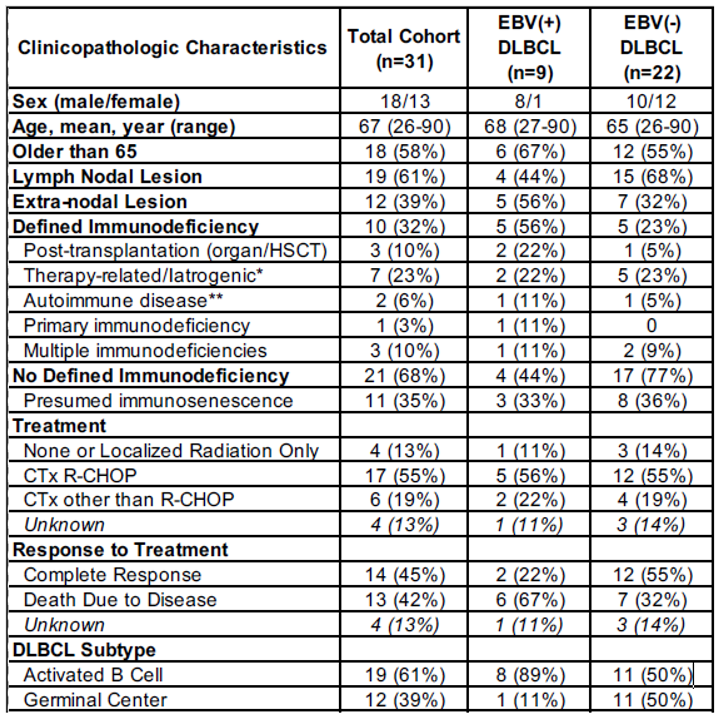
3.1. Clinicopathologic Characteristics of Study Cohort
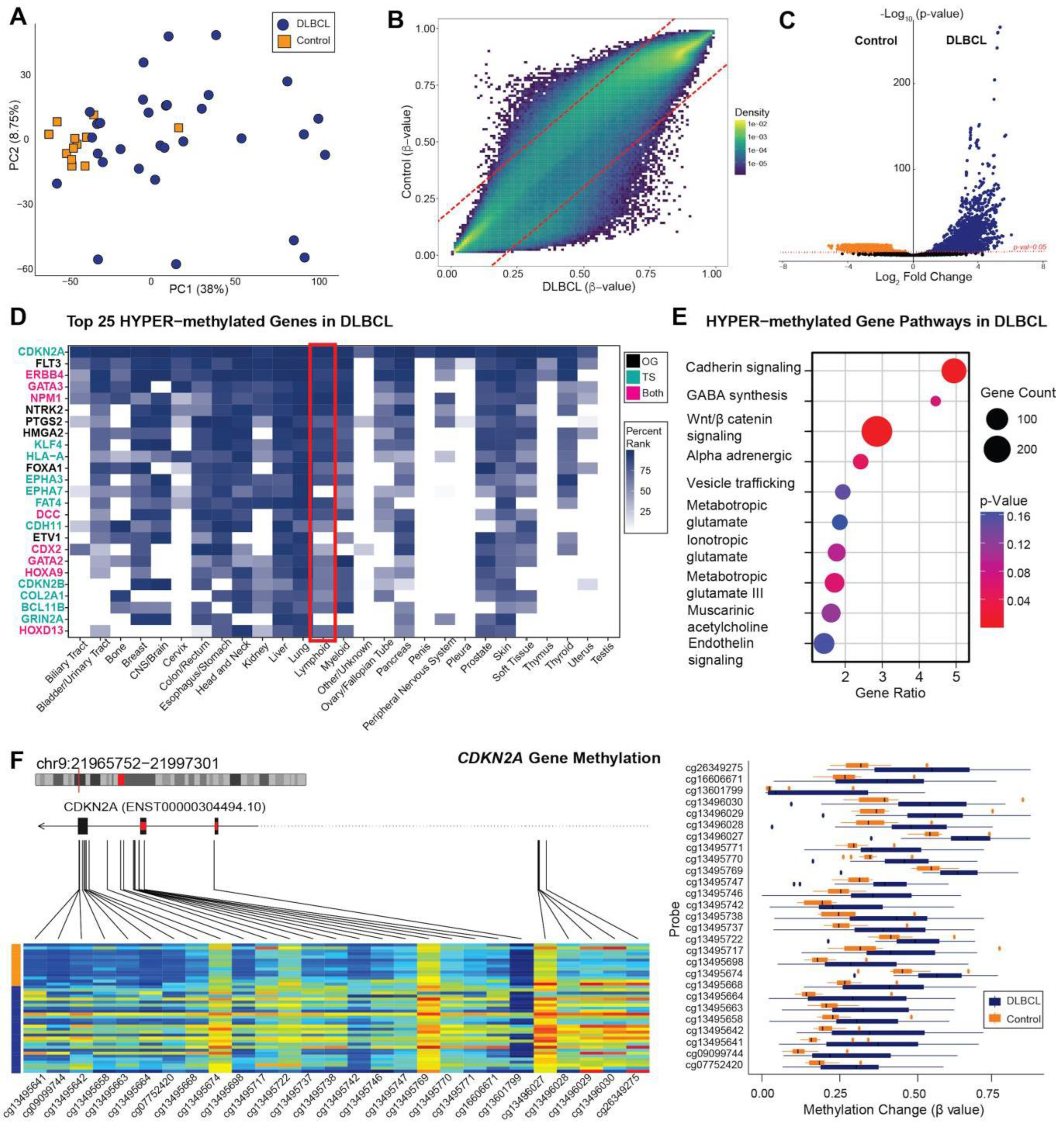
3.2. Global DNA Methylation Analysis of Clinical Cohort
3.3. Methylome Patterns: All DLBCL versus Control Cases
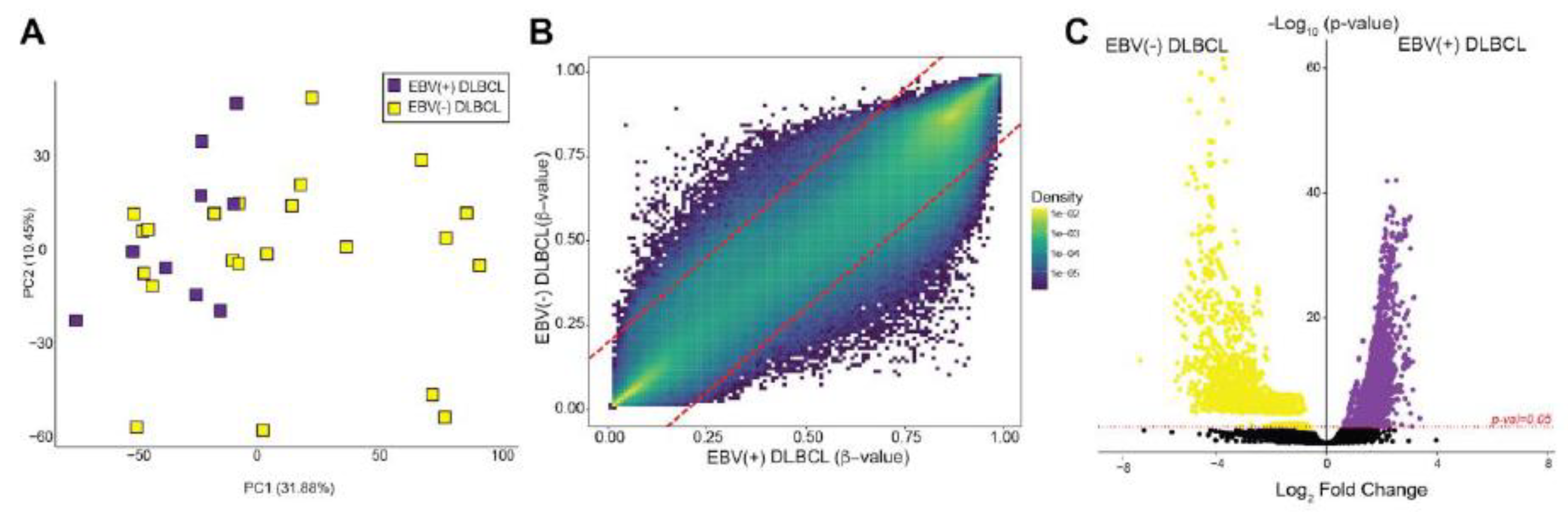
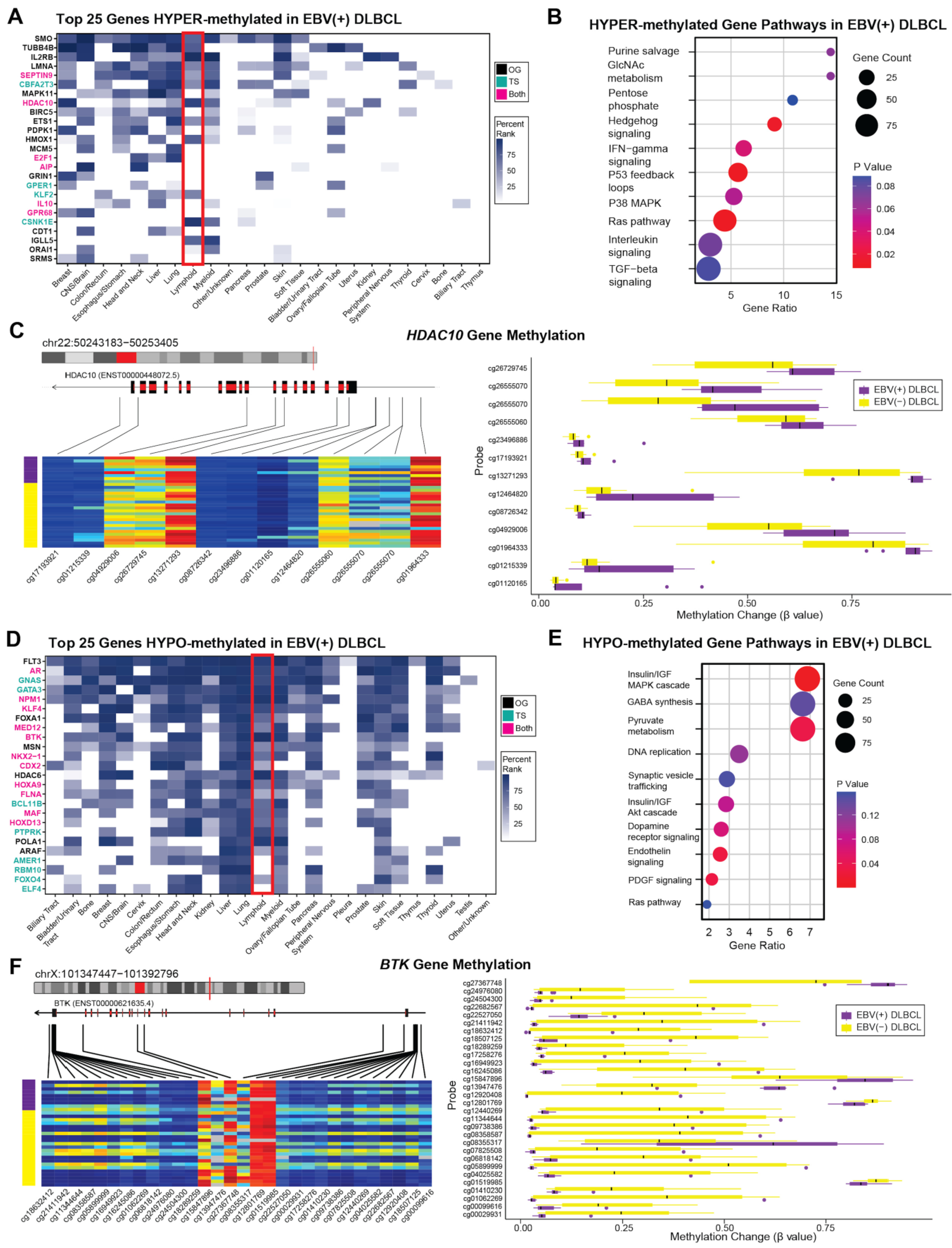
3.4. Methylome Patterns: EBV(+) DLBCL versus EBV(-) DLBCL
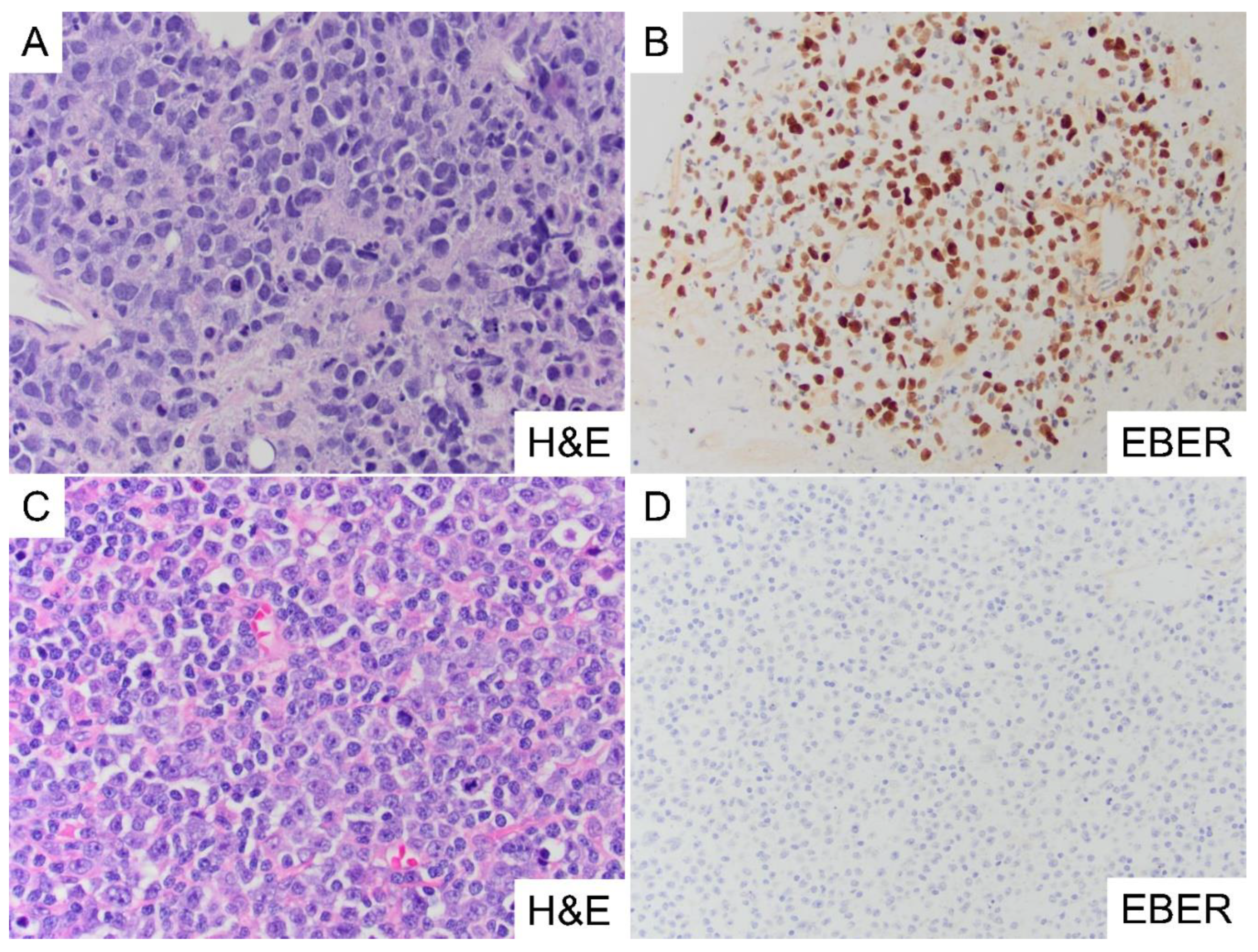
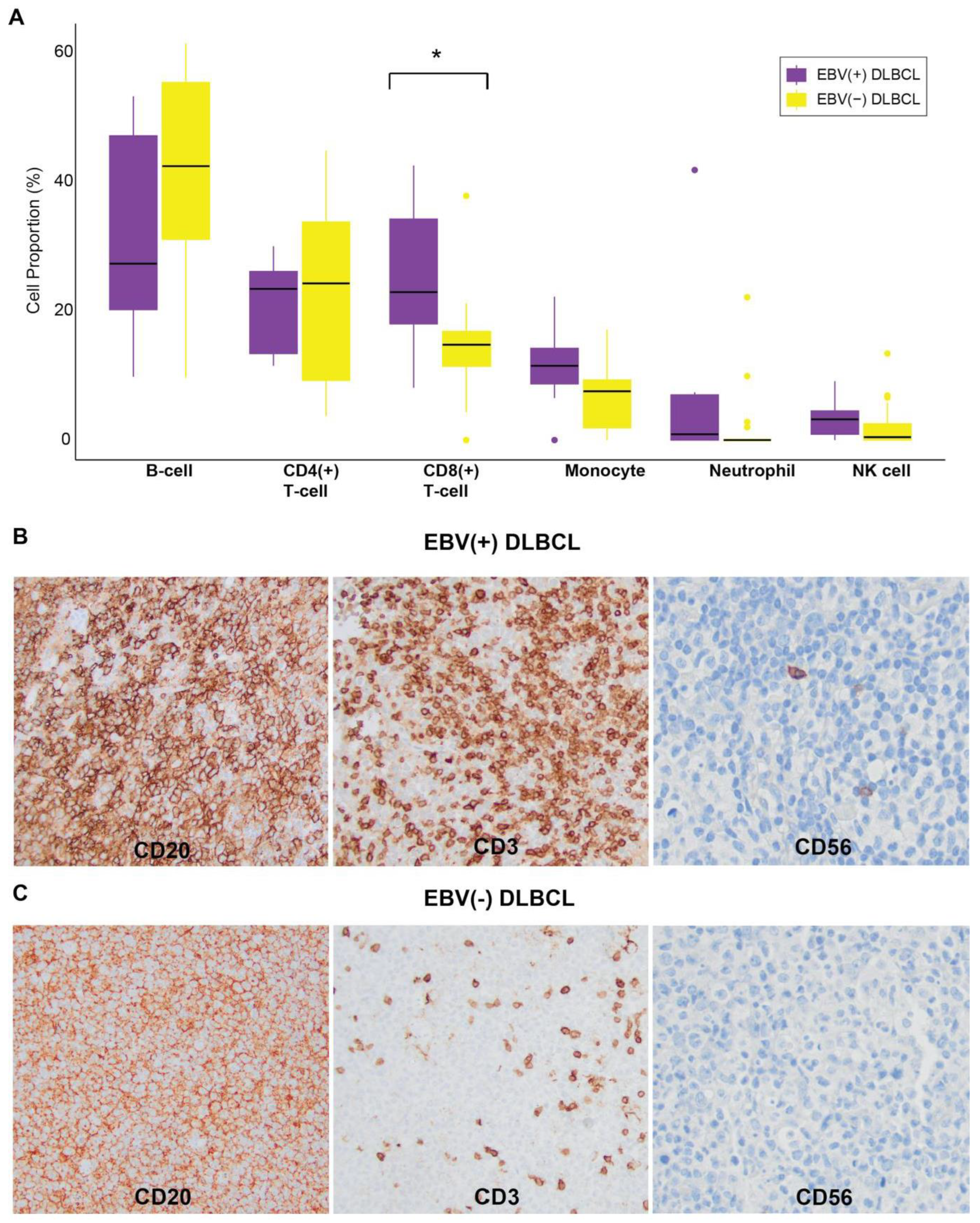
3.5. Immune Cell Composition in EBV(+) DLBCL
4. Discussion
5. Conclusions
Supplementary Materials
Funding
References
- Ahn, J.S.; Yang, D.H.; Duk Choi, Y.; Jung, S.H.; Yhim, H.Y.; Kwak, J.Y.; Sung Park, H.; Shin, M.G.; Kim, Y.K.; Kim, H.J.; Lee, J.J. Clinical Outcome of Elderly Patients with Epstein–Barr Virus Positive Diffuse Large B-Cell Lymphoma Treated with a Combination of Rituximab and CHOP Chemotherapy. American Journal of Hematology 2013, 88, 774–779. [Google Scholar] [CrossRef]
- Ok, C.Y.; Li, L.; Xu-Monette, Z.Y.; Visco, C.; Tzankov, A.; Manyam, G.C.; Montes-Moreno, S.; Dybaer, K.; Chiu, A.; Orazi, A.; Zu, Y.; Bhagat, G.; Chen, J.; Richards, K.L.; Hsi, E.D.; Choi, W.W. L.; Van Krieken, J.H.; Huh, J.; Ai, W.; Ponzoni, M.; Ferreri, A.J. M.; Farnen, J.P.; Møller, M.B.; Bueso-Ramos, C.E.; Miranda, R.N.; Winter, J.N.; Piris, M.A.; Medeiros, L.J.; Young, K.H. Prevalence and Clinical Implications of Epstein-Barr Virus Infection in de Novo Diffuse Large B-Cell Lymphoma in Western Countries. Clinical Cancer Research 2014, 20, 2338–2349. [Google Scholar] [CrossRef]
- Lu, T.X.; Liang, J.H.; Miao, Y.; Fan, L.; Wang, L.; Qu, X.Y.; Cao, L.; Gong, Q.X.; Wang, Z.; Zhang, Z.H.; Xu, W.; Li, J.Y. Epstein-Barr Virus Positive Diffuse Large B-Cell Lymphoma Predict Poor Outcome, Regardless of the Age. Scientific reports 2015, 5. [Google Scholar] [CrossRef]
- Editorial Board., W.C. of. Haematolymphoid Tumours [Internet; Beta Version Ahead of Print]; International Agency for Research on Cancer: Lyon (France), 2022.
- Ohashi, A.; Kato, S.; Okamoto, A.; Inaguma, Y.; Satou, A.; Tsuzuki, T.; Emi, N.; Okamoto, M.; Nakamura, S. Reappraisal of Epstein–Barr Virus (EBV) in Diffuse Large B-Cell Lymphoma (DLBCL): Comparative Analysis between EBV-Positive and EBV-Negative DLBCL with EBV-Positive Bystander Cells. Histopathology 2017, 71, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmann-Laeisz, C.; Szczepanowski, M.; Borchert, A.; Brüggemann, M.; Klapper, W. Epstein-Barr Virus-Negative Diffuse Large B-Cell Lymphoma Hosts Intra- and Peritumoral B-Cells with Activated Epstein-Barr Virus. Virchows Arch 2015, 466, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Hofscheier, A.; Ponciano, A.; Bonzheim, I.; Adam, P.; Lome-Maldonado, C.; Vela, T.; Cortes, E.; Ortiz-Hidalgo, C.; Fend, F.; Quintanilla-Martinez, L. Geographic Variation in the Prevalence of Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma of the Elderly: A Comparative Analysis of a Mexican and a German Population. Mod Pathol 2011, 24, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Mundo, L.; Del Porro, L.; Granai, M.; Siciliano, M.C.; Mancini, V.; Santi, R.; Marcar, L.; Vrzalikova, K.; Vergoni, F.; Di Stefano, G.; Schiavoni, G.; Segreto, G.; Onyango, N.; Nyagol, J.A.; Amato, T.; Bellan, C.; Anagnostopoulos, I.; Falini, B.; Leoncini, L.; Tiacci, E.; Lazzi, S. Frequent Traces of EBV Infection in Hodgkin and Non-Hodgkin Lymphomas Classified as EBV-Negative by Routine Methods: Expanding the Landscape of EBV-Related Lymphomas. Modern Pathology 2020, 33, 2407–2421. [Google Scholar] [CrossRef]
- Volaric, A.K.; Fedoriw, Y. Epstein-Barr Virus-Associated B-Cell Lymphoproliferative Disorders and Lymphomas: Diagnostic Overlaps and Defining Features. Human Pathology 2024, 105697. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, R.; Xie, Z. The Roles of DNA Methylation on the Promotor of the Epstein–Barr Virus (EBV) Gene and the Genome in Patients with EBV-Associated Diseases. Applied Microbiology and Biotechnology 2022, 106, 4413–4426. [Google Scholar] [CrossRef]
- Saha, A.; Jha, H.C.; Upadhyay, S.K.; Robertson, E.S. Epigenetic Silencing of Tumor Suppressor Genes during in Vitro Epstein-Barr Virus Infection. Proceedings of the National Academy of Sciences of the United States of America 2015, 112, E5199–E5207. [Google Scholar] [CrossRef]
- Ghosh Roy, S.; Robertson, E.S.; Saha, A. Epigenetic Impact on EBV Associated B-Cell Lymphomagenesis. Biomolecules 2016, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R. M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat Genet 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Kaur, D.; Horvath, S.; Zhou, W. Comparative Epigenome Analysis Using Infinium DNA Methylation BeadChips. Brief Bioinform 2023, 24, bbac617. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tang, J.; Li, N.; Zhao, Y.; Ai, R.; Zhang, K.; Wang, M.; Du, W.; Wang, W. Integrative Analysis with Expanded DNA Methylation Data Reveals Common Key Regulators and Pathways in Cancers. npj Genomic Med 2019, 4, 1–11. [Google Scholar] [CrossRef]
- Berglund, A.; Yamoah, K.; Eschrich, S.A.; Falahat, R.; Mulé, J.J.; Kim, S.; Matta, J.; Dutil, J.; Ruiz-Deya, G.; Ortiz Sanchez, C.; Wang, L.; Park, H.; Banerjee, H.N.; Lotan, T.; Barry, K.H.; Putney, R.M.; Kim, S.J.; Gwede, C.; Kresovich, J.K.; Kim, Y.; Lin, H.; Dhillon, J.; Chakrabarti, R.; Park, J.Y. Epigenome-wide Association Study of Prostate Cancer in African American Men Identified Differentially Methylated Genes. Cancer Medicine 2024, 13, e70044. [Google Scholar] [CrossRef]
- Zhou, W.; Triche, T.J., Jr; Laird, P.W.; Shen, H. SeSAMe: Reducing Artifactual Detection of DNA Methylation by Infinium BeadChips in Genomic Deletions. Nucleic Acids Research 2018, 46, e123. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA Methylation Landscape of Cancer. Trends in Genetics 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Besselink, N.; Keijer, J.; Vermeulen, C.; Boymans, S.; de Ridder, J.; van Hoeck, A.; Cuppen, E.; Kuijk, E. The Genome-Wide Mutational Consequences of DNA Hypomethylation. Sci Rep 2023, 13, 6874. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Hypomethylation in Cancer Cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Chambwe, N.; Kormaksson, M.; Geng, H.; De, S.; Michor, F.; Johnson, N.A.; Morin, R.D.; Scott, D.W.; Godley, L.A.; Gascoyne, R.D.; Melnick, A.; Campagne, F.; Shaknovich, R. Variability in DNA Methylation Defines Novel Epigenetic Subgroups of DLBCL Associated with Different Clinical Outcomes. Blood 2014, 123, 1699–1708. [Google Scholar] [CrossRef]
- Drillenburg, P.; Pals, S.T. Cell Adhesion Receptors in Lymphoma Dissemination. Blood 2000, 95, 1900–1910. [Google Scholar] [CrossRef]
- Ashton-Key, M.; Cowley, G.P.; Smith, M.E. Cadherins in Reactive Lymph Nodes and Lymphomas: High Expression in Anaplastic Large Cell Lymphomas. Histopathology 1996, 28, 55–59. [Google Scholar] [CrossRef]
- Yu, W.; Yang, L.; Li, T.; Zhang, Y. Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target. Front Oncol 2019, 9, 989. [Google Scholar] [CrossRef]
- Yu, S.; Han, R.; Gan, R. The Wnt/β-Catenin Signalling Pathway in Haematological Neoplasms. Biomarker Research 2022, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Fang, J.; Ye, F.; Zhang, S.; Huang, H.; Hou, J.; Wang, T. Diffuse Large B-Cell Lymphoma Promotes Endothelial-to-Mesenchymal Transition via WNT10A/Beta-Catenin/Snail Signaling. Front Oncol 2022, 12, 871788. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, A.; Kamran, H.; Akhter, A.; Seno, R.; Torlakovic, E.E.; Roshan, T.M.; Shabani-Rad, M.-T.; Elyamany, G.; Minoo, P.; Stewart, D. Identification of Potential Therapeutic Targets for Plasmablastic Lymphoma Through Gene Expression Analysis: Insights into RAS and Wnt Signaling Pathways. Modern Pathology 2023, 36, 100198. [Google Scholar] [CrossRef] [PubMed]
- Jardin, F.; Jais, J.-P.; Molina, T.-J.; Parmentier, F.; Picquenot, J.-M.; Ruminy, P.; Tilly, H.; Bastard, C.; Salles, G.-A.; Feugier, P.; Thieblemont, C.; Gisselbrecht, C.; de Reynies, A.; Coiffier, B.; Haioun, C.; Leroy, K. Diffuse Large B-Cell Lymphomas with CDKN2A Deletion Have a Distinct Gene Expression Signature and a Poor Prognosis under R-CHOP Treatment: A GELA Study. Blood 2010, 116, 1092–1104. [Google Scholar] [CrossRef]
- Cobbers, J.M. J. L.; Wolter, M.; Reifenberger, J.; Ring, G.U.; Jessen, F.; An, H.; Niederacher, D.; Schmidt, E.E.; Ichimura, K.; Floeth, F.; Kirsch, L.; Borchard, F.; Louis, D.N.; Collins, V.P.; Reifenberger, G. Frequent In Activation of CDKN2A and Rare Mutation of TP53 in PCNSL. Brain Pathol 2006, 8, 263–276. [Google Scholar] [CrossRef]
- Maura, F.; Dodero, A.; Carniti, C.; Bolli, N.; Magni, M.; Monti, V.; Cabras, A.; Leongamornlert, D.; Abascal, F.; Diamond, B.; Rodriguez-Martin, B.; Zamora, J.; Butler, A.; Martincorena, I.; Tubio, J.M. C.; Campbell, P.J.; Chiappella, A.; Pruneri, G.; Corradini, P. CDKN2A Deletion Is a Frequent Event Associated with Poor Outcome in Patients with Peripheral T-Cell Lymphoma Not Otherwise Specified (PTCL-NOS). Haematologica 2021, 106, 2918–2926. [Google Scholar] [CrossRef]
- Laharanne, E.; Chevret, E.; Idrissi, Y.; Gentil, C.; Longy, M.; Ferrer, J.; Dubus, P.; Jouary, T.; Vergier, B.; Beylot-Barry, M.; Merlio, J.-P. CDKN2A–CDKN2B Deletion Defines an Aggressive Subset of Cutaneous T-Cell Lymphoma. Modern Pathology 2010, 23, 547–558. [Google Scholar] [CrossRef]
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; LeBrun, D.P. Inactivation of the CDKN2A Tumor-Suppressor Gene by Deletion or Methylation Is Common at Diagnosis in Follicular Lymphoma and Associated with Poor Clinical Outcome. Clinical Cancer Research 2014, 20, 1676–1686. [Google Scholar] [CrossRef]
- Gaudio, F.; Dicataldo, M.; Di Giovanni, F.; Cazzato, G.; d’Amati, A.; Perrone, T.; Masciopinto, P.; Laddaga, F.E.; Musto, P.; Maiorano, E.; Ingravallo, G. Prognostic Role of CDKN2A Deletion and P53 Expression and Association With MIPIb in Mantle Cell Lymphoma. Clinical Lymphoma Myeloma and Leukemia 2023, 23, 599–605. [Google Scholar] [CrossRef]
- Salaverria, I.; Akasaka, T.; Gesk, S.; Szczepanowski, M.; Burkhardt, B.; Harder, L.; Damm-Welk, C.; Oschlies, I.; Klapper, W.; Dyer, M.J. S.; Siebert, R. The CBFA2T3/ACSF3 Locus Is Recurrently Involved in IGH Chromosomal Translocation t(14;16)(Q32;Q24) in Pediatric B-Cell Lymphoma with Germinal Center Phenotype. Genes Chromosomes Cancer 2012, 51, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Cheney, K.M.; Neilsen, P.M.; Schulz, R.B.; Callen, D.F. CBFA2T3–ZNF651, like CBFA2T3–ZNF652, Functions as a Transcriptional Corepressor Complex. FEBS Letters 2010, 584, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Micci, F.; Thorsen, J.; Panagopoulos, I.; Nyquist, K.B.; Zeller, B.; Tierens, A.; Heim, S. High-Throughput Sequencing Identifies an NFIA/CBFA2T3 Fusion Gene in Acute Erythroid Leukemia with t(1;16)(P31;Q24). Leukemia 2013, 27, 980–982. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, M.; Li, S.; Niu, J.; Xue, J.; Li, J.; Li, X. Identification of Hub Genes and Key Pathways Associated with Peripheral T-Cell Lymphoma. CURR MED SCI 2020, 40, 885–899. [Google Scholar] [CrossRef]
- Sahakian, E.; Shah, B.D.; Powers, J.; Deng, S.; Merino, O.; Gill, A.S.; Rock-Klotz, J.; Woan, K.V.; Vazquez, L.; Wang, H.; Chen-Kiang, S.; Tao, J.; Villagra, A.; Pinilla-Ibarz, J.; Sotomayor, E.M. The Opposing Role of Histone Deacetylase 10 (HDAC10) and HDAC11 in Proliferation/Survival of Mantle Cell Lymphoma (MCL) and Chronic Lymphocytic Leukemia (CLL). Blood 2011, 118, 1363. [Google Scholar] [CrossRef]
- Wen, W.; Zhang, W.-L.; Tan, R.; Zhong, T.-T.; Zhang, M.-R.; Fang, X.-S. Progress in Deciphering the Role of P53 in Diffuse Large B-Cell Lymphoma: Mechanisms and Therapeutic Targets. Am J Cancer Res 2024, 14, 3280–3293. [Google Scholar] [CrossRef]
- Bakkebø, M.; Huse, K.; Hilden, V.I.; Smeland, E.B.; Oksvold, M.P. TGF-β-Induced Growth Inhibition in B-Cell Lymphoma Correlates with Smad1/5 Signalling and Constitutively Active P38 MAPK. BMC Immunology 2010, 11, 57. [Google Scholar] [CrossRef]
- Chen, G.; Ghosh, P.; Osawa, H.; Sasaki, C.Y.; Rezanka, L.; Yang, J.; O’Farrell, T.J.; Longo, D.L. Resistance to TGF-Β1 Correlates with Aberrant Expression of TGF-β Receptor II in Human B-Cell Lymphoma Cell Lines. Blood 2007, 109, 5301–5307. [Google Scholar] [CrossRef]
- Lebman, D.A.; Edmiston, J.S. The Role of TGF-β in Growth, Differentiation, and Maturation of B Lymphocytes. Microbes and Infection 1999, 1, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK Pathway for Cancer Therapy: From Mechanism to Clinical Studies. Sig Transduct Target Ther 2023, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, V.; Navarro, L.; Sample, C.E.; David, M.; Sung, S.; Swaminathan, S. The Epstein-Barr Virus SM Protein Induces STAT1 and Interferon-Stimulated Gene Expression. J Virol 2003, 77, 3690–3701. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Mauser, A.; Wong, A.; Ting, J.P.; Kenney, S.C. Inhibition of IFN-Gamma Signaling by an Epstein-Barr Virus Immediate-Early Protein. Immunity 2001, 15, 787–799. [Google Scholar] [CrossRef]
- Powers, J.; Lienlaf, M.; Perez-Villarroel, P.; Deng, S.; Knox, T.; Villagra, A.; Sahakian, E. Expression and Function of Histone Deacetylase 10 (HDAC10) in B Cell Malignancies. Methods Mol Biol 2016, 1436, 129–145. [Google Scholar] [CrossRef]
- Wu, C.; Song, Q.; Gao, S.; Wu, S. Targeting HDACs for Diffuse Large B-Cell Lymphoma Therapy. Sci Rep 2024, 14, 289. [Google Scholar] [CrossRef]
- Mieland, A.O.; Petrosino, G.; Dejung, M.; Chen, J.-X.; Fulzele, A.; Mahmoudi, F.; Tu, J.-W.; Mustafa, A.-H. M.; Zeyn, Y.; Hieber, C.; Bros, M.; Schnöder, T.M.; Heidel, F.H.; Najafi, S.; Oehme, I.; Hofmann, I.; Schutkowski, M.; Hilscher, S.; Kosan, C.; Butter, F.; Bhatia, S.; Sippl, W.; Krämer, O.H. The Protein Deacetylase HDAC10 Controls DNA Replication in Malignant Lymphoid Cells. Leukemia 2025, 39, 1756–1768. [Google Scholar] [CrossRef]
- Medina, K.L. Flt3 Signaling in B Lymphocyte Development and Humoral Immunity. Int J Mol Sci 2022, 23, 7289. [Google Scholar] [CrossRef]
- Tobón, G.J.; Renaudineau, Y.; Hillion, S.; Cornec, D.; Devauchelle-Pensec, V.; Youinou, P.; Pers, J.-O. The Fms-like Tyrosine Kinase 3 Ligand, a Mediator of B Cell Survival, Is Also a Marker of Lymphoma in Primary Sjögren’s Syndrome. Arthritis Rheum 2010, 62, 3447–3456. [Google Scholar] [CrossRef]
- Yang, J.; Li, D.; Zhou, J. Histone Deacetylase 6 as a Therapeutic Target in B Cell-Associated Hematological Malignancies. Front Pharmacol 2020, 11, 971. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, J. Nucleophosmin1 (NPM1) Abnormality in Hematologic Malignancies, and Therapeutic Targeting of Mutant NPM1 in Acute Myeloid Leukemia. Ther Adv Hematol 2020, 11, 2040620719899818. [Google Scholar] [CrossRef]
- Wang, H.; Guo, H.; Yang, J.; Liu, Y.; Liu, X.; Zhang, Q.; Zhou, K. Bruton Tyrosine Kinase Inhibitors in B-Cell Lymphoma: Beyond the Antitumour Effect. Experimental Hematology & Oncology 2022, 11, 60. [Google Scholar] [CrossRef]
- Mehra, S.; Nicholls, M.; Taylor, J. The Evolving Role of Bruton’s Tyrosine Kinase Inhibitors in B Cell Lymphomas. Int J Mol Sci 2024, 25, 7516. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhu, Y.; Cheng, Y.; Hou, J.; Jin, F.; Li, M.; Jia, W.; Cheng, Z.; Xing, H.; Liu, M.; Han, T. BTK Kinase Activity Is Dispensable for the Survival of Diffuse Large B-Cell Lymphoma. J Biol Chem 2022, 298, 102555. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Jeffrey, M.L.; Li, Y.; Li, J.; Young, K.H. Genetic Alterations and Their Clinical Implications in DLBCL. Nature Reviews. Clinical Oncology 2019, 16, 634–652. [Google Scholar] [CrossRef]
- Loo, S.K.; Ab. Hamid, S.S.; Musa, M.; Wong, K.K. DNMT1 Is Associated with Cell Cycle and DNA Replication Gene Sets in Diffuse Large B-Cell Lymphoma. Pathology - Research and Practice 2018, 214, 134–143. [Google Scholar] [CrossRef]
- Pal Singh, S.; Dammeijer, F.; Hendriks, R.W. Role of Bruton’s Tyrosine Kinase in B Cells and Malignancies. Molecular Cancer 2018, 17, 57. [Google Scholar] [CrossRef]
- Tang, C.-L.; Li, X.-Z.; Zhou, T.; Deng, C.-M.; Jiang, C.-T.; Zhang, Y.-M.; Liao, Y.; Wang, T.-M.; He, Y.-Q.; Xue, W.-Q.; Jia, W.-H.; Zheng, X.-H. EBV DNA Methylation Profiles and Its Application in Distinguishing Nasopharyngeal Carcinoma and Nasal NK/T-Cell Lymphoma. Clinical Epigenetics 2024, 16, 11. [Google Scholar] [CrossRef]
- Healy, J.A.; Dave, S.S. The Role of EBV in the Pathogenesis of Diffuse Large B Cell Lymphoma. Current topics in microbiology and immunology 2015, 390, 315–337. [Google Scholar] [CrossRef]
- Liu, F.; Tian, S.; Liu, Q.; Deng, Y.; He, Q.; Shi, Q.; Chen, G.; Xu, X.; Yuan, J.; Nakamura, S.; Karube, K.; Wang, Z. Comparison of Genomic Alterations in Epstein–Barr Virus-Positive and Epstein–Barr Virus-Negative Diffuse Large B-Cell Lymphoma. Cancer Medicine 2024, 13, e6995. [Google Scholar] [CrossRef]
- Chapman, J.R.; Bouska, A.C.; Zhang, W.; Alderuccio, J.P.; Lossos, I.S.; Rimsza, L.M.; Maguire, A.; Yi, S.; Chan, W.C.; Vega, F.; Song, J.Y. EBV-Positive HIV-Associated Diffuse Large B Cell Lymphomas Are Characterized by JAK/STAT (STAT3) Pathway Mutations and Unique Clinicopathologic Features. British journal of haematology 2021, 194, 870–878. [Google Scholar] [CrossRef]
- Frontzek, F.; Staiger, A.M.; Wullenkord, R.; Grau, M.; Zapukhlyak, M.; Kurz, K.S.; Horn, H.; Erdmann, T.; Fend, F.; Richter, J.; Klapper, W.; Lenz, P.; Hailfinger, S.; Tasidou, A.; Trautmann, M.; Hartmann, W.; Rosenwald, A.; Quintanilla-Martinez, L.; Ott, G.; Anagnostopoulos, I.; Lenz, G. Molecular Profiling of EBV Associated Diffuse Large B-Cell Lymphoma. Leukemia 2023, 37, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Melnick, A. The Epigenetic Basis of Diffuse Large B-Cell Lymphoma. Semin Hematol 2015, 52, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.L.; Reyes-Garau, D.; Armengol, M.; Fernández-Serrano, M.; Roué, G. Recent Advances in the Targeting of Epigenetic Regulators in B-Cell Non-Hodgkin Lymphoma. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in Cancer. New England Journal of Medicine 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Hassler, M.R.; Schiefer, A.-I.; Egger, G. Combating the Epigenome: Epigenetic Drugs against Non-Hodgkin’s Lymphoma. Epigenomics 2013, 5, 397–415. [Google Scholar] [CrossRef]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D.; Briem, E.; Zhang, K.; Irizarry, R.A.; Feinberg, A.P. Increased Methylation Variation in Epigenetic Domains across Cancer Types. Nat Genet 2011, 43, 768–775. [Google Scholar] [CrossRef]
- Fiches, G.N.; Zhou, D.; Kong, W.; Biswas, A.; Ahmed, E.H.; Baiocchi, R.A.; Zhu, J.; Santoso, N. Profiling of Immune Related Genes Silenced in EBV-Positive Gastric Carcinoma Identified Novel Restriction Factors of Human Gammaherpesviruses. PLoS Pathog 2020, 16, e1008778. [Google Scholar] [CrossRef]
- Shaknovich, R.; Geng, H.; Johnson, N.A.; Tsikitas, L.; Cerchietti, L.; Greally, J.M.; Gascoyne, R.D.; Elemento, O.; Melnick, A. DNA Methylation Signatures Define Molecular Subtypes of Diffuse Large B-Cell Lymphoma. Blood 2010, 116, e81–e89. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Inman, G.J.; Allday, M.J. Resistance to TGF-Β1 Correlates with a Reduction of TGF-β Type II Receptor Expression in Burkitt’s Lymphoma and Epstein–Barr Virus-Transformed B Lymphoblastoid Cell Lines. Journal of General Virology 2000, 81, 1567–1578. [Google Scholar] [CrossRef]
- Cheng, F.; Zheng, B.; Wang, J.; Zhao, G.; Yao, Z.; Niu, Z.; He, W. Histone Deacetylase 10, a Potential Epigenetic Target for Therapy. Biosci Rep 2021, 41, BSR20210462. [Google Scholar] [CrossRef]
- Cerchietti, L. Genetic Mechanisms Underlying Tumor Microenvironment Composition and Function in Diffuse Large B-Cell Lymphoma. Blood 2024, 143, 1101–1111. [Google Scholar] [CrossRef]
- Cioroianu, A.I.; Stinga, P.I.; Sticlaru, L.; Cioplea, M.D.; Nichita, L.; Popp, C.; Staniceanu, F. Tumor Microenvironment in Diffuse Large B-Cell Lymphoma: Role and Prognosis. Analytical Cellular Pathology 2019, 2019, 8586354. [Google Scholar] [CrossRef]
- He, M.; Liu, M.; Yuan, J.; Lv, J.; Li, W.; Yan, Q.; Tang, Y.; Wang, L.; Guo, L.; Liu, F. Spatial Transcriptomics Reveals Tumor Microenvironment Heterogeneity in EBV Positive Diffuse Large B Cell Lymphoma. Sci Rep 2025, 15, 15878. [Google Scholar] [CrossRef]
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





