Submitted:
18 August 2025
Posted:
19 August 2025
You are already at the latest version
Abstract
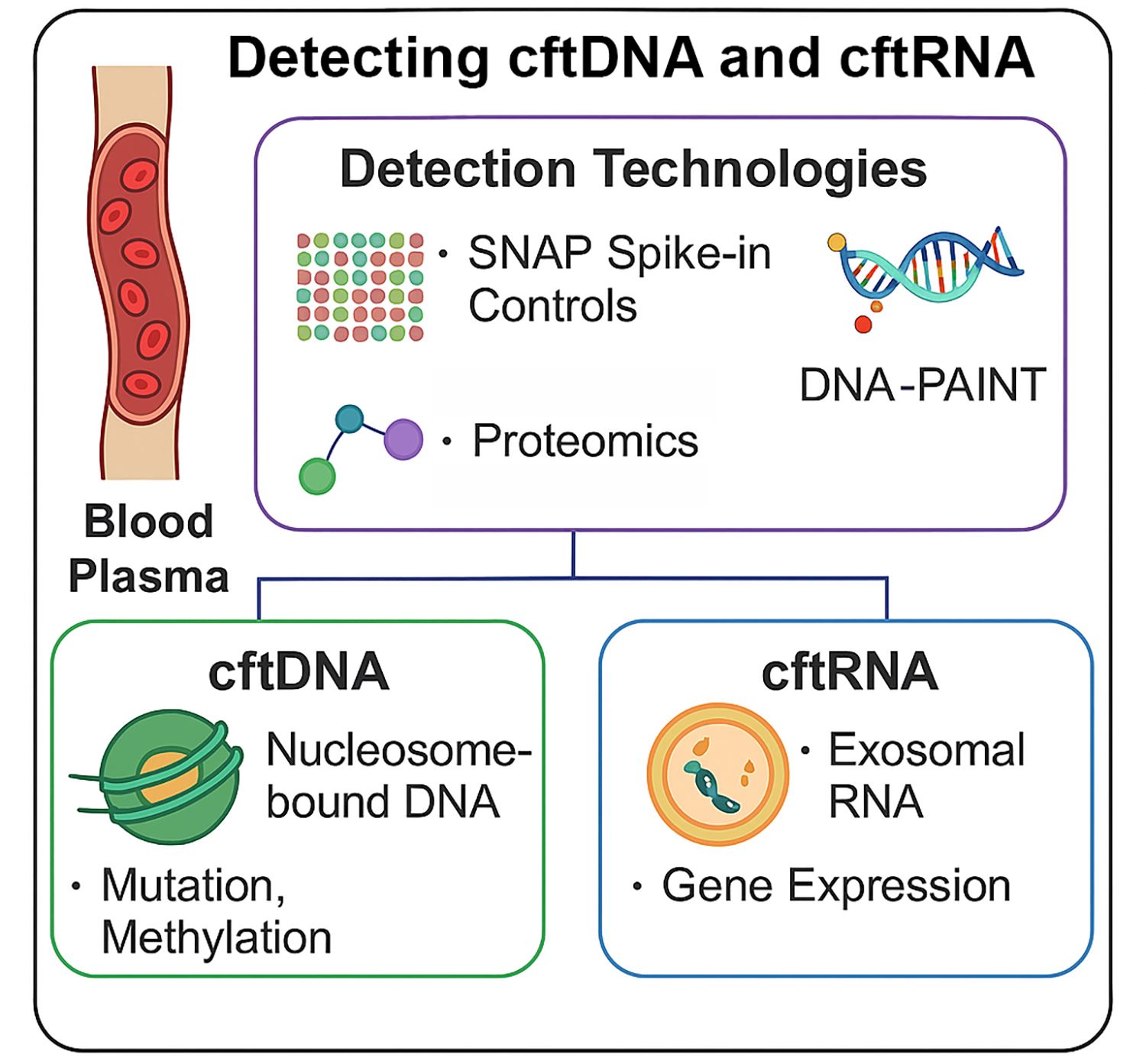
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Detection Technologies for cftDNA
3.1.1. Quantifying Nucleosome-Bound cftDNA Using Epicypher SNAP Spike-in Controls
3.1.2. DNA Methylation Detection
3.1.3. Fragmentomics and Size-Selective Enrichment
3.1.4. Mutation Detection: Digital PCR and NGS Panels
3.1.5. DNA-PAINT in Cancer Detection
-
High Sensitivity and Multiplexing:DNA-PAINT can detect DNA sequences at extremely low concentrations (femtomolar range), making it ideal for identifying rare cancer-associated mutations in liquid biopsies. It also supports multiplexing, allowing simultaneous detection of multiple oncogenic mutations, methylation sites, or genomic rearrangements in the same sample [51].
-
Point Mutation and Methylation Detection:Recent developments enable DNA-PAINT to identify single-nucleotide variants and methylation patterns characteristic of cancer DNA [50]. Methylated DNA often has a higher melting temperature than unmethylated DNA, and pattern recognition approaches like those used in the GRAIL platform can be applied to DNA-PAINT datasets for classification of cancer type and stage [50].
-
Single-Molecule Resolution for Amplified DNA:DNA-PAINT can visualize amplified DNA sequences, chromosomal rearrangements, and telomeric repeats with high spatial precision, providing both quantitative and structural insights into tumor DNA architecture [51].
-
Exosome and Tissue-of-Origin Profiling:When applied to exosome-derived nucleic acids, DNA-PAINT can be combined with advanced proteomics and proximity-barcoding methods that profile the protein cargo of exosomes to identify their tissue of origin [52]. This integration allows determination of where cftDNA originated, adding an important layer of diagnostic specificity [52].
-
Advantages Over Other Methods:Compared to fluorescence in situ hybridization (FISH) or PCR-based assays, DNA-PAINT offers superior spatial resolution, lower background noise, and reduced need for amplification, minimizing the risk of introducing errors [51]. The method’s flexibility enables adaptation for DNA, RNA, or protein targets, making it a versatile addition to the cancer biomarker detection toolkit [51].
3.2. Detection Technologies for cftRNA
3.2.1. RNA Stabilization and Isolation
3.2.2. Exosomal RNA Enrichment
3.2.3. Reverse Transcription and Amplification Technologies
3.2.4. RNA-Seq and Digital PCR Applications
3.2.5. Circular and Non-Coding RNAs
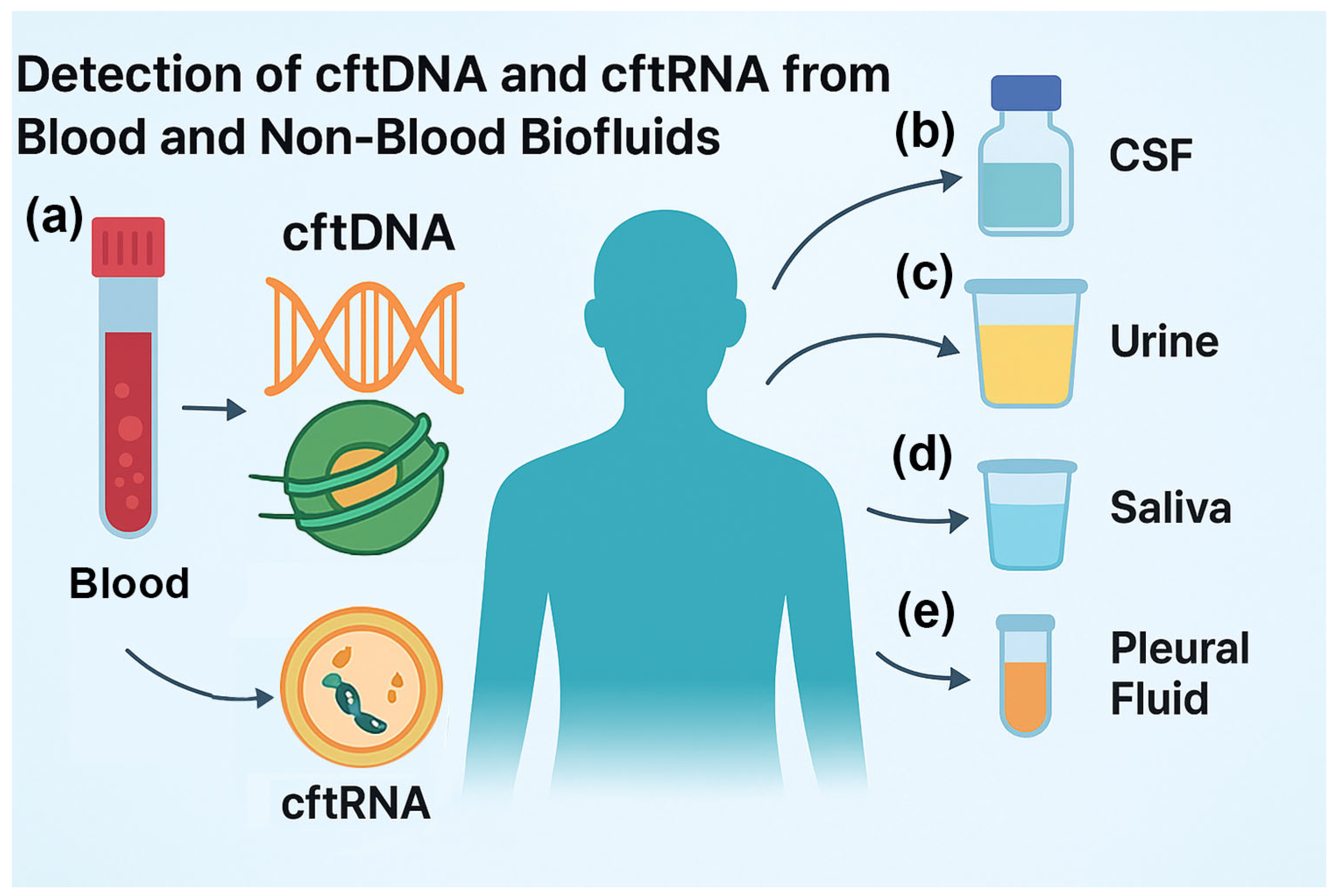
3.3. cftDNA and cftRNA from Non-Blood Body Fluids
3.3.1. cftDNA and cftRNA from Cerebrospinal fluid (CSF) for CNS Tumor Detection and Monitoring
3.3.2. cftDNA and cftRNA from Urine for Bladder, Kidney, and Prostate Cancer
3.3.3. cftDNA and cftRNA from Saliva for Cancers of the Aerodigestive Tract
3.3.4. cftDNA and cftRNA from Pleural Fluid for Malignant Cancers
3.3.5. Advantages of Non-Blood Fluids
- Higher local cftDNA concentration when sampling near the tumor site (e.g., CSF for CNS tumors, urine for bladder cancer) [79].
- Utility for inaccessible tumors, where tissue biopsy is high-risk or impractical [79].
- Enhanced tumor heterogeneity profiling, as sampling from multiple fluids may capture distinct subclonal populations [101].
- Facilitation of serial monitoring, particularly with easily collectible fluids such as urine or saliva [97].
4. Discussion
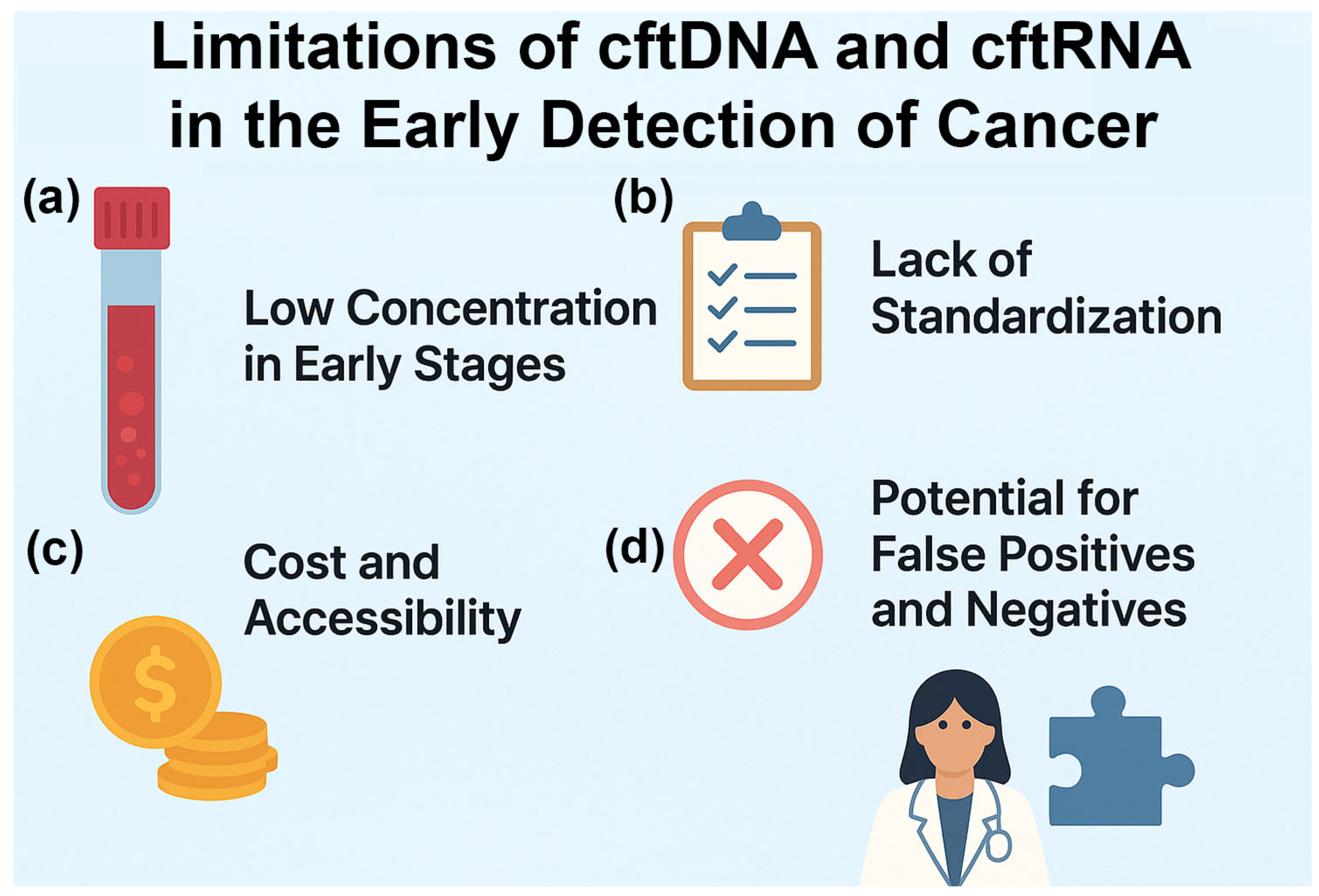
5. Limitations and Future Directions
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cftDNA | Cell free tumor DNA |
| cftRNA | Cell free tumor RNA |
| cfDNA | Cell free DNA (not necessarily from tumor) |
| cfRNA | Cell free RNA (not necessarily from tumor) |
| UMI | Unique molecular identifier |
| ddPCR | Droplet digital polymerase chain reaction |
| WGS | Whole genome sequencing |
| PTM | Post translational modifications |
| DNA-PAINT | DNA points accumulation for imaging in nanoscale topography |
| FISH | Fluorescent in situ hybridization |
| EV | Extracellular vesicle |
| RT-qPCR | Reverse transcriptase quantitative polymerase chain reaction |
| circRNA | Circular RNA |
| lncRNA | Long noncoding RNA |
| FRET | Fluorescence resonance energy transfer |
| MRD | Minimal residual disease |
| CSF | Cerebral spinal fluid |
| HNSCC | Head and neck small cell carcinoma |
| VAF | Variable allele frequency |
References
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef]
- Milner, D.A., Jr.; Lennerz, J.K. Technology and Future of Multi-Cancer Early Detection. Life (Basel) 2024, 14. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, S.; Xia, C.; Yu, H.; Shi, S.; Chen, K.; He, Y.; Deng, C.; Jin, H.; Liu, J.; et al. Liquid biopsy-based multi-cancer early detection: an exploration road from evidence to implementation. Sci Bull (Beijing) 2025. [Google Scholar] [CrossRef]
- Madar, S.; Amor, R.E.; Furman-Assaf, S.; Friedman, E. Innovative Approaches to Early Detection of Cancer-Transforming Screening for Breast, Lung, and Hard-to-Screen Cancers. Cancers (Basel) 2025, 17. [Google Scholar] [CrossRef]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Goncalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes (Basel) 2021, 12. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Loy, C.; Ahmann, L.; De Vlaminck, I.; Gu, W. Liquid Biopsy Based on Cell-Free DNA and RNA. Annu Rev Biomed Eng 2024, 26, 169–195. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Shehzad, A.; Khan, S.A.; Miran, W.; Khan, S.; Lee, Y.S. Diagnostic and Therapeutic Potential of Circulating-Free DNA and Cell-Free RNA in Cancer Management. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, K.; Kurian, N.S.; Goswami, H.M.; Rishi, K.D.; Veldore, V.H. Exploring the clinical utility of liquid biopsy with cfDNA in cancer: A systematic review. J Liq Biopsy 2024, 5, 100150. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Grzybowski, A.T.; Shah, R.N.; Richter, W.F.; Ruthenburg, A.J. Native internally calibrated chromatin immunoprecipitation for quantitative studies of histone post-translational modifications. Nat Protoc 2019, 14, 3275–3302. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Oberhofer, A.; Gabriel, S.; Polatoglou, E.; Randeu, H.; Uhlig, C.; Pfister, H.; Mayer, Z.; Holdenrieder, S. New Perspectives on the Importance of Cell-Free DNA Biology. Diagnostics (Basel) 2022, 12. [Google Scholar] [CrossRef]
- Saha, S.; Araf, Y.; Promon, S.K. Circulating tumor DNA in cancer diagnosis, monitoring, and prognosis. J Egypt Natl Canc Inst 2022, 34, 8. [Google Scholar] [CrossRef] [PubMed]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019, 10, 1930. [Google Scholar] [CrossRef]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Clausen, F.B.; Jorgensen, K.; Wardil, L.W.; Nielsen, L.K.; Krog, G.R. Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects. PLoS One 2023, 18, e0282332. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.A.; Chung, J.H.; Kennedy, M.; Hughes, J.D.; Chennagiri, N.; Lieber, D.S.; Fendler, B.; Young, L.; Zhao, M.; Coyne, M.; et al. Analytical Validation of a Hybrid Capture-Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA. J Mol Diagn 2018, 20, 686–702. [Google Scholar] [CrossRef]
- Cabus, L.; Lagarde, J.; Curado, J.; Lizano, E.; Perez-Boza, J. Current challenges and best practices for cell-free long RNA biomarker discovery. Biomark Res 2022, 10, 62. [Google Scholar] [CrossRef]
- Tang, Y.T.; Huang, Y.Y.; Zheng, L.; Qin, S.H.; Xu, X.P.; An, T.X.; Xu, Y.; Wu, Y.S.; Hu, X.M.; Ping, B.H.; et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med 2017, 40, 834–844. [Google Scholar] [CrossRef]
- Bryce, A.H.; Thiel, D.D.; Seiden, M.V.; Richards, D.; Luan, Y.; Coignet, M.; Zhang, Q.; Zhang, N.; Hubbell, E.; Kurtzman, K.N.; et al. Performance of a Cell-Free DNA-Based Multi-cancer Detection Test in Individuals Presenting With Symptoms Suspicious for Cancers. JCO Precis Oncol 2023, 7, e2200679. [Google Scholar] [CrossRef]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020, 369. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016, 34, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors (Basel) 2018, 18. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.O.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat Protoc 2019, 14, 2749–2780. [Google Scholar] [CrossRef]
- Wilson, S.L.; Shen, S.Y.; Harmon, L.; Burgener, J.M.; Triche, T., Jr.; Bratman, S.V.; De Carvalho, D.D.; Hoffman, M.M. Sensitive and reproducible cell-free methylome quantification with synthetic spike-in controls. Cell Rep Methods 2022, 2, 100294. [Google Scholar] [CrossRef]
- Bauml, J.M.; Li, B.T.; Velcheti, V.; Govindan, R.; Curioni-Fontecedro, A.; Dooms, C.; Takahashi, T.; Duda, A.W.; Odegaard, J.I.; Cruz-Guilloty, F.; et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer 2022, 166, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Jungmann, R.; Avendano, M.S.; Woehrstein, J.B.; Dai, M.; Shih, W.M.; Yin, P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods 2014, 11, 313–318. [Google Scholar] [CrossRef]
- van Wee, R.; Filius, M.; Joo, C. Completing the canvas: advances and challenges for DNA-PAINT super-resolution imaging. Trends Biochem Sci 2021, 46, 918–930. [Google Scholar] [CrossRef]
- Diaz, I.M.; Nocon, A.; Held, S.A.E.; Kobilay, M.; Skowasch, D.; Bronkhorst, A.J.; Ungerer, V.; Fredebohm, J.; Diehl, F.; Holdenrieder, S.; et al. Pre-Analytical Evaluation of Streck Cell-Free DNA Blood Collection Tubes for Liquid Profiling in Oncology. Diagnostics (Basel) 2023, 13. [Google Scholar] [CrossRef]
- Feng, B.; Liu, T.; Feng, Y.; Huang, J.; Qin, J. From sensitivity to public health: integrating cfMeDIP-seq into early breast cancer detection strategies. J Transl Med 2024, 22, 1083. [Google Scholar] [CrossRef]
- Tanaka, Y.; Mizuguchi, R.; Koseki, N.; Suzuki, H.; Suzuki, T. Quality assessment of enzymatic methyl-seq library constructed using crude cell lysate. Biochem Biophys Res Commun 2024, 696, 149488. [Google Scholar] [CrossRef]
- Huang, J.; Wang, L. Cell-Free DNA Methylation Profiling Analysis-Technologies and Bioinformatics. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; An, Y.; Wang, B.; Liang, G. Comparative analysis of SDC2 and SEPT9 methylation tests in the early detection of colorectal cancer: a systematic review and meta-analysis. Front Med (Lausanne) 2024, 11, 1460233. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, P.; Liao, Q.; Huang, Z. Association of DNA methylation of RASSF1A and SHOX2 with lung cancer risk: A systematic review and meta-analysis. Medicine (Baltimore) 2024, 103, e40042. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhao, Y.; Hou, X.; Li, N.; Liu, H.; Zhang, X.; Xu, X. SHOX2 and RASSF1A methylation in diagnosing malignant pleural effusion induced by lung cancer. Clin Chim Acta 2025, 572, 120273. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Zhang, L.; Xing, Y.; Zhu, Y.; Mao, K.; Wang, M.; Xuan, K.; Yang, Y.; Lo, R.; Luo, B.; et al. Clinical significance of urine-based automated detection of GSTP1 methylation for the diagnosis of suspected prostate cancer patients. Transl Androl Urol 2025, 14, 1204–1213. [Google Scholar] [CrossRef]
- Mharrach, I.; Aqerrout, M.; Tadlaoui, K.A.; Laraqui, A.; Ghazzaly, A.E.; Ennaji, M.M. Methylation status of GSTP1 and APC promoter genes as potential biomarkers for early detection and tumor aggressiveness in prostate cancer: insights from a Moroccan cohort. Mol Biol Rep 2025, 52, 711. [Google Scholar] [CrossRef]
- Qi, T.; Pan, M.; Shi, H.; Wang, L.; Bai, Y.; Ge, Q. Cell-Free DNA Fragmentomics: The Novel Promising Biomarker. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Gupta, R.; Othman, T.; Chen, C.; Sandhu, J.; Ouyang, C.; Fakih, M. Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer. Oncologist 2020, 25, 235–243. [Google Scholar] [CrossRef]
- Qi, Z.; Tokuhiro, S.; Odegaard, J.I.; Wienke, S.; Karnoub, M.; Feng, W.; Shiga, R.; Smit, E.F.; Goto, Y.; De Langen, A.J.; et al. Analytical and Clinical Validation of the Plasma-Based Guardant360 CDx Test for Assessing HER2 (ERBB2) Mutation Status in Patients with Non-Small-Cell Lung Cancer for Treatment with Trastuzumab Deruxtecan in DESTINY-Lung01/02. J Mol Diagn 2025, 27, 119–129. [Google Scholar] [CrossRef]
- Isla, D.; Alvarez, R.; Arnal, M.; Arriola, E.; Azkarate, A.; Azkona, E.; Garcia-Campelo, R.; Garrido, P.; Nadal, E.; Ortega, A.L.; et al. Detection of genomic alterations in liquid biopsies from patients with non-small cell lung cancer using FoundationOne Liquid CDx: a cost-effectiveness analysis. J Med Econ 2024, 27, 1379–1387. [Google Scholar] [CrossRef]
- Dunwell, T.L.; Dailey, S.C.; Ottestad, A.L.; Yu, J.; Becker, P.W.; Scaife, S.; Richman, S.D.; Wood, H.M.; Slaney, H.; Bottomley, D.; et al. Adaptor Template Oligo-Mediated Sequencing (ATOM-Seq) is a new ultra-sensitive UMI-based NGS library preparation technology for use with cfDNA and cfRNA. Sci Rep 2021, 11, 3138. [Google Scholar] [CrossRef] [PubMed]
- Streubel, A.; Stenzinger, A.; Stephan-Falkenau, S.; Kollmeier, J.; Misch, D.; Blum, T.G.; Bauer, T.; Landt, O.; Am Ende, A.; Schirmacher, P.; et al. Comparison of different semi-automated cfDNA extraction methods in combination with UMI-based targeted sequencing. Oncotarget 2019, 10, 5690–5702. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Reichl, A.; Tinnefeld, P. Kinetic Referencing Allows Identification of Epigenetic Cytosine Modifications by Single-Molecule Hybridization Kinetics and Superresolution DNA-PAINT Microscopy. ACS Nano 2024, 18, 1496–1503. [Google Scholar] [CrossRef]
- Piantanida, L.; Li, I.T.S.; Hughes, W.L. Advancements in DNA-PAINT: applications and challenges in biological imaging and nanoscale metrology. Nanoscale 2025, 17, 14016–14034. [Google Scholar] [CrossRef]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat Commun 2019, 10, 3854. [Google Scholar] [CrossRef]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur J Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Kremenska, Y.; Nair, V.M.; Kremenskoy, M.; Joseph, B.; Kurochkin, I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013, 1, e201. [Google Scholar] [CrossRef] [PubMed]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol 2021, 23, 631–641. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Brandao, B.B.; Thomou, T.; Altindis, E.; Kahn, C.R. Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep 2022, 38, 110277. [Google Scholar] [CrossRef]
- Roshani, M.; Baniebrahimi, G.; Mousavi, M.; Zare, N.; Sadeghi, R.; Salarinia, R.; Sheida, A.; Molavizadeh, D.; Sadeghi, S.; Moammer, F.; et al. Exosomal long non-coding RNAs: novel molecules in gastrointestinal cancers' progression and diagnosis. Front Oncol 2022, 12, 1014949. [Google Scholar] [CrossRef]
- Kocabey, S.; Chiarelli, G.; Acuna, G.P.; Ruegg, C. Ultrasensitive and multiplexed miRNA detection system with DNA-PAINT. Biosens Bioelectron 2023, 224, 115053. [Google Scholar] [CrossRef] [PubMed]
- Capone, I.; Bozzi, F.; Dagrada, G.P.; Verderio, P.; Conca, E.; Busico, A.; Testi, M.A.; Monti, V.; Duca, M.; Proto, C.; et al. Targeted RNA-sequencing analysis for fusion transcripts detection in tumor diagnostics: assessment of bioinformatic tools reliability in FFPE samples. Explor Target Antitumor Ther 2022, 3, 582–597. [Google Scholar] [CrossRef]
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol 2019, 20, 213. [Google Scholar] [CrossRef]
- Kim, J.; Kang, C.; Shin, S.; Hohng, S. Rapid quantification of miRNAs using dynamic FRET-FISH. Commun Biol 2022, 5, 1072. [Google Scholar] [CrossRef]
- Urbanek, M.O.; Nawrocka, A.U.; Krzyzosiak, W.J. Small RNA Detection by in Situ Hybridization Methods. Int J Mol Sci 2015, 16, 13259–13286. [Google Scholar] [CrossRef]
- Shaba, E.; Vantaggiato, L.; Governini, L.; Haxhiu, A.; Sebastiani, G.; Fignani, D.; Grieco, G.E.; Bergantini, L.; Bini, L.; Landi, C. Multi-Omics Integrative Approach of Extracellular Vesicles: A Future Challenging Milestone. Proteomes 2022, 10. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; He, L.; Du, J.; Li, L.; Bie, Z.; Li, Y.; Xu, X.; Zhou, W.; Wu, X.; et al. Preservation of cfRNA in cytological supernatants for cfDNA & cfRNA double detection in non-small cell lung cancer patients. Cancer Med 2024, 13, e70197. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.; Bai, L.; Hong, M.; Ouyang, J.; Wang, R.; Zhang, X.; Chen, P. A Comprehensive Review on Circulating cfRNA in Plasma: Implications for Disease Diagnosis and Beyond. Diagnostics (Basel) 2024, 14. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Jeyhani, M.; Thillainadesan, G.; Sheng, M.; Wunsche, R.; Dayarathna, T.; Cimolai, K.; Weng, H.; Jerzak, K.J.; Liu, S.K.; et al. Next Generation Aqueous Two-Phase System for Gentle, Effective, and Timely Extracellular Vesicle Isolation and Transcriptomic Analysis. J Extracell Vesicles 2025, 14, e70058. [Google Scholar] [CrossRef]
- Du, K.; Sun, X.; Tang, X.; Xu, H.; Duan, P.; Xu, Q.; Sang, M. [Effects of storage temperature and time on quality of plasma exosomes extracted by ExoQuick(TM) and Umibio kits]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2020, 36, 330–336. [Google Scholar]
- Gemoll, T.; Rozanova, S.; Roder, C.; Hartwig, S.; Kalthoff, H.; Lehr, S.; ElSharawy, A.; Habermann, J.K. Protein Profiling of Serum Extracellular Vesicles Reveals Qualitative and Quantitative Differences After Differential Ultracentrifugation and ExoQuick(TM) Isolation. J Clin Med 2020, 9. [Google Scholar] [CrossRef]
- Ghazi, B.; Harmak, Z.; Rghioui, M.; Kone, A.S.; El Ghanmi, A.; Badou, A. Decoding the secret of extracellular vesicles in the immune tumor microenvironment of the glioblastoma: on the border of kingdoms. Front Immunol 2024, 15, 1423232. [Google Scholar] [CrossRef] [PubMed]
- Yekula, A.; Yekula, A.; Muralidharan, K.; Kang, K.; Carter, B.S.; Balaj, L. Extracellular Vesicles in Glioblastoma Tumor Microenvironment. Front Immunol 2019, 10, 3137. [Google Scholar] [CrossRef]
- Reclusa, P.; Laes, J.F.; Malapelle, U.; Valentino, A.; Rocco, D.; Gil-Bazo, I.; Rolfo, C. EML4-ALK translocation identification in RNA exosomal cargo (ExoALK) in NSCLC patients: a novel role for liquid biopsy. Transl Cancer Res 2019, 8, S76–S78. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Rasmussen, M.H.; Christensen, M.H.; Frydendahl, A.; Maretty, L.; Andersen, C.L.; Besenbacher, S. Evaluating Bioinformatics Processing of Somatic Variant Detection in cfDNA Using Targeted Sequencing with UMIs. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.M.; Yeung, M.H.Y.; Wong, A.N.N.; Tsang, H.F.; Yu, A.C.S.; Yim, A.K.Y.; Wong, S.C.C. Targeted Sequencing Approach and Its Clinical Applications for the Molecular Diagnosis of Human Diseases. Cells 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Bochicchio, M.T.; Petiti, J.; Berchialla, P.; Izzo, B.; Giugliano, E.; Ottaviani, E.; Errichiello, S.; Rege-Cambrin, G.; Venturi, C.; Luciano, L.; et al. Droplet Digital PCR for BCR-ABL1 Monitoring in Diagnostic Routine: Ready to Start? Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Check, M.H.; Ernst, S.E.; Sfanos, K.S. Use of Droplet Digital PCR for Consistent Detection of TMPRSS2:ERG Gene Fusion Transcripts Initiated In Vitro. Prostate 2025. [Google Scholar] [CrossRef]
- Grudda, T.; Jiang, C.; Lo, C.M.; Skinner, N.E.; Sulkowski, M.S.; Balagopal, A.; Thio, C.L. High-dimensional droplet digital PCR of multiple hepatitis B splice variants in serum, liver, and tissue culture. J Virol Methods 2025, 336, 115155. [Google Scholar] [CrossRef]
- Feng, X.Y.; Zhu, S.X.; Pu, K.J.; Huang, H.J.; Chen, Y.Q.; Wang, W.T. New insight into circRNAs: characterization, strategies, and biomedical applications. Exp Hematol Oncol 2023, 12, 91. [Google Scholar] [CrossRef]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going circular: history, present, and future of circRNAs in cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef]
- Werner, B.; Warton, K.; Ford, C.E. Transcending Blood-Opportunities for Alternate Liquid Biopsies in Oncology. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Xiao, F.; Lv, S.; Zong, Z.; Wu, L.; Tang, X.; Kuang, W.; Zhang, P.; Li, X.; Fu, J.; Xiao, M.; et al. Cerebrospinal fluid biomarkers for brain tumor detection: clinical roles and current progress. Am J Transl Res 2020, 12, 1379–1396. [Google Scholar]
- Otsuji, R.; Fujioka, Y.; Hata, N.; Kuga, D.; Hatae, R.; Sangatsuda, Y.; Nakamizo, A.; Mizoguchi, M.; Yoshimoto, K. Liquid Biopsy for Glioma Using Cell-Free DNA in Cerebrospinal Fluid. Cancers (Basel) 2024, 16. [Google Scholar] [CrossRef]
- Crucitta, S.; Pasqualetti, F.; Gonnelli, A.; Ruglioni, M.; Luculli, G.I.; Cantarella, M.; Ortenzi, V.; Scatena, C.; Paiar, F.; Naccarato, A.G.; et al. IDH1 mutation is detectable in plasma cell-free DNA and is associated with survival outcome in glioma patients. BMC Cancer 2024, 24, 31. [Google Scholar] [CrossRef]
- Fontanilles, M.; Marguet, F.; Beaussire, L.; Magne, N.; Pepin, L.F.; Alexandru, C.; Tennevet, I.; Hanzen, C.; Langlois, O.; Jardin, F.; et al. Cell-free DNA and circulating TERT promoter mutation for disease monitoring in newly-diagnosed glioblastoma. Acta Neuropathol Commun 2020, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Cantor, E.; Wierzbicki, K.; Tarapore, R.S.; Ravi, K.; Thomas, C.; Cartaxo, R.; Nand Yadav, V.; Ravindran, R.; Bruzek, A.K.; Wadden, J.; et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro Oncol 2022, 24, 1366–1374. [Google Scholar] [CrossRef]
- Shi, L.; Tang, J.; Tao, H.; Guo, L.; Wu, W.; Wu, H.; Liu, Z.; Tong, L.; Wu, W.; Li, H.; et al. Detection of EGFR Mutations in Cerebrospinal Fluid of EGFR-Mutant Lung Adenocarcinoma With Brain Metastases. Front Oncol 2021, 11, 622142. [Google Scholar] [CrossRef]
- Riviere-Cazaux, C.; Dong, X.; Mo, W.; Kumar, R.; Dai, C.; Carlstrom, L.P.; Munoz-Casabella, A.; Ghadimi, K.; Nesvick, C.L.; Andersen, K.M.; et al. Longitudinal Glioma Monitoring via Cerebrospinal Fluid Cell-Free DNA. Clin Cancer Res 2025, 31, 881–889. [Google Scholar] [CrossRef]
- Shaw, L.M.; Arias, J.; Blennow, K.; Galasko, D.; Molinuevo, J.L.; Salloway, S.; Schindler, S.; Carrillo, M.C.; Hendrix, J.A.; Ross, A.; et al. Appropriate use criteria for lumbar puncture and cerebrospinal fluid testing in the diagnosis of Alzheimer's disease. Alzheimers Dement 2018, 14, 1505–1521. [Google Scholar] [CrossRef]
- Fitzpatrick, A.; Iravani, M.; Mills, A.; Childs, L.; Alaguthurai, T.; Clifford, A.; Garcia-Murillas, I.; Van Laere, S.; Dirix, L.; Harries, M.; et al. Assessing CSF ctDNA to Improve Diagnostic Accuracy and Therapeutic Monitoring in Breast Cancer Leptomeningeal Metastasis. Clin Cancer Res 2022, 28, 1180–1191. [Google Scholar] [CrossRef]
- Peng, H.; Pan, M.; Zhou, Z.; Chen, C.; Xing, X.; Cheng, S.; Zhang, S.; Zheng, H.; Qian, K. The impact of preanalytical variables on the analysis of cell-free DNA from blood and urine samples. Front Cell Dev Biol 2024, 12, 1385041. [Google Scholar] [CrossRef] [PubMed]
- Lih, C.J.; Harrington, R.D.; Sims, D.J.; Harper, K.N.; Bouk, C.H.; Datta, V.; Yau, J.; Singh, R.R.; Routbort, M.J.; Luthra, R.; et al. Analytical Validation of the Next-Generation Sequencing Assay for a Nationwide Signal-Finding Clinical Trial: Molecular Analysis for Therapy Choice Clinical Trial. J Mol Diagn 2017, 19, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.Y.; Smith, K.S.; Kumar, R.; Paul, L.; Bihannic, L.; Lin, T.; Maass, K.K.; Pajtler, K.W.; Chintagumpala, M.; Su, J.M.; et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell 2021, 39, 1519–1530. [Google Scholar] [CrossRef]
- Tse, R.T.; Zhao, H.; Wong, C.Y.; Cheng, C.K.; Kong, A.W.; Peng, Q.; Chiu, P.K.; Ng, C.F.; Teoh, J.Y. Urinary Cell-Free DNA in Bladder Cancer Detection. Diagnostics (Basel) 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Abou Alaiwi, S.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat Med 2020, 26, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Xiao, Y.; Yan, S.; Zhu, Y.; Ren, S. Cell-free DNA in the management of prostate cancer: Current status and future prospective. Asian J Urol 2023, 10, 298–316. [Google Scholar] [CrossRef] [PubMed]
- Dermody, S.M.; Bhambhani, C.; Swiecicki, P.L.; Brenner, J.C.; Tewari, M. Trans-Renal Cell-Free Tumor DNA for Urine-Based Liquid Biopsy of Cancer. Front Genet 2022, 13, 879108. [Google Scholar] [CrossRef]
- Huang, F.F.; Di, X.F.; Bai, M.H. Analysis of urine cell-free DNA in bladder cancer diagnosis by emerging bioactive technologies and materials. Front Bioeng Biotechnol 2024, 12, 1458362. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, S.; Das, B.C. Saliva as a potential non-invasive liquid biopsy for early and easy diagnosis/prognosis of head and neck cancer. Transl Oncol 2024, 40, 101827. [Google Scholar] [CrossRef]
- Rigotti, P.; Polizzi, A.; Quinzi, V.; Blasi, A.; Lombardi, T.; Lo Muzio, E.; Isola, G. Cell-Free DNA as a Prognostic Biomarker in Oral Carcinogenesis and Oral Squamous Cell Carcinoma: A Translational Perspective. Cancers (Basel) 2025, 17. [Google Scholar] [CrossRef]
- Mattox, A.K.; D'Souza, G.; Khan, Z.; Allen, H.; Henson, S.; Seiwert, T.Y.; Koch, W.; Pardoll, D.M.; Fakhry, C. Comparison of next generation sequencing, droplet digital PCR, and quantitative real-time PCR for the earlier detection and quantification of HPV in HPV-positive oropharyngeal cancer. Oral Oncol 2022, 128, 105805. [Google Scholar] [CrossRef]
- Swarup, N.; Cheng, J.; Choi, I.; Heo, Y.J.; Kordi, M.; Aziz, M.; Arora, A.; Li, F.; Chia, D.; Wei, F.; et al. Multi-faceted attributes of salivary cell-free DNA as liquid biopsy biomarkers for gastric cancer detection. Biomark Res 2023, 11, 90. [Google Scholar] [CrossRef]
- Chang, S.C.; Wei, Y.F.; Chen, C.Y.; Lai, Y.C.; Hu, P.W.; Hung, J.C.; Chang, C.Y. Profiling Cell-Free DNA from Malignant Pleural Effusion for Oncogenic Driver Mutations in Patients with Treatment-Naive Stage IV Adenocarcinoma: A Multicenter Prospective Study. Mol Diagn Ther 2024, 28, 803–810. [Google Scholar] [CrossRef]
- Guo, Z.; Xie, Z.; Shi, H.; Du, W.; Peng, L.; Han, W.; Duan, F.; Zhang, X.; Chen, M.; Duan, J.; et al. Malignant pleural effusion supernatant is an alternative liquid biopsy specimen for comprehensive mutational profiling. Thorac Cancer 2019, 10, 823–831. [Google Scholar] [CrossRef]
- Zhang, N.; Li, Y.; Zhang, H.; Dong, Y.; Zhang, C.; Du, W.; Long, C.; Xing, X.; Li, K.; Liu, Z.; et al. Performance of SHOX2 and RASSF1A methylation assay in supernatants and matched cell pellets for the diagnosis of malignant pleural effusion. Clin Chim Acta 2024, 553, 117699. [Google Scholar] [CrossRef]
- Na, M.J. Diagnostic tools of pleural effusion. Tuberc Respir Dis (Seoul) 2014, 76, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: the future of cancer early detection. J Transl Med 2023, 21, 118. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, Z.; Altaf, F.; Khanzada, M.; Safi, A.; Asghar, Z.; Warraich, D.; Shah, S. Liquid biopsies for early detection and monitoring of cancer: advances, challenges, and future directions. Ann Med Surg (Lond) 2025, 87, 3244–3253. [Google Scholar] [CrossRef] [PubMed]
| Method | Short description | References |
| CAPP-Seq | Cancer Personalized Profiling by deep Sequencing; Targeted hybrid-capture NGS selector approach designed for ultrasensitive, broad patient-coverage quantitation of ctDNA (mutation, indel, rearrangement, CNV). | [22] |
| iDES | Integrated digital error suppression — molecular barcodes (UMIs) + in-silico background filtering to reduce NGS errors and lower limit of detection. | [23] |
| BEAMing | Beads, Emulsion, Amplification, Magnetics; Emulsion PCR + bead capture and flow cytometry readout for ultra-sensitive detection of known hotspot mutations (digital PCR class). | [24] |
| ddPCR | Droplet digital PCR; Partitioned PCR (droplets) for absolute quantitation of known single nucleotide variants (SNVs) and small insertion-deletions (indels); highly sensitive for low variant allele frequencies (VAFs) and orthogonal validation. | [25,26] |
| sWGS | Shallow whole-genome sequencing; Low-coverage WGS to detect genome-wide copy number alterations and tumor fraction estimation from cfDNA. | [27] |
| Fragmentomics (DELFI) | Genome-wide cfDNA fragmentation profiling (fragment size, end-motifs, nucleosome signals) used with machine learning (ML) to detect and localize cancer. | [27] |
| cfMeDIP-seq | DNA methylome profiling; Immunoprecipitation-based enrichment for methylated cfDNA enabling low-input methylome profiling and tissue-specific cancer signals (bisulfite-free options). | [28,29] |
| cfMeDIP-spike | Synthetic spike-in DNA controls (variable length/GC/CpG) for normalization and absolute quantification in cfMeDIP workflows. | [30] |
| FSE-EME | Fragment size selection & end-motif enrichment; Size-selection/enrichment for mono-nucleosomal-sized fragments or defined end motifs to enrich tumor-derived cfDNA prior to library prep. | [27] |
| cfDNA NGS panels | Commercial comprehensive cfDNA NGS panels (Guardant360, FoundationOne® Liquid CDx); Clinically validated, large targeted panels for therapy selection, monitoring and companion diagnostics; include hybrid-capture, UMIs, and bioinformatic QC. | [31,32] |
| SM-AFD | Single-molecule / amplification-free detection (emerging); Experimental approaches that minimize amplification bias (single-molecule sequencing, nanopore/PacBio with specialized prep) for direct detection of fragmented cfDNA. | [27,28] |
| DNA-PAINT | Super-resolution, single-molecule imaging that uses transient DNA hybridization to detect point mutations, methylation patterns, and amplified sequences at femtomolar sensitivity. | [33,34] |
| Method | Short description | References |
| EV-RNA-seq | Exosome / extracellular vesicle (EV) isolation + exo-RNA sequencing; Enrichment of EVs (ultracentrifugation, size-exclusion, precipitation, commercial kits) followed by RNA extraction and sequencing. | [53] |
| Targeted RT-qPCR | ddPCR for miRNAs and fusion transcripts; Sensitive quantification of defined transcripts using RT-qPCR or ddPCR on exoRNA or total plasma RNA. | [53,54] |
| UMI-RNA-seq | RNA-Seq with UMIs and low-input library preps; Low-input RNA-Seq protocols optimized for extracellular RNA to avoid amplification bias. | [54] |
| EV-proteomics | Exosomal proteomics for tissue-of-origin determination; Mass-spectrometry proteomic profiling of exosome cargo proteins to infer tissue/cell-type origin. | [55,56] |
| circRNA / lncRNA profiling | Circular RNAs and lncRNAs in exosomes are resistant to degradation and can yield cancer-specific biomarkers. | [57] |
| DNA-PAINT-miRNA | DNA-PAINT and DNA-origami nanoarrays for amplification-free detection of miRNAs in fluids/exosomes. | [58] |
| Targeted Fus-RNA-seq | Fusion transcript detection (targeted RNA panels); Targeted RNA assays for fusion transcript detection in plasma/exosomes. | [59,60] |
| Ago-FISH, FRET-FISH | Ago-FISH, FRET-FISH and other single-molecule RNA detection; Single-molecule RNA detection methods for multiplexed, amplification-free detection of short RNAs. | [61,62] |
| exo-RNA + proteomics | Integration: multi-omic exo-RNA + proteomics pipelines; Combined workflows analyzing exosomal RNA, DNA and protein cargo for integrated biomarker signatures. | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





